Significance
Evolution acts on mutations that naturally arise within the genome and are shaped both by intrinsic genomic features and by the cellular environment. We catalog the mutations arising in a heterozygous diploid yeast during passaging in vitro and in the mammalian host. We establish genome-wide mutation rates and reveal that “microscale” changes (base substitutions and short-track recombination events) are the primary drivers of microevolution, although chromosomal-level changes also occur in specific host environments. Our results define mutation hotspots, including those adjoining recombination tracts, and indicate that many mutations are purged from the population due to purifying selection. Together, these data provide a high-resolution picture of how the heterozygous diploid genome of a fungal pathogen undergoes evolution over short time scales.
Keywords: microevolution, Candida albicans, aneuploidy, LOH, diploid species
Abstract
Candida albicans is a heterozygous diploid yeast that is a commensal of the human gastrointestinal tract and a prevalent opportunistic pathogen. Here, whole-genome sequencing was performed on multiple C. albicans isolates passaged both in vitro and in vivo to characterize the complete spectrum of mutations arising in laboratory culture and in the mammalian host. We establish that, independent of culture niche, microevolution is primarily driven by de novo base substitutions and frequent short-tract loss-of-heterozygosity events. An average base-substitution rate of ∼1.2 × 10−10 per base pair per generation was observed in vitro, with higher rates inferred during host infection. Large-scale chromosomal changes were relatively rare, although chromosome 7 trisomies frequently emerged during passaging in a gastrointestinal model and was associated with increased fitness for this niche. Multiple chromosomal features impacted mutational patterns, with mutation rates elevated in repetitive regions, subtelomeric regions, and in gene families encoding cell surface proteins involved in host adhesion. Strikingly, de novo mutation rates were more than 800-fold higher in regions immediately adjacent to emergent loss-of-heterozygosity tracts, indicative of recombination-induced mutagenesis. Furthermore, genomes showed biased patterns of mutations suggestive of extensive purifying selection during passaging. These results reveal how both cell-intrinsic and cell-extrinsic factors influence C. albicans microevolution, and provide a quantitative picture of genome dynamics in this heterozygous diploid species.
Microbial evolution studies have provided detailed insights into genome dynamics and the factors that shape evolution (1–5). Most of these studies have focused on haploid or homozygous diploid genomes, yet there is now an increasing need to also examine genome variation in heterozygous genomes (4, 6–9). While sexual reproduction plays an important role in accelerating genome variation in eukaryotic species (5, 10, 11), mitotic populations can still generate diversity by a number of mechanisms, including de novo base substitutions, loss-of-heterozygosity (LOH) events, insertions/deletions (indels), and chromosomal-level changes, such as the acquisition of different aneuploid states. Many of these events have been defined in model eukaryotes and are also recognized in the human genome as major drivers of somatic mosaicism and cancer development (12, 13).
In this study, we provide a high-resolution analysis of genome microevolution in the heterozygous diploid yeast Candida albicans. This species is an opportunistic human pathogen responsible for both a variety of debilitating mucosal infections and life-threatening systemic infections (14, 15). A common resident of the human microbiota, C. albicans can become pathogenic in individuals that are immunocompromised or undergo prolonged antibiotic use (15–17). The genome consists of eight chromosomes, with the reference isolate SC5314 harboring ∼70,000 heterozygous positions representing ∼0.5% of the 14.3-Mb genome (18–20). Heterozygous regions of the genome can undergo LOH, the frequency of which increases in response to environmental stressors (21–24). Isolates can also experience large-scale genomic changes, including the emergence of aneuploid forms (24–27). C. albicans isolates therefore display extensive genomic plasticity due to a variety of events, including acquisition of SNPs and indels, LOH events, and changes in gene and chromosome copy number (22, 24, 27–32).
Here, we define the complete spectrum of mutations accompanying microevolution in C. albicans, using deep sequencing on multiple clinical isolates to precisely determine the mutations arising during both in vitro and in vivo passaging. Our experiments reveal that microevolution is driven by widespread, small-scale genetic changes, overwhelmingly represented by de novo base substitutions and short-tract LOH (SLOH) events. In contrast, large-scale genomic changes are rare, although both long-tract LOH (LLOH) events and the acquisition of supernumerary chromosomes were observed, with the latter associated with an increase in commensal fitness. We also identify hypermutable sites within the genome, including repetitive and telomeric regions, and show that DNA recombination events are themselves highly mutagenic and are accompanied by a large number of de novo mutations. Genetic events leading to gains of heterozygosity (GOH) and LOH occurred at similar frequencies so that global heterozygosity levels were, for the most part, stably maintained during microevolution. Finally, we note that mutational patterns infer a dominant role for purifying selection, with most mutations that alter protein-coding sequences likely purged from the genome during infection of the mammalian host.
Results
Microevolution of C. albicans Diploid Genomes.
