Significance
Cell adhesion is important for the survival of living organisms and is mediated by the adhesion molecules that bridge the membranes of adjacent cells. Cell adhesion molecules usually have long flexible ectodomains, and their structures have been studied extensively in the past decades. However, due to technical limitations, how these long molecules are assembled between membranes and what are the mechanisms of forming cell adhesion interfaces have not been well understood. Here, we combine electron microscopy with other biophysical methods to investigate the structure of cell–cell adhesion mediated by Sdk molecules and generate three-dimensional views of the adhesion interfaces in situ, thus revealing the architecture and the potential mechanism of Sdk-mediated cell adhesion at molecular level.
Keywords: cell adhesion, Sidekick, electron tomography, IgSF, cell–cell interaction
Abstract
Cell–cell adhesion is important for cell growth, tissue development, and neural network formation. Structures of cell adhesion molecules have been widely studied by crystallography, revealing the molecular details of adhesion interfaces. However, due to technical limitations, the overall structure and organization of adhesion molecules at cell adhesion interfaces has not been fully investigated. Here, we combine electron microscopy and other biophysical methods to characterize the structure of cell–cell adhesion mediated by the cell adhesion molecule Sidekick (Sidekick-1 and Sidekick-2) and obtain 3D views of the Sidekick-mediated adhesion interfaces as well as the organization of Sidekick molecules between cell membranes by electron tomography. The results suggest that the Ig-like domains and the fibronectin III (FnIII) domains of Sidekicks play different roles in cell adhesion. The Ig-like domains mediate the homophilic transinteractions bridging adjacent cells, while the FnIII domains interact with membranes, resulting in a tight adhesion interface between cells that may contribute to the specificity and plasticity of cell–cell contacts during cell growth and neural development.
Cell–cell interactions are important for the survival of living organisms. Cell adhesion is a major type of cell–cell contact for maintaining multicellular structures and is mediated by cell adhesion molecules that bridge the membranes of adjacent cells through either homophilic or heterophilic interactions (1, 2). To date, a large number of cell adhesion molecules have been identified and are involved in many biological processes including cell growth, migration, and cellular network formation and play multiple roles beyond connecting cells (3–5). Although cell adhesion molecules exhibit abundant functional diversities, their major structural features are less diversified, suggesting the evolutionary relations among these molecules (6). For example, many adhesion molecules belong to the Ig superfamily (IgSF), which usually contain a number of Ig-like domains as well as other types of domains, such as fibronectin type II or III domain (FnII or III). The total number of domains varies in different cases and, surprisingly, some adhesion molecules have more than 10 extracellular domains, resulting in long flexible molecules (6, 7). It is unclear why a large number of domains is required for adhesion molecules and how these domains contribute to the specificity and plasticity of cell–cell contacts. To address these questions, structural studies of adhesion interfaces are necessary; however, current imaging techniques cannot provide high-resolution details of adhesion interfaces. Due to the development of electron microscopy (EM) in recent years, electron tomography (ET) has become a powerful technique to generate 3D views of biological specimens such as organelles, cells, or tissues (8–10), therefore providing a practical tool to visualize the cell–cell contacts in 3D, which would shed light on the architecture and the molecular mechanisms of cell–cell interactions (11, 12).
Sidekick (Sdk) proteins are the members of the IgSF. Their extracellular portion contains six N-terminal Ig-like domains followed by 13 FnIII domains, which may be one of the largest ectodomains in the IgSF (13). It has been shown that Sdk is required for eye pattern formation of Drosophila (14) and, similarly, their vertebrate homologs, Sdk1 and Sdk2, can promote lamina-specific connectivity in the development of retina (15). Recently, Sdk2 has been shown to be important for the formation of the retinal circuit that detects differential motions (16). Besides their neuronal functions, Sdks have also been reported to function in nonneuronal systems such as kidney, heart, intestines, and stomach and are involved in the development of organs as well as in pathogenesis pathways (17, 18).
The roles of cell adhesion molecules in neural systems have been widely investigated as they are essential for the formation of neural networks where billions of neurons are connected with specificity and plasticity (19, 20). It has been shown that adhesion molecules are not only a scaffold for neuronal contacts, they may also contribute to the specificity of neuron connectivity (3, 13, 21). Sdks have been reported to have homophilic adhesion activities and are concentrated at synapses as the determinants of laminar-specific synaptic connectivity in vertebrate retina (15, 22, 23). Along with Sdks, Dscam, DscamL1, and contactins, which are also IgSF molecules with homophilic adhesion activities, are found having similar functions in determining the laminar specificities in the retina development (24, 25). Therefore, these molecules have been proposed as IgSF codes to define the specificity of lamina targeting through homophilic interactions (25). Although the functions of Sdks have been studied in both neuronal and nonneuronal systems, the overall structure and organization of Sdk molecules in adhesion interfaces remain unclear.
Here, we characterize the ectodomains of mouse Sdks by electron microscopy and investigate the roles of the Ig-like domains and the FnIII domains of Sdks in forming adhesion interfaces. Moreover, we examine the architecture of the Sdk-mediated cell adhesion with the high pressure freezing and freeze substitution (HPF-FS) method and generated 3D views of the adhesion interfaces using electron tomography.
Results
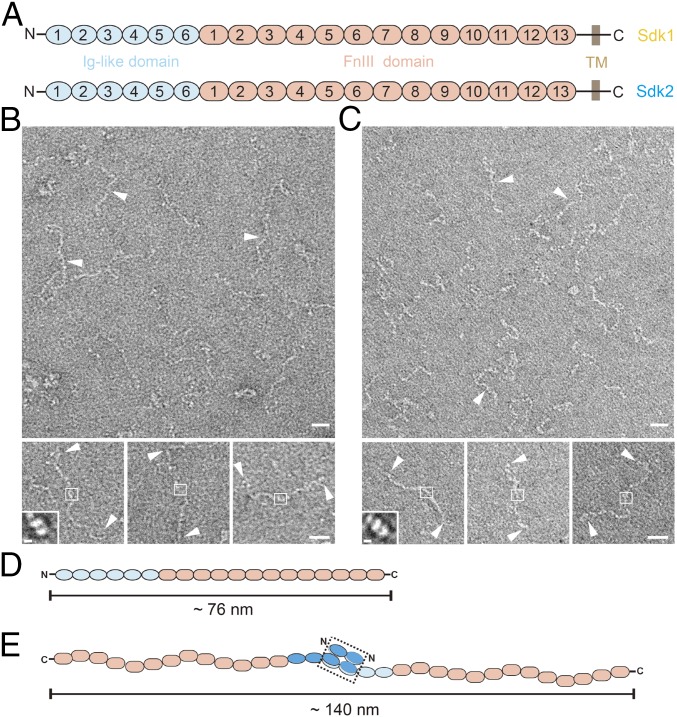
The Ectodomains of Sdks Adopt Flexible Conformations and Form Homophilic Dimers.
The ectodomains of both Sdk1 and Sdk2, which contain six Ig-like domains and 13 FnIII domains (Fig. 1A), were expressed in HEK293 cells. Size exclusion chromatography (SEC) showed that the ectodomains eluted much earlier than expected, suggesting that Sdks may exist as homophilic dimers in solution (SI Appendix, Fig. S1). To visualize the conformation of Sdks directly, the purified proteins were negatively stained and examined by electron microscopy. EM images showed that both Sdk1 and Sdk2 ectodomains adopt a linear shape with multiple conformations (Fig. 1 B and C). According to the average length of individual Ig-like domain and FnIII domain (∼4 nm), the length of a fully extended Sdk ectodomain was around 76 nm (Fig. 1D). However, the EM images also showed that the length of both Sdk1 and Sdk2 ectodomains were around 140 nm, suggesting they were forming homophilic dimers (Fig. 1 B–E). It has been reported that the homophilic adhesion of Sdk molecules occurred at the N-terminal Ig-like domains (22, 23), therefore we boxed out the central regions of the Sdk dimers in the EM images and performed 2D averaging (Fig. 1 B and C). The resulting images revealed the low-resolution contours of the domains involved in forming homophilic dimers of Sdks (Fig. 1 B and C, Insets).
Fig. 1.
The ectodomains of Sdk1 and Sdk2 form homophilic dimers with flexible conformations. (A) Domain organization of Sdk1 and Sdk2. (B) Negatively stained EM images of Sdk1 ectodomain (white arrowheads, Upper) and the selected Sdk1 particles (white arrowheads, Lower) as homophilic dimers with flexible conformations. The potential dimeric heads of Sdk1 are shown in white squares. A 2D class averaged image of Sdk1 dimeric head is shown in Inset. (C) Negatively stained EM images of Sdk2 ectodomain (white arrowheads, Upper) and the selected Sdk2 particles (white arrowheads, Lower) as homophilic dimers with flexible conformations. The potential dimeric heads of Sdk2 are shown in white squares. A 2D class averaged image of Sdk2 dimeric head is shown in Inset. (D) A schematic model of the extended Sdk ectodomain. (E) A schematic model of the extended Sdk homophilic dimer. The dimerized horseshoe heads are shown in a black dashed rectangle. (Scale bars: B and C, 20 nm; B and C, Insets, 5 nm.)
Due to the highly flexible conformation of Sdks, determining the intact structures of Sdk ectodomains would be difficult for both crystallography and EM single-particle reconstruction. Therefore, we expressed the four N-terminal Ig-like domains of mouse Sdk1 and Sdk2 in baculovirus-infected insect cells. Purified proteins were screened for crystallization, and the crystals of both Sdk1 Ig1∼4 and Sdk2 Ig1∼4 were obtained and refined to 1.6 Å and 2.4 Å resolution, respectively (SI Appendix, Fig. S2 and Table S1). These structures are similar to the crystal structures of Sdk1 Ig1∼4 and Sdk2 Ig1∼4 published recently (23). The crystal structures showed that the four N-terminal domains of Sdks fold into typical Ig-like domains that adopt a horseshoe-shaped conformation (SI Appendix, Fig. S2), as found in structures of other cell adhesion molecules such as hemolin and contactin (26, 27). Crystal structures also showed that both Sdk1 Ig1∼4 and Sdk2 Ig1∼4 form homophilic dimers (SI Appendix, Fig. S2), which is consistent with the SEC results and EM images (Fig. 1). The Sdk homophilic interactions occur between Ig1 and Ig2, which are in agreement with previous mutagenesis and structural studies (22, 23). The overall shape of the dimeric structures of Sdk Ig1 to Ig4 resembles the 2D averaging results from EM (Fig. 1 B and C, Insets), suggesting that Ig1 and Ig2 might be the only contacting domains mediating Sdk homophilic dimerization.
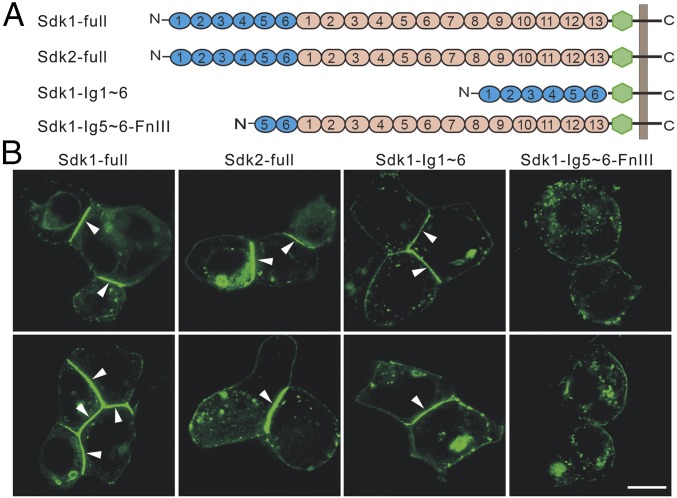
Sdks Mediate Cell Adhesion Through the N-Terminal Ig-Like Domains.