We selected four clinical isolates of C. albicans (SC5314, P78048, P76055, and P57055) for microevolution experiments. These isolates belong to three major C. albicans clades (I, II, and III), originate from bloodstream infections, and are heterozygous at the mating type-like locus (MTLa/α), except for P78048, which is MTLα/α (Dataset S1). Each strain has a highly heterozygous genome with no chromosomal aneuploidies, and heterozygous positions represent 0.41–0.55% of these genomes corresponding to an average of ∼70,000 positions in the ∼14-Mb genome (Dataset S2) (24). The four isolates have been shown to be virulent in both mouse and insect models of systemic infection (24, 33), exhibit normal morphology patterns, and are susceptible to antifungal agents (24). Generation times for these isolates in YPD medium at 30 °C range between 88 and 95 min (Dataset S3).
Each of the four isolates were passaged both in vitro and in three different murine models in vivo (Fig. 1A). The latter included two commensal models of gastrointestinal (GI) colonization using either a standard diet (SD) that requires antibiotics for C. albicans colonization (34), or a purified diet (PD) that does not require antibiotics (35). For GI colonization, fungal cells were collected from fecal pellets after 42 d (n = 2–3). No differences were observed in colonization levels during microevolution, as determined by fungal burdens in fecal pellets from both GI colonization models (SI Appendix, Fig. S1 A and B), with one exception (P78048 GI SD isolate D). A model of systemic infection was also utilized in which fungal cells were introduced into the bloodstream and recovered from the kidney, the major murine organ targeted by C. albicans (36). For systemic infections, five sequential passages were performed in which cells were isolated from infected kidneys 3 d postinfection and used for infection of new hosts (n = 2). Fungal burdens in the kidneys did not show any obvious trends during microevolution (SI Appendix, Fig. S1C). For comparison, in vitro passaging was performed daily under standard laboratory conditions (YPD medium, 30 °C) and isolates collected after 80 d (∼600 generations, n = 1–2). Genomes of evolved isolates were analyzed by Illumina ultradeep sequencing (∼185× coverage, 97.7% of SC5314 assembly covered by reads) (Dataset S2). Using high-depth read alignments and stringent variant calling with both Haplotype Caller (Genome Analysis Toolkit) and Pilon (37), SNPs, heterozygous positions, and indels were identified for each isolate (Dataset S2), and a number of these were further validated, as described below. Quality metrics for identified variants are included in SI Appendix, Fig. S1 D and E.
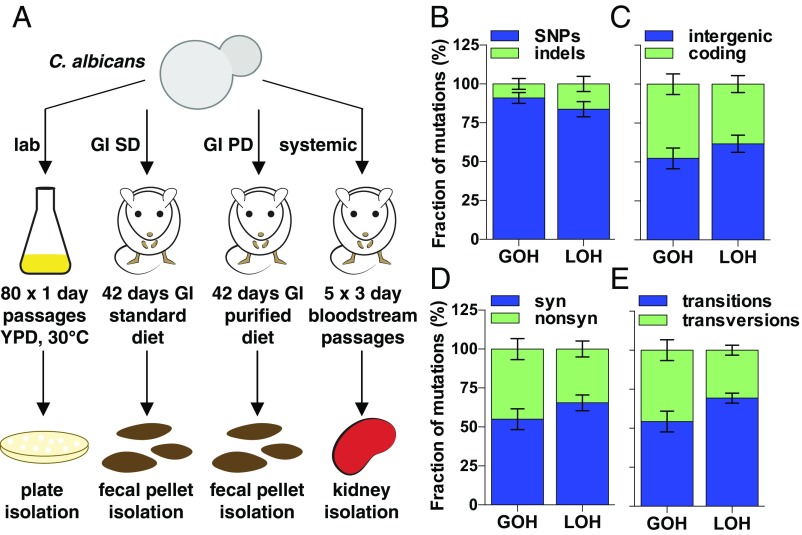
Fig. 1.
Microevolution of C. albicans genomes. (A) Schematic of in vitro and in vivo microevolution experiments. (B) Distribution of SNPs and indels, (C) intergenic and coding mutations, (D) synonymous and nonsynonymous mutations, and (E) transitions and transversions. Data are averaged across all microevolution experiments. Note that panels indicate mutations resulting from either GOH or LOH events.
Common Patterns of Microvariation in C. albicans Genomes.
A detailed analysis of microevolved isolates was performed to determine the spectrum of nucleotide changes in each genome. A total of 564 mutations were identified across the 28 microevolved isolates in the four lineages. All 564 mutations were individually evaluated using the Integrative Genomics Viewer (IGV) (38) and 63 sites were additionally verified using an allele-specific fluorescent PCR technology (KASP genotyping). Of these, 55 positions (87%) matched the genotypes from genome sequencing and included SNPs and indels in both genic and intergenic regions (SI Appendix, Fig. S1F and Dataset S4).
Mutations were subdivided into those leading to GOH and LOH, and also classified as resulting from indels or changes in SNPs. For example, in vitro passaging for 600 generations revealed a total of 31 mutations in four microevolved lineages (4, 5, 7, and 15 mutations in each). The 31 mutations involved 25 SNPs (17 transitions and 8 transversions) and 6 indels (4 insertions and 2 deletions). These are comprised of 8 GOH events (due to 7 de novo base substitutions and 1 indel) and 11 LOH events (due to gene conversion impacting 18 SNPs and 5 indels). Nineteen of the mutations occurred in intergenic regions and 12 in coding sequences, of which there were 7 synonymous and 5 nonsynonymous mutations.
Microevolution consistently involved more changes in SNPs (either de novo mutations or LOH) than events due to indels. Thus, from the 564 mutations, 523 (93%) involved changes in SNPs and 41 (7%) involved indels (Fig. 1B). For GOH events, the average ratio of base substitutions to indels was 1:0.17, which is lower than ratios reported for Saccharomyces cerevisiae (∼1:0.03) (39–41), suggesting that C. albicans experiences proportionally higher rates of indels to de novo substitutions than S. cerevisiae. An average of 48% of GOH mutations and 38% of LOH mutations occurred in coding regions and involved mostly synonymous mutations (55% and 65%, respectively) (Fig. 1 C and D). Nonsynonymous mutations predicted to disrupt ORF function were rare; only 14 nonsense mutations and 5 readthrough mutations occurred across the 28 evolved isolates, and 17 of these were the direct result of three very large LOH events that occurred in two isolates (Dataset S5). De novo base substitutions (GOH SNPs) in evolved isolates were the result of a higher fraction of transitions than transversions, with an average Ts/Tv ratio of 1.3:1, which is lower than the 2:1 ratio reported for model yeast genomes (40, 42) (Fig. 1E and SI Appendix, Fig. S2A). These mutational patterns were consistent across microevolved isolates and were independent of genetic background and the passaging environment.
Purifying Selection Shapes the Evolution of C. albicans Genomes.
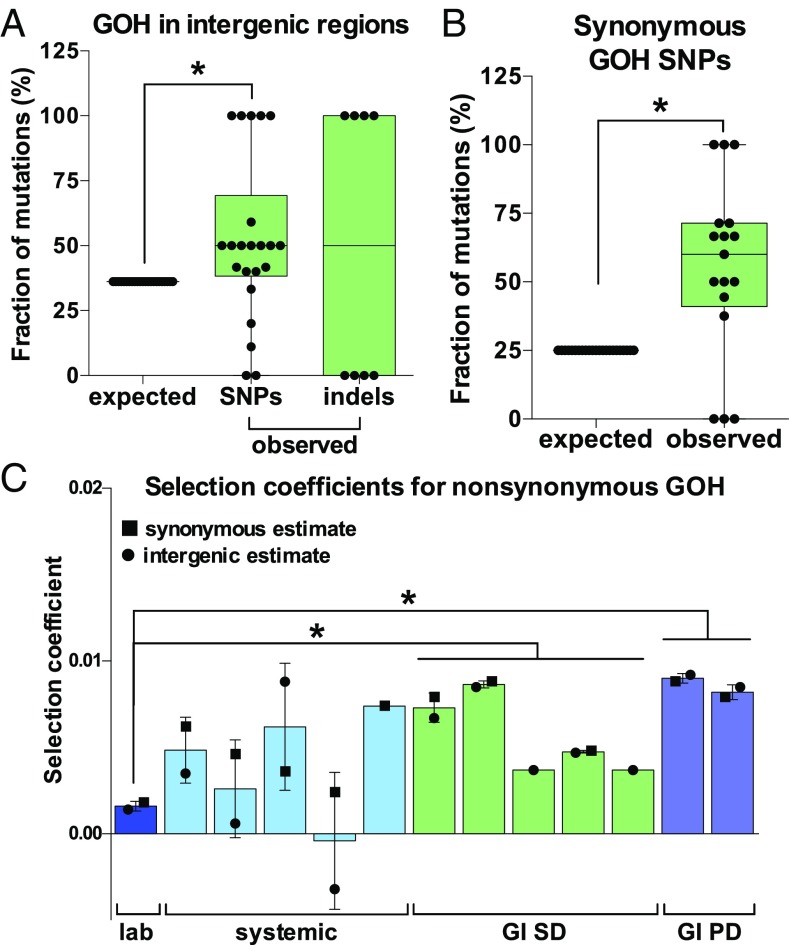
In the absence of bottlenecks, new mutations with deleterious effects may be purged from the population via purifying selection, and we therefore tested whether mutational patterns in our dataset showed evidence for selection. If occurring randomly, mutations will accumulate in intergenic and coding regions at frequencies proportional to their representation in the genome (39, 40, 43). In our experiments, 48% (59 of 123) of all de novo base substitutions (GOH SNPs) and 62% (8 of 13) of GOH indels were present in intergenic regions, even though these regions account for only 36.2% of the genome (P < 0.05) (Fig. 2A). Moreover, none of the indels (0 of 5) found in coding sequences resulted in frameshifts (i.e., all were a multiple of three nucleotides), whereas only one-eighth of indels observed in intergenic regions consisted of multiples of three nucleotides (P < 0.05, difference between indels in intergenic and coding regions is significant using a binomial distribution) (SI Appendix, Fig. S2B). The fraction of synonymous to nonsynonymous mutations also differed from that expected by chance: ∼25% of coding substitutions are expected to be synonymous if mutations occur randomly (39, 43), yet 52% (36 of 69) of base substitutions were synonymous in our dataset (P < 0.05) (Fig. 2B). This suggests that selection frequently acts to limit the accumulation of mutations that alter the protein-coding sequence.