Before structural characterization of the Sdk mediated cell adhesion, we generated a series of constructs of Sdks with a GFP insertion between the ectodomain and the transmembrane domain and monitored the cell adhesion of the transfected HEK293 cells by confocal fluorescence microscopy (Fig. 2). The results showed that cell adhesion could be observed by transfecting both full-length Sdk1 and Sdk2 as well as a Sdk1 mutant containing only the N-terminal Ig-like domains (Ig 1∼6), where the adhesion interfaces were highlighted between adjacent cells (Fig. 2B). By contrast, a mutant of Sdk1 without the N-terminal four Ig-like domains failed to form adhesion interfaces between cells (Fig. 2B), thus confirming the importance of the Ig-like domains in Sdk homophilic adhesion, consistent with the EM and crystallographic data shown above as well as the published results (22, 23). It is noteworthy that the fluorescent intensities at the adhesion interfaces induced by Sdks were much brighter than the nonadhesion regions on the cell surface, implying that Sdk molecules might be densely packed or assembled in the adhesion interfaces (Fig. 2B).
Fig. 2.
Cell adhesion mediated by Sdks. (A) A diagram of the Sdk constructs with GFP (green) generated for cell adhesion assays. (B) Cell–cell adhesion monitored by fluorescence microscopy. HEK293 cells transfected with Sdk1, Sdk2, Sdk1 Ig1∼6 form adhesion interfaces (white arrowheads) between cells, whereas HEK293 cells transfected with Sdk1 Ig5∼6-FnIII1∼13 has no adhesion between cells. (Scale bar: 10 μm.)
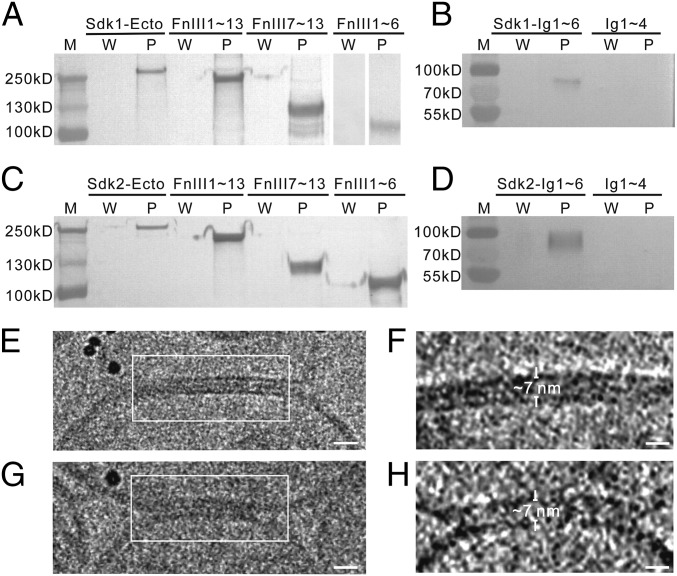
The FnIII Domains of Sdks Interact with Lipid Membranes.
According to the EM images shown above, the purified Sdk ectodomains were highly flexible with multiple conformations and the fully extended homophilic dimers of Sdk were ∼140 nm in length (Fig. 1E), therefore it would be expected that the intermembrane spacing may vary for the adhesion interfaces. However, the confocal images showed that the adhesion interfaces formed by Sdks exhibited a roughly uniform distribution for the distance between adjacent cell membranes (Fig. 2B), implying that these long flexible molecules may adopt relatively rigid conformations in adhesion interfaces. Among the six Ig-like domains of Sdks, the four N-terminal Ig-like domains form a trans-homophilic dimer as revealed by the crystal structures, therefore they should contribute to the intermembrane spacing between cells. The FnIII domains, which make roughly two-thirds of the length of Sdks, may also regulate the spacing between cell membranes. However, the exact roles of the Ig-like domain and the FnIII domain in defining the intermembrane distance are unclear. To address this issue, we evaluated the potential interactions of Sdks with lipid membranes using liposome pull-down assays, which had been used to examine the protein–lipid interactions (28, 29). The liposomes were prepared with phosphatidylcholines, which is a major component of cell membrane, especially on the exoplasmic leaflet (29–31). Surprisingly, we found that the ectodomains of both Sdk1 and Sdk2 could be pulled down by liposomes (Fig. 3 A and C). Then, we constructed a series of Sdk truncation mutants for liposome pull-down assays. The results showed that the fragments of both Sdk1- and Sdk2-containing FnIII1∼13, FnIII1∼6, or FnIII7∼13 could be pulled down by liposomes (Fig. 3 A and C). These results are similar to the EphA2 receptor where the FnII domain interacts with the lipid membrane (28). By contrast, the fragments including only the Ig-like domain 1∼4, which form the horseshoe heads of Sdks, had no detectable binding for liposomes (Fig. 3 B and D). Interestingly, the fragments containing the Ig-like domain 1∼6 also showed binding affinities with liposomes, suggesting that the Ig-like domain 5–6 might also interact with lipid membrane (Fig. 3 B and D). Taken together, these data suggested that a large portion of Sdk ectodomains, especially the FnIII domains, might associate with lipids, perhaps by “lying down” on the membrane surface, thus reducing the intermembrane spacing of adhesion interfaces.
Fig. 3.
The FnIII domains of Sdks interact with liposomes. The supernatants after washing (W) and the liposome pellets (P) are assayed by Western blot (A–D). (A) The ectodomain and the FnIII domain-only fragments (Sdk1 FnIII1∼13, Sdk1 FnIII1∼6, Sdk1 FnIII7∼13) of Sdk1 can be pulled down by liposomes. (B) Sdk1 Ig1∼6 can be pulled down by liposomes, while Sdk1 Ig1∼4 has no interaction with liposomes. (C) The ectodomain and the FnIII domain-only fragments (Sdk2 FnIII1∼13, Sdk2 FnIII1∼6, Sdk2 FnIII7∼13) of Sdk2 can be pulled down by liposomes. (D) Sdk2 Ig1∼6 can be pulled down by liposomes, while the Sdk2 Ig1∼4 has no interaction with liposomes. (E and G) Cryo-images of adhesion interfaces (white square) formed between the Sdk1 ectodomain-incorporated liposomes. (F and H) Tomographic slices of the adhesion interfaces indicated in E and G, respectively. The intermembrane distances are indicated and labeled. (Scale bars: E and G, 15 nm; F and H, 10 nm.)