Fig. 2.
Selection shapes microevolution of C. albicans genomes. (A) Frequency of observed and expected GOH mutations in intergenic regions (these regions represent 36.2% of the C. albicans genome). GOH SNP mutations represent de novo base substitutions. (B) Observed and expected fractions of synonymous GOH SNPs in coding regions during microevolution experiments (∼25% of base substitutions are expected to be synonymous if random events). (C) Selection coefficients for nonsynonymous GOH SNPs calculated based on the number of observed vs. expected nonsynonymous substitutions. Expected nonsynonymous substitutions were estimated for each microevolution experiment based on observed synonymous substitutions (■) or observed intergenic substitutions (●). Only isolates for which both nonsynonymous and synonymous/intergenic substitutions were observed were included in this analysis. For each panel, data points represent independently evolved isolates and asterisks indicate significant differences (t test, *P < 0.05).
We also estimated the fraction of mutations impacted by selection by examining how many nonsynonymous mutations should occur during microevolution based on the number of synonymous or intergenic mutations observed. Assuming an even distribution of mutational events, selection effectively removed an average of 71–79% of the nonsynonymous mutations predicted to occur. Selection coefficients for nonsynonymous mutations in evolved isolates averaged 0.0053 (0.0047 and 0.0059, using estimates based on observed intergenic or synonymous mutations, respectively). Selection coefficients were highest for isolates passaged in the GI tract (P < 0.0005) (Fig. 2C). Taken together, these results establish that C. albicans isolates display a larger proportion of synonymous and intergenic mutations than that expected by chance, implying that purifying selection removes a large proportion of mutations impacting protein-coding genes during passaging.
Impact of Strain Background and Environment on C. albicans Mutation Frequencies.
Mutation frequencies were compared between the four clinical isolates and across culture conditions to examine cell-intrinsic and cell-extrinsic factors impacting microevolution. Strains passaged in vitro displayed an average rate of 1.17 × 10−10 base substitutions per base pair per generation (Fig. 3A). These de novo substitution rates are similar to those reported for asexual populations of both S. cerevisiae and Schizosaccharomyces pombe (39–41, 44) (SI Appendix, Fig. S2C). Mutations in C. albicans cells passaged in vitro reflected mutational patterns common to all microevolution experiments, with more SNPs than indels, and fewer coding mutations than expected by chance (SI Appendix, Fig. S2D). Isolates passaged in vitro displayed an average LOH rate of 1.61 × 10−10 per base pair per generation corresponding to 6.2 × 10−8 events per heterozygous position per generation (Fig. 3A).
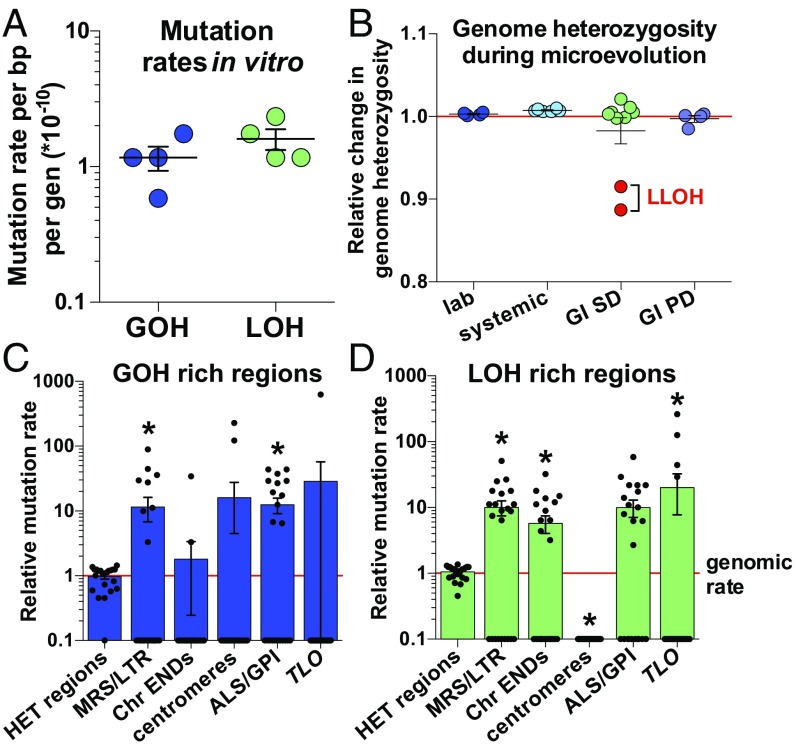
Fig. 3.