The Ectodomains of Sdks Introduce Tight Adhesion Between Lipid Membranes.
To visualize the Sdk adhesion interfaces directly, we used liposomes to model the cell adhesion formation by attaching the purified Sdk1 ectodomain fused with a His tag at the C terminus to liposomes prepared with DOPC and DOGS-NTA nickel lipids, therefore the binding between His tag and DOGS-NTA lipid could mimic the cellular expressed Sdk molecule on the cell surface (32), which is anchored on the plasma membrane through a transmembrane helix. The Sdk-incorporated liposomes were frozen by a plunge freezer at liquid nitrogen temperature and loaded onto an electron microscope for imaging and cryo-electron tomographic data collection. The cryo-EM images showed that the adhesion interfaces were formed between liposomes by the addition of purified Sdk1 ectodomain (Fig. 3 E and G). To measure the intermembrane distances and visualize the adhesion interfaces in 3D, tomographic reconstructions were calculated and the results showed that the Sdk1-mediated adhesion interfaces between liposomes had a uniformly distributed spacing of ∼7 nm (Fig. 3 F and H), much shorter than the total length of the Sdk1 dimer, which is ∼140 nm (Fig. 1), thus supporting the adhesion model proposed above where the FnIII domains of Sdk molecules were lying down on cell surface. Moreover, consistent with the fluorescent images, the dark tomographic densities between the two adjacent membranes suggested that Sdk1 ectodomain could be tightly packed in the interfaces (Fig. 3 F and H). Unfortunately, the individual Sdk molecules forming the adhesion connections between membranes could not be identified unambiguously in the tomograms, probably due to the low contrast of cryo-imaging conditions.
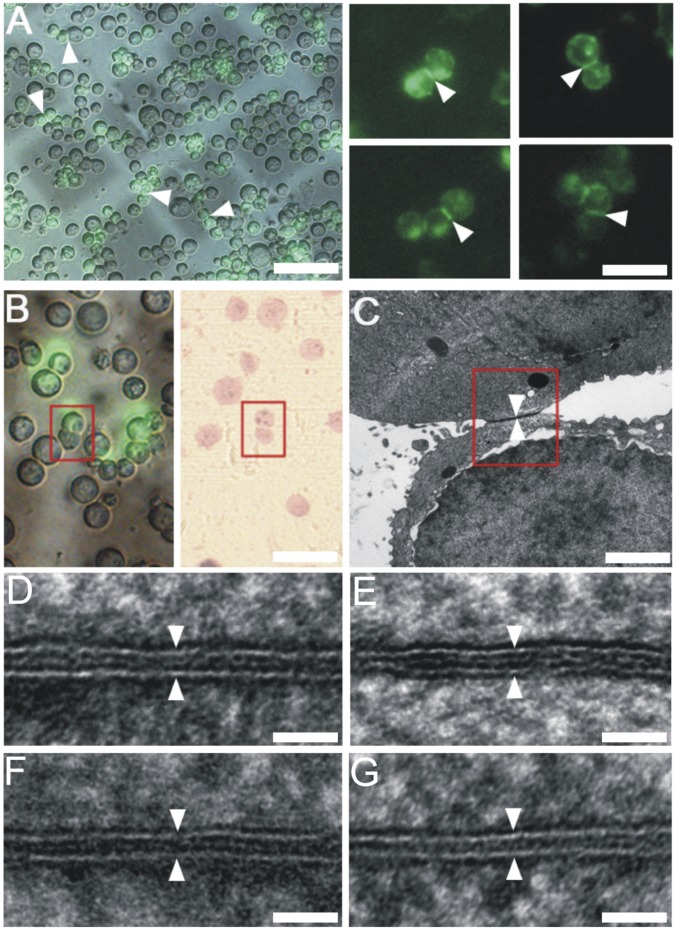
Sdks Mediate Tight Adhesion Between Cell Membranes.
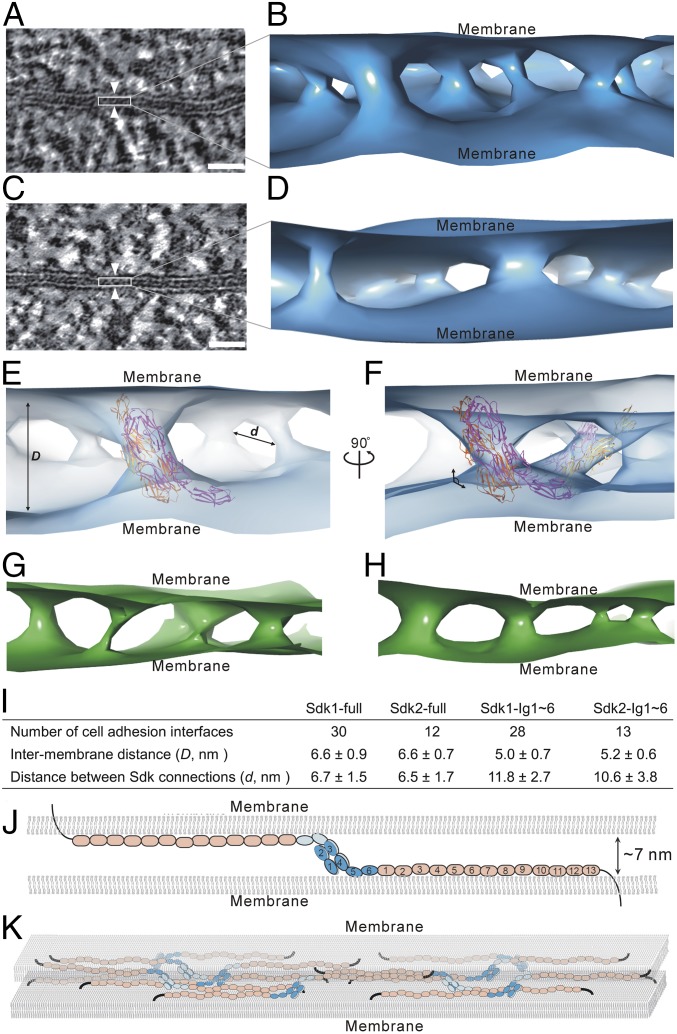
To validate the liposome adhesion results and visualize the cell adhesion interfaces directly in situ, we transfected HEK293 cells with the full-length Sdk1 and Sdk2 as well as two Ig-like domain-only mutants, Sdk1 Ig1∼6 and Sdk2 Ig1∼6. The transfected cells were grown on sapphire discs for HPF-FS (33), then the plastic embedded samples were sectioned and imaged by electron microscopy (SI Appendix, Fig. S3A). The formation of the Sdk-mediated adhesion interfaces was also validated by the immune-gold labeling experiments (SI Appendix, Fig. S3B). To locate the Sdk-mediated adhesion interfaces, we first checked cell adhesion formation under a fluorescence microscope and marked the positions of the potential cell adhesion interfaces on sapphire discs (Fig. 4A and SI Appendix, Fig. S3A) (34). The markers were then relocated on the plastic-embedded resin block for trimming and sectioning, thus correlating the fluorescent targets with EM (Fig. 4 A and B and SI Appendix, Fig. S3A) (12). This procedure ensured that the Sdk-mediated adhesion interfaces identified by fluorescent imaging could be located specifically on thin sections for EM. The resulting EM images showed that the lipid bilayers of cell membrane as well as the adhesion interfaces were seen clearly with better contrast comparing to the cryo-imaging results (Fig. 4 and SI Appendix, Figs. S4 and S5). Indeed, the adhesion interfaces between cell membranes are much narrower than the length of the purified Sdk dimers. The averaged intermembrane distances (D) were ∼7 nm for the full-length Sdk1 (Figs. 4D and 5I and SI Appendix, Fig. S4) and Sdk2 (Figs. 4F and 5I and SI Appendix, Fig. S4), similar to the distances found in the liposome-based adhesion models (Fig. 3). The intermembrane distances of the interfaces formed by the Sdk Ig-like domain-only fragments were ∼5 nm (Figs. 4 E and G and 5I and SI Appendix, Fig. S5), slightly narrower than the spacing formed by the full-length Sdks (Fig. 5I). This is not surprising as the FnIII domains may contribute somewhat to the intermembrane spacing even if they are lying down on cell surface.
Fig. 4.
Correlative light-electron microscopy and EM images of the Sdk-mediated cell adhesion interfaces. (A) A merged fluorescence-light microscopic image showing the formation of cell adhesion (Left; white arrowheads) between the Sdk1-transfected cells on a sapphire disk with carbon marker (gray). Fluorescent images of the Sdk1-mediated adhesion interfaces (white arrowheads) formed between the adherent cell pairs on sapphire disk (Right). (B) A Sdk1-mediated cell adhesion pair identified by the fluorescence-light imaging (Left; red box) can be located on the plastic-embedded sections (Right; red box) prepared after HPF preservation for EM. (C) An electron micrograph of a Sdk1-mediated cell adhesion interface located by the correlative light-electron microscopy from B (red box). (D) An electron micrograph of a cell adhesion interface (white arrowheads) mediated by the full-length Sdk1. (E) An electron micrograph of a cell adhesion interface (white arrowheads) mediated by the Sdk1 Ig1∼6. (F) An electron micrograph of a cell adhesion interface (white arrowheads) mediated by the full-length Sdk2. (G) An electron micrograph of a cell adhesion interface (white arrowheads) mediated by the Sdk2 Ig1∼6. (Scale bars: A, Left, 100 μm; A, Right and B, 50 μm; C, 2 μm; D–G, 25 nm.)
Fig. 5.
Architecture of Sdk-mediated cell–cell adhesion revealed by electron tomography. (A) A tomographic slice of a cell adhesion interface (white arrowheads) formed by Sdk1. (B) An isosurface of tomographic density showing the connections and organization of Sdk1 molecules between cell membranes. (C) A tomographic slice of a cell adhesion interface (white arrowheads) formed by Sdk2. (D) An isosurface of tomographic density showing the connections and organization of Sdk2 molecules between cell membranes. (E) Fitting of the crystal structure of Sdk1 Ig1∼5 homophilic dimer (magenta and orange; PDB ID code 5k6w) into the tomographic density (light blue) of Sdk1 in the adhesion interface. (F) Another view of the fitting of Sdk1 Ig1∼5 into the tomographic density (light blue) showing the tilted positions of Sdk1 horseshoe heads (magenta and orange) to cell membranes. (G) An isosurface of tomographic density (green) showing the connections and organization of Sdk1 Ig1∼6 molecules between cell membranes. (H) An isosurface of tomographic density (green) showing the connections and organization of Sdk2 Ig1∼6 molecules between cell membranes. (I) The averaged intermembrane distances (D) and the distances between the Sdk connections (d) in the Sdk mediated adhesion interfaces. (J) A structural model of a homophilic pair of Sdk between cell membranes. (K) A model of the cell adhesion interface mediated by Sdk molecules. (Scale bars: 25 nm.)
According to the liposome-binding assays (Fig. 3), the FnIII domains and the Ig-like domain 5–6 of Sdks could associate with the cell membrane, suggesting that the horseshoe heads of Sdks would be the key component to define the spacing between adjacent membranes. The crystal structures show that the N-terminal horseshoe dimer of Sdk is about 8∼9 nm in length (SI Appendix, Fig. S2), suggesting that it may need to be tilted between membranes to fit the intermembrane spacing that is around 7 nm (Fig. 5). In fact, according to the crystal structure that includes the Ig-like domain 1∼5 of Sdk1 (23), there is a tilt angle about 130° between Ig4 and Ig5 (Fig. 5J), consistent with the potential orientation of the Sdk horseshoe dimer in the adhesion interfaces. Taken together, the results regarding the horseshoe head, the Ig-like domains, and the FnIII domains provide a structural model of cell–cell adhesion mediated by Sdks (Fig. 5J).