Mutation rates in C. albicans and the impact of chromosomal features on mutation rates. (A) GOH and LOH mutation rates during in vitro growth. Rates include both SNP and indel events. (B) Fluctuations in genome heterozygosity during microevolution relative to starting heterozygosity levels (red line). Significant decreases in heterozygosity are only observed for two isolates that underwent LLOH events (red symbols). (C and D) GOH (C) and LOH (D) mutation rates in specific regions relative to whole-genome rates. These include heterozygous (HET) regions, repeat regions (MRS and LTR), Chr END regions (final 10 kb of each chromosome arm), centromeres, genes encoding ALS and GPI-linked proteins, and TLO genes. Mutations include both changes in SNPs and indel events. Data points represent independently evolved isolates, and asterisks indicate significant differences relative to whole-genome mutation frequencies (t test, *P < 0.05).
In contrast to in vitro mutation rates, in vivo rates cannot be accurately determined as precise generation times have not been defined in the host. Estimates of 0.09 generations per hour during systemic infection (31) and 0.14 generations per hour during GI colonization (45) are only two- to threefold slower than in vitro growth in rich medium. To avoid using these estimates, we calculated mutation frequencies relative to total passage time. The standard “laboratory” strain SC5314 displayed the lowest mutation frequencies (both for GOH and LOH) as frequencies in the other lineages were 1.5- to 6-fold higher (SI Appendix, Fig. S2E). The environment also potentially impacted the mutation frequency; strains grown in vivo (both GI and systemic models) showed GOH frequencies that were 4.9- to 11-fold higher than those in vitro (SI Appendix, Fig. S2F). LOH frequencies were also higher in vivo than in vitro (6- to 8.3-fold) (SI Appendix, Fig. S2F). These results suggest that C. albicans cells exhibit higher mutation frequencies when passaged in the host relative to in vitro culture.
Genome Heterozygosity Levels Are Often Maintained During Microevolution.
Given that LOH events are frequently observed in C. albicans (22, 24, 25, 46–48), we examined if genome heterozygosity levels changed during passaging. We observed that LOH and GOH frequencies were balanced in most microevolved lineages (SI Appendix, Fig. S2G). Thus, genome heterozygosity levels in the four lineages were often maintained during passaging, with heterozygosity levels within −1.5 to +2.1% of starting levels (Fig. 3B). The striking exceptions to this pattern were two isolates that experienced very large (>0.27 Mb) LOH events and therefore showed concomitant decreases (−8.5% and −11.3%) in heterozygous positions across their genomes. These results establish that, in the absence of large LOH events, mitotic recombination events (driving LOH) and GOH events (resulting from both de novo base substitutions and indels) often impact similar numbers of positions so that overall heterozygosity levels can be stably maintained.
Defining Hypervariable Regions Within the C. albicans Genome.
To determine the impact of genomic context on mutation rates, we compared the frequency of GOH and LOH mutations arising at different chromosomal features, including centromeres, terminal chromosome regions, subtelomeric TLO genes, ALS (agglutinin-like sequence) genes, other glycosylphosphatidylinositol (GPI)-linked genes, and annotated repeat regions (Datasets S6 and S7). Several of these features have been associated with higher mutation rates in C. albicans and model organisms (4, 49–55). For each feature, we examined the frequency of SNP and indel mutations relative to the genome average. We found that LOH (but not GOH) frequencies were elevated at the ends of chromosomes (5.8-fold increase within the 10-kb terminal regions, P = 0.01), in line with previous observations that C. albicans telomeric regions are highly dynamic (56) (Fig. 3 C and D). In contrast, centromeric regions displayed decreased LOH mutation frequencies relative to the genome average. This contrasts with the high mutation rates observed at S. cerevisiae centromeres (53); however, the 3- to 4.5-kb regional centromeres present in C. albicans are much larger than the point centromeres (often <400 bp) found in the model yeast (57, 58).