Visualization of the Sdk-Mediated Cell Adhesion by Electron Tomography.
To verify the structural model of the Sdk-mediated cell adhesion, we reconstructed tomograms of the adhesion interfaces formed by the full-length Sdk1 and Sdk2 (Fig. 5 A and C and SI Appendix, Fig. S6) as well as the Ig-like domain-only mutants, Sdk1 Ig1∼6 and Sdk2 Ig1∼6, and generated the 3D views of the adhesion interfaces (Fig. 5 and SI Appendix, Fig. S6). As expected, the HPF-prepared specimens exhibited better contrast than cryo-samples: The individual Sdk connections between cell membranes can be identified in the tomograms (Fig. 5 B and D), thus providing in situ visualization of the architecture of the adhesion interfaces mediates by Sdks. Although the atomic details of individual Sdk connections between cell membranes cannot be seen due to resolution limitation, 3D visualization of the adhesion interfaces is still informative. The volumes of the Sdk connections in the interfaces show some variations, which might be caused by the reagents used during the HPF-FS sample preparation. The averaged diameter of the individual Sdk connections between membranes is about 4.7 ± 1.6 nm. According to the crystal structures, the size of a Sdk horseshoe head dimer is about ∼8 nm in length and ∼5 nm in diameter (SI Appendix, Fig. S2), therefore matching the size of the individual Sdk connections in the tomograms (Fig. 5 E and F). The Sdk horseshoe dimer can be fitted into the Sdk connection density reasonably well and generate a molecular model for the adhesion interface (Fig. 5 E and F). A noticeable feature in the tomograms is that most of the Sdk connections in the interfaces are not perpendicular to cell membrane, but with a tilted angle of about 130° instead (Fig. 5F). This is consistent with the model suggested by the EM images and the crystal structures (Fig. 5J). Previous studies have shown that myelin P0 may interact with the cell membrane through the exposed tryptophan residues (35). Interestingly, according to the crystal structure of Sdk1 Ig1∼5 (23), there is a conserved tryptophan residue Trp553 (corresponds to Trp473 in PDB ID code 5k6w) for both Sdk1 (Trp553) and Sdk2 (Trp497) near the C-terminal end of Ig5, therefore similar membrane interactions may occur for Sdk molecules on the cell surface. In addition, protrusions on membrane surface can be found in some areas in the tomograms, which could be associated with the cis clusters of Sdk molecules on cell surface, but details are not available at the current resolution.
In parallel, the tomograms of the adhesion interfaces formed by the Sdk Ig-like domain-only mutants are also calculated (Fig. 5 G and H), revealing the adhesion connections between cell membranes. Indeed, the intermembrane distances formed by the Sdk Ig-like domain-only mutants are shorter than that formed by the intact Sdk ectodomain (Fig. 5I), which is expected since the FnIII domains may contribute a little to the intermembrane spacing although they are lying down on the cell surface. However, the densities of the adhesion connections by these mutants (Fig. 5 G and H) are much worse than those in the interfaces formed by the full-length Sdks (Fig. 5 B and D), suggesting that the FnIII domains might also play a role in stabilizing the adhesion interfaces, although they do not form trans interactions between cells. Assuming the FnIII domains lie down on the membrane surface, their densities could be merged within cell membrane densities and, therefore, were not resolved at the resolution of the tomographic reconstructions (Fig. 5 B and D). Nevertheless, the separation between the individual Sdk connections (d) in the interfaces can be measured, showing that the average distance between the full-length Sdk connections is about 6.5 nm (Fig. 5 E and I). By contrast, the average separation between the connections formed by the Ig-like domain-only mutants is around 11 nm with larger variations (Fig. 5I), suggesting that the FnIII domains might not only associate with cell membranes, but also regulate the distribution of Sdk molecules in the interfaces, probably through the cis interactions among Sdk molecules (36), which may lead to a Sdk network on cell surface and contribute to the stability and plasticity of adhesion interfaces between cells (Fig. 5K).
Discussion
Cell adhesion molecules such as IgSF molecules usually have a long extracellular portion containing multiple domains. The N-terminal domains usually form homophilic pairs mediating the trans interaction between cell membranes. For the IgSF adhesion molecules, the N-terminal Ig-like domains could adopt either linear or the horseshoe-like conformation for trans interactions. A potential advantage of the horseshoe-shaped conformation is that it may provide relatively larger binding interfaces with higher specificity and selectivity (SI Appendix, Fig. S2). In the case of Dscam, its N-terminal Ig-like domains form a unique S-shaped conformation (dual horseshoe) with a larger dimerization interface comparing to the horseshoe-shaped conformation, hence providing the structural bases for the dimerization of a large number of isoforms (37). Therefore, the number and the conformation of the N-terminal Ig-like domains may correlate with the selectivity and specificity of the IgSF adhesion molecules.
Sdks, Dscams, and contactins are the IgSF adhesion molecules playing important roles in establishing sublaminar specificity in the retina (15, 25) and have been proposed as cell surface recognition codes for forming precise synaptic connectivity in the nervous system (25). Crystal structures of these molecules have revealed the molecular details of the N-terminal Ig-like domains as well as some Fn domains (23, 37–39). Although these N-terminal domains adopt a conserved horseshoe-shaped conformation, their dimerization patterns are different. The dimers of Sdks are formed through the interactions between Ig1 and Ig2 of the monomers, while the dimerization of Dscam is formed by the interactions from Ig2, Ig3, and Ig7 (37). Different dimerization patterns are also found for other horseshoe-shaped heads (27, 39, 40), suggesting that the dimerization modes may also be relevant to the functional activities of IgSF adhesion molecules.
A common feature shared among many IgSF adhesion molecules is that they usually contain different types of domains (6). The Ig-like domains usually locate at the N terminus and are involved in homophilic or heterophilic interactions by forming trans interactions between cell membranes. The FnII or FnIII domains are also frequently found in the IgSF adhesion molecules, and the number of the Fn domains varies in different cases (13). However, the roles of the Fn domains in cell adhesion have not been clearly defined, although it has been suggested that Fn domains may be involved in the cis interactions and facilitate the cluster formation in adhesion interfaces (36). Similar to other IgSF adhesion molecules, the N-terminal Ig-like domains of Sdks mediate the trans-homophilic adhesion. A notable feature of Sdk molecules is that they have 13 FnIII domains, which may be one of the largest FnIII fragments among IgSF adhesion molecules and makes two-thirds of the Sdk ectodomains (6). The extended Sdk ectodomain is roughly 70–80 nm in length, and the EM images show that the isolated Sdk ectodomains exhibit highly flexible conformations, it would be difficult to imagine how these long, flexible molecules could mediate stable interactions between cell membranes. However, the EM results show that the adhesion interfaces mediated by Sdks are rather narrow with a constant intermembrane distance that is much shorter than the length of Sdk ectodomains, suggesting that a large part of Sdk ectodomain may not contribute to the intermembrane spacing directly. Indeed, our data also show that the FnIII domains are associated with membranes and probably lying down on the membrane surface, acting as anchors for Sdks on cell membranes. In the meantime, the FnIII domains may also mediate cis interactions among Sdk molecules and lead to a network formation on cell surface, which may stabilize the adhesion between membranes (Fig. 5K). Moreover, we are also trying to identify potential 2D patterns of Sdk in the interfaces, but the current data do not show any obvious 2D pattern in the interfaces, although some small local patterns may exist. It is unclear whether any regular 2D patterns are required for forming functional adhesion interfaces or the packing density of Sdk molecules in the interfaces could be important for adhesion.
Since a large number of IgSF adhesion molecules have been identified, especially in neural systems, the major questions about cell adhesion are how these molecules are organized in the interfaces and how they regulate the specificity and plasticity of adhesion. The results obtained here for Sdks may provide a generic model to address these questions. Although adhesion molecules may have multiple domains and flexible conformations, they could lead to compact adhesion interfaces with constant intermembrane spacing, and different types of domains are playing different roles in establishing adhesion interfaces. The Ig-like domains might behave as “hooks” to mediate the trans-homophilic pair formation, while the Fn domains act as “anchors” associating with membrane and also introduce cis interactions among each other, resulting in a tightly packed stable interface. The number and the conformation of the Ig-like domains may reflect the specificity and selectivity of adhesion interactions, while the number of the Fn domains may be relevant to the plasticity or stability of adhesion interfaces. Therefore, the combination of different types of domains might be evolved as structural codes of IgSF adhesion molecules for establishing precise cell–cell contacts with specificity and plasticity.
Materials and Methods
The ectodomain of mouse Sdk1, Sdk2, and their truncation mutants were expressed in both insect cells and HEK293 cells, and the purified proteins were applied for structural and biochemical experiments.
Further experimental details can be found in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the National Center for Protein Science Shanghai (Electron Microscopy and Integrated Laser Microscopy systems) for their instrumental support and technical assistance. We thank beam line BL17U of the Shanghai Synchrotron Radiation Facility and BL19U1 of the National Facility for Protein Science Shanghai for X-ray crystallographic data collection. This work is supported by Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB08020102, National Natural Science Foundation of China Grants 31470735 and 31670747, and the Chinese Academy of Sciences Facility-based Open Research Program (to Y. He).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystal structures of the four N-terminal Ig-like domains of mouse Sdk1 and mouse Sdk2 have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5XWX and 5XX0, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801810115/-/DCSupplemental.
References
- 1.Walsh FS, Doherty P. Neural cell adhesion molecules of the immunoglobulin superfamily: Role in axon growth and guidance. Annu Rev Cell Dev Biol. 1997;13:425–456. doi: 10.1146/annurev.cellbio.13.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Rougon G, Hobert O. New insights into the diversity and function of neuronal immunoglobulin superfamily molecules. Annu Rev Neurosci. 2003;26:207–238. doi: 10.1146/annurev.neuro.26.041002.131014. [DOI] [PubMed] [Google Scholar]
- 3.Carrillo RA, et al. Control of synaptic connectivity by a network of Drosophila IgSF cell surface proteins. Cell. 2015;163:1770–1782. doi: 10.1016/j.cell.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sytnyk V, Leshchyns’ka I, Schachner M. Neural cell adhesion molecules of the immunoglobulin superfamily regulate synapse formation, maintenance, and function. Trends Neurosci. 2017;40:295–308. doi: 10.1016/j.tins.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Benson DL, Schnapp LM, Shapiro L, Huntley GW. Making memories stick: Cell-adhesion molecules in synaptic plasticity. Trends Cell Biol. 2000;10:473–482. doi: 10.1016/s0962-8924(00)01838-9. [DOI] [PubMed] [Google Scholar]
- 6.Vogel C, Teichmann SA, Chothia C. The immunoglobulin superfamily in Drosophila melanogaster and Caenorhabditis elegans and the evolution of complexity. Development. 2003;130:6317–6328. doi: 10.1242/dev.00848. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Wu Y. Understanding the functional roles of multiple extracellular domains in cell adhesion molecules with a coarse-grained model. J Mol Biol. 2017;429:1081–1095. doi: 10.1016/j.jmb.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He W, He Y. Electron tomography for organelles, cells, and tissues. Methods Mol Biol. 2014;1117:445–483. doi: 10.1007/978-1-62703-776-1_20. [DOI] [PubMed] [Google Scholar]