Repeat sequences are common in fungal genomes and have been associated with mobile elements and gene regulation (59, 60). However, studies in model yeast have tended to exclude analysis of repeat regions to simplify genome analyses (39, 44). C. albicans is unusual among Candida species in that it contains nine large major repeat sequence (MRS) elements that span ∼1.7% of the genome and are linked to chromosome translocations and chromosome-length polymorphisms (56, 60–63). We found that microevolved C. albicans lineages exhibited significant differences in mutation frequencies between repeat and nonrepeat regions. For example, MRS elements and long terminal repeat (LTR) retrotransposons showed GOH and LOH frequencies that were 11.5- and 10-fold higher than the genome average, respectively (P < 0.04) (Fig. 3 C and D). Importantly, a number of these mutations were validated by KASP genotyping, confirming that they arose during microevolution (Dataset S4).
Genes encoding GPI-linked cell wall proteins and ALS family genes are rich in internal tandem repeats that can vary in number, and thereby contribute to allelic diversity and phenotypic variation (64–67). In line with these observations, we found that GPI and ALS gene families accumulated mutations at higher frequencies than the genome average (12.5- and 10-fold higher GOH and LOH rates, respectively, P < 0.005) (Fig. 3 C and D). These results establish that numerous chromosomal features, including genomic repeats, telomeric regions, ALS genes, and GPI-linked cell wall genes, undergo evolution faster than the rest of the genome.
We also compared the distribution of mutations arising in heterozygous versus homozygous regions of the genome, while noting that certain LOH events in C. albicans have been shown to promote adaptation (30, 68, 69). Heterozygous and homozygous regions were mapped in each of the four parental strains based on the density of heterozygous positions per 5-kb window (24). Using this metric, we found that heterozygous regions varied between 69.5% and 84.2% of the genome (Dataset S8). Comparison of the frequency of mutations between heterozygous and homozygous regions revealed that these regions accumulated GOH and LOH mutations in line with their relative abundance (Fig. 3 C and D). For example, 84.2% of the P78048 genome is represented by heterozygous regions and 80.2% of all mutations in evolved derivatives of this lineage occurred in heterozygous regions. We therefore did not observe any bias in the pattern of either de novo mutations or gene-conversion events toward either heterozygous or homozygous regions of the genome.
Microevolution Is Punctuated by Frequent SLOH Events and Small Indels.
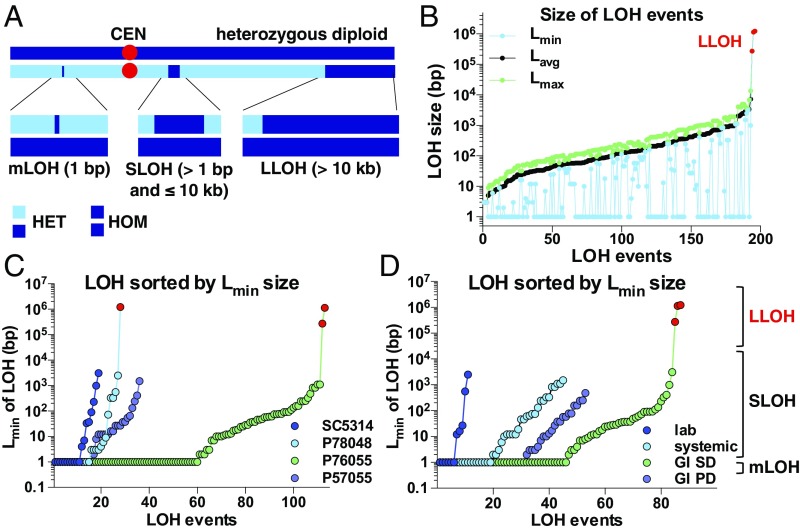
A wide variety of LOH events have been observed in C. albicans, with elevated rates reported during exposure to stress, antifungal treatment, DNA damage, and host passage (22, 31, 32, 70, 71). To characterize these events during microevolution, we divided LOH tracts into three categories based on length and the number of heterozygous positions affected: (i) microLOH (mLOH) events that involve loss of single heterozygous positions, (ii) SLOH events that involve loss of two or more heterozygous positions yet cover short (≤10 kb) genomic regions, and (iii) LLOH events that are >10 kb and often affect hundreds of heterozygous positions (Fig. 4A). The relative frequency of mLOH and SLOH events observed during microevolution was similar across experiments, with the minimum size of these events ranging between 1 and 3,090 bp (Lmin size) (Fig. 4B). Thus, isolates underwent an average of 52.3% mLOH and 46.4% SLOH events during passaging (Fig. 4 C and D). The average size of LOH tracts (Lavg) varied between 222 bp (P57055) and 889 bp (SC5314) for the four strain backgrounds and impacted between 1.6 and 3.2 heterozygous positions (excluding LLOH events) (SI Appendix, Fig. S3 A and B). In contrast to frequent mLOH and SLOH events, only three LLOH events occurred in two microevolved isolates (P76055 and P78048 passaged in the GI SD model) and involved tracts of 273–1,230 kb that extended to the ends of the chromosomes.