- 9.Studer D, Humbel BM, Chiquet M. Electron microscopy of high pressure frozen samples: Bridging the gap between cellular ultrastructure and atomic resolution. Histochem Cell Biol. 2008;130:877–889. doi: 10.1007/s00418-008-0500-1. [DOI] [PubMed] [Google Scholar]
- 10.Oikonomou CM, Jensen GJ. Cellular electron cryotomography: Toward structural biology in situ. Annu Rev Biochem. 2017;86:873–896. doi: 10.1146/annurev-biochem-061516-044741. [DOI] [PubMed] [Google Scholar]
- 11.He W, Cowin P, Stokes DL. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–113. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka H, et al. Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin. Cell Rep. 2012;2:101–110. doi: 10.1016/j.celrep.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Rudenko G. Dynamic control of synaptic adhesion and organizing molecules in synaptic plasticity. Neural Plast. 2017;2017:6526151. doi: 10.1155/2017/6526151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen DN, Liu Y, Litsky ML, Reinke R. The sidekick gene, a member of the immunoglobulin superfamily, is required for pattern formation in the Drosophila eye. Development. 1997;124:3303–3312. doi: 10.1242/dev.124.17.3303. [DOI] [PubMed] [Google Scholar]
- 15.Yamagata M, Weiner JA, Sanes JR. Sidekicks: Synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002;110:649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- 16.Krishnaswamy A, Yamagata M, Duan X, Hong YK, Sanes JR. Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature. 2015;524:466–470. doi: 10.1038/nature14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaufman L, et al. Up-regulation of the homophilic adhesion molecule sidekick-1 in podocytes contributes to glomerulosclerosis. J Biol Chem. 2010;285:25677–25685. doi: 10.1074/jbc.M110.133959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufman L, et al. The homophilic adhesion molecule sidekick-1 contributes to augmented podocyte aggregation in HIV-associated nephropathy. FASEB J. 2007;21:1367–1375. doi: 10.1096/fj.06-7191com. [DOI] [PubMed] [Google Scholar]
- 19.Thalhammer A, Cingolani LA. Cell adhesion and homeostatic synaptic plasticity. Neuropharmacology. 2014;78:23–30. doi: 10.1016/j.neuropharm.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Zinn K, Özkan E. Neural immunoglobulin superfamily interaction networks. Curr Opin Neurobiol. 2017;45:99–105. doi: 10.1016/j.conb.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan L, et al. Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell. 2015;163:1756–1769. doi: 10.1016/j.cell.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi K, Kaufman L, Ross MD, Klotman PE. Definition of the critical domains required for homophilic targeting of mouse sidekick molecules. FASEB J. 2005;19:614–616. doi: 10.1096/fj.04-2947fje. [DOI] [PubMed] [Google Scholar]
- 23.Goodman KM, et al. Molecular basis of sidekick-mediated cell-cell adhesion and specificity. eLife. 2016;5:e19058. doi: 10.7554/eLife.19058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamagata M, Sanes JR. Dscam and sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- 25.Yamagata M, Sanes JR. Expanding the Ig superfamily code for laminar specificity in retina: Expression and role of contactins. J Neurosci. 2012;32:14402–14414. doi: 10.1523/JNEUROSCI.3193-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su XD, et al. Crystal structure of hemolin: A horseshoe shape with implications for homophilic adhesion. Science. 1998;281:991–995. doi: 10.1126/science.281.5379.991. [DOI] [PubMed] [Google Scholar]
- 27.Mörtl M, Sonderegger P, Diederichs K, Welte W. The crystal structure of the ligand-binding module of human TAG-1 suggests a new mode of homophilic interaction. Protein Sci. 2007;16:2174–2183. doi: 10.1110/ps.072802707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chavent M, Seiradake E, Jones EY, Sansom MS. Structures of the EphA2 receptor at the membrane: Role of lipid interactions. Structure. 2016;24:337–347. doi: 10.1016/j.str.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julkowska MM, Rankenberg JM, Testerink C. Liposome-binding assays to assess specificity and affinity of phospholipid-protein interactions. Methods Mol Biol. 2013;1009:261–271. doi: 10.1007/978-1-62703-401-2_24. [DOI] [PubMed] [Google Scholar]
- 30.Fliesler SJ, Anderson RE. Chemistry and metabolism of lipids in the vertebrate retina. Prog Lipid Res. 1983;22:79–131. doi: 10.1016/0163-7827(83)90004-8. [DOI] [PubMed] [Google Scholar]
- 31.Li G, et al. Efficient replacement of plasma membrane outer leaflet phospholipids and sphingolipids in cells with exogenous lipids. Proc Natl Acad Sci USA. 2016;113:14025–14030. doi: 10.1073/pnas.1610705113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y, Jensen GJ, Bjorkman PJ. Cryo-electron tomography of homophilic adhesion mediated by the neural cell adhesion molecule L1. Structure. 2009;17:460–471. doi: 10.1016/j.str.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald KL. Out with the old and in with the new: Rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma. 2014;251:429–448. doi: 10.1007/s00709-013-0575-y. [DOI] [PubMed] [Google Scholar]
- 34.McDonald K, et al. “Tips and tricks” for high-pressure freezing of model systems. Methods Cell Biol. 2010;96:671–693. doi: 10.1016/S0091-679X(10)96028-7. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro L, Doyle JP, Hensley P, Colman DR, Hendrickson WA. Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin. Neuron. 1996;17:435–449. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 36.Kunz B, et al. Axonin-1/TAG-1 mediates cell-cell adhesion by a cis-assisted trans-interaction. J Biol Chem. 2002;277:4551–4557. doi: 10.1074/jbc.M109779200. [DOI] [PubMed] [Google Scholar]
- 37.Sawaya MR, et al. A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms. Cell. 2008;134:1007–1018. doi: 10.1016/j.cell.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolaienko RM, et al. Structural basis for interactions between contactin family members and protein-tyrosine phosphatase receptor type G in neural tissues. J Biol Chem. 2016;291:21335–21349. doi: 10.1074/jbc.M116.742163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouyain S, Watkins DJ. The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules. Proc Natl Acad Sci USA. 2010;107:2443–2448. doi: 10.1073/pnas.0911235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H, Focia PJ, He X. Homophilic adhesion mechanism of neurofascin, a member of the L1 family of neural cell adhesion molecules. J Biol Chem. 2011;286:797–805. doi: 10.1074/jbc.M110.180281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.