Fig. 4.
Microevolution is punctuated by frequent SLOH events. (A) Schematic of different types of LOH events, including mLOH (loss of single heterozygous positions), SLOH (<10 kb and involve loss of two or more heterozygous positions), and LLOH (>10 kb and affect hundreds of heterozygous positions). (B) Distribution of LOH events showing the Lmin, Lavg, and Lmax size for each LOH event. (C and D) Lmin size distribution of LOH events, including mLOH, SLOH, and LLOH (shown in red), for each lineage (C) and each niche (D). For B–D, data points represent independent LOH events.
We also analyzed the 41 indels that occurred in the 28 evolved isolates (SI Appendix, Fig. S2 H and I), noting that indels in S. cerevisiae are biased toward insertions in haploid cells and toward deletions in diploid cells (39, 40). In the evolved C. albicans lineages, indels were represented by similar numbers of insertions (21 events) and deletions (20 events). Indel sizes averaged 3.3 bp for in vitro-evolved isolates, whereas indels were two- to threefold larger for in vivo-passaged isolates (SI Appendix, Fig. S2I), although no significant size differences were found between different lineages or niches for either LOH events or indels (P > 0.05). Together, these results provide a comprehensive analysis of LOH and indel events in C. albicans and highlight that SLOH, most of which involve homozygosis of only single heterozygous positions, are the most common LOH event during microevolution.
LOH Events Are Frequently Associated with MRS Repeat and Telomeric Regions.
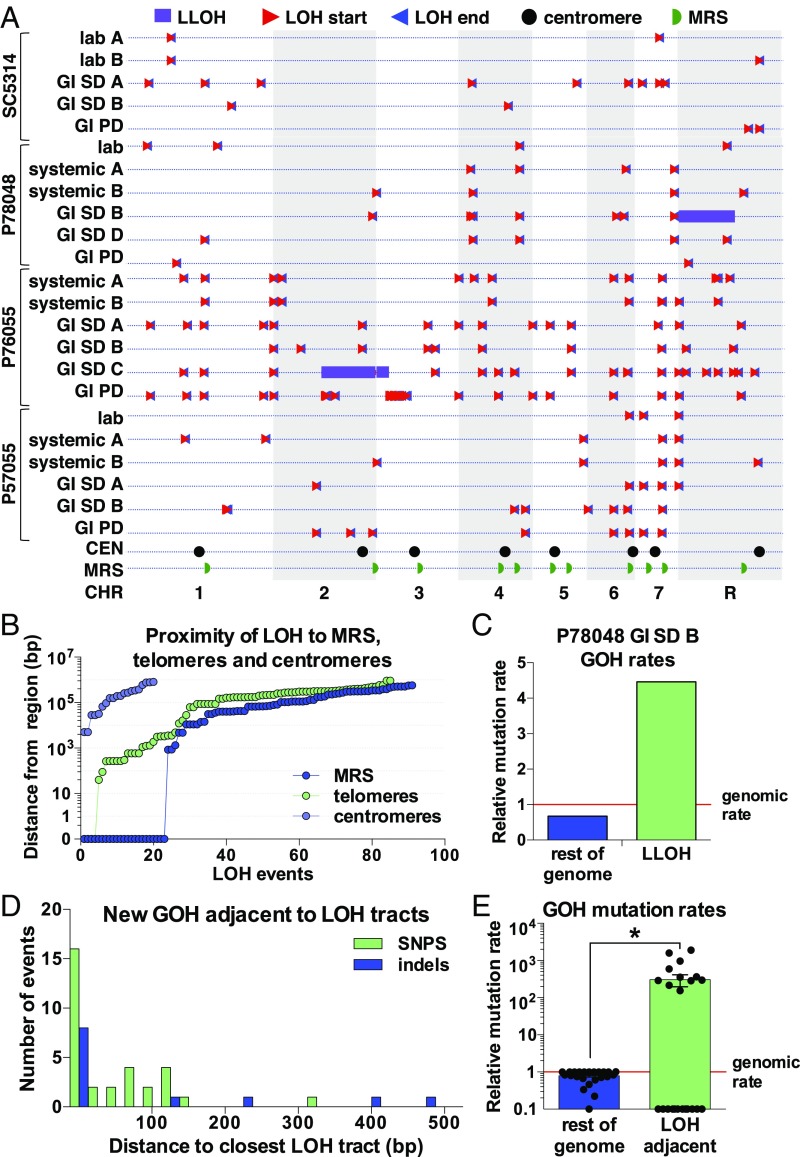
To examine the distribution of LOH events relative to chromosomal features, the 196 LOH tracts were mapped within the C. albicans genome (Fig. 5A) and their frequency determined per 0.2-Mb window (SI Appendix, Fig. S3C). Mapping the distance of each LOH to the closest chromosomal feature revealed that a large subset of LOH tracts (21%) arose either within MRS regions or in the 1-kb tracts adjacent to MRS or telomeric regions (Fig. 5B). MRS elements were the main hotspot for these recombination events, with the start sites for 12% of all LOH tracts being located at these elements. Not all MRS tracts showed the same propensity for recombination; MRS6 and MRS7b displayed the highest LOH frequencies and MRS2, -3, and -5 displayed the lowest LOH frequencies (SI Appendix, Fig. S3C). In contrast, no LOH events were detected within 5 kb of the centromeres (Fig. 5B). These findings establish that MRS and telomeric regions are hotspots for recombination in C. albicans.
Fig. 5.
Relationship between LOH location and different genomic regions. (A) Chromosomal location of all LOH events (using Lmin) with triangles marking the start (red) and end (blue) of each event. Location of centromeres (CEN) and MRS regions are shown. (B) Proximity of LOH events to the closest genomic feature, including MRS regions, telomeres (Chr ENDs or TLO genes), and centromeres. Each LOH event is uniquely mapped to the closest of these features on the same chromosome arm. Distances equal to 0 indicate an LOH start site inside the respective genomic region. (C) GOH rates (including SNPs and indels) in the duplicated LLOH region and in the rest of the genome in the P78048 GI SD B isolate, relative to whole genome GOH rates. Only GOH SNPs (base substitutions) were observed in the duplicated LLOH region. (D) Number of GOH events (SNPs and indels per 25 bp) observed within 500 bp of LOH tracts in microevolved isolates. (E) GOH rates (including SNPs and indels) in regions adjacent to LOH tracts (within 500 bp) and in the rest of the genome, relative to whole genome GOH rates. Data points represent independently evolved isolates, asterisks indicate significant differences relative to whole genome mutation frequencies (t test, *P < 0.05).
New Heterozygous SNPs Emerge Within a Large LOH Tract.
Three large LOH tracts (0.27–1.23 Mbp) were formed during passaging and involved two isolates evolved in the GI (antibiotic-treated) model with the SD. LOH events involved the terminal regions of chromosome (Chr) 2 and Chr 3 in P76055 (GI SD isolate C) and Chr R in P78048 (GI SD isolate B) (Fig. 5A and SI Appendix, Fig. S4A). LOH occurred via truncation of chromosome arms in P76055 GI SD isolate C, as the resulting LOH tracts were monosomic for both Chr 2 and Chr 3 (SI Appendix, Fig. S4B). In contrast, LOH likely involved break-induced replication (BIR) or interhomolog crossing-over in P78048 GI SD isolate B, as the chromosome was still disomic for the emergent homozygous region (SI Appendix, Fig. S4B). These events affected thousands of heterozygous positions (as well as tens of indels) that were present in the parental genomes. In total, the three LLOH led to the homozygosis of 5,419 sites in coding regions, 2,135 of which resulted in nonsynonymous changes: 14 produced nonsense mutations and 3 resulted in readthrough mutations (SI Appendix, Fig. S4C and Dataset S5). Interestingly, the LLOH region in P78048 GI SD isolate B showed the reemergence of multiple heterozygous positions due to de novo base substitutions (GOH SNPs) within the LOH tract. In fact, a 6.6-fold higher rate of GOH events was detected within this LLOH tract than was evident in the rest of the genome (Fig. 5C). The high frequency of de novo mutations is consistent with an elevated rate of base substitutions emerging during BIR, as shown for S. cerevisiae (72).
Impact of LOH Events on Mutational Patterns.
Chromosomal cross-overs can induce de novo mutations in DNA regions close to the cross-over in a wide variety of species (4, 49, 50, 73, 74). We therefore examined whether the regions flanking emergent LOH tracts in C. albicans isolates showed altered mutation rates relative to the genome average. Strikingly, LOH-adjacent sites appeared highly enriched for mutations; 44 of 136 GOH events (32 de novo base substitutions and 12 indels) were located within 500 bp of emergent LOH tracts (Fig. 5D). In fact, 36 GOH events were located within just 100 bp of LOH tracts. Thus, 32.4% of all GOH events (26% of base substitutions and 92.3% of indels) were found in regions close to new LOH tracts, even though these regions represent only ∼1.3% of the genome. GOH rates in LOH-adjacent regions (500-bp up/down of LOH tract) were therefore 840-fold higher than in the rest of the genome (Fig. 5E).
The pattern of GOH events adjacent to LOH tracts differed from that in the rest of the genome, with indels particularly enriched in LOH-adjacent regions (SI Appendix, Fig. S4D). In contrast, GOH mutations arising in TLO genes, centromeric regions, or telomeric regions were not closely associated with LOH (SI Appendix, Fig. S4D). We further note that de novo base substitutions in LOH-adjacent regions showed a Ts/Tv ratio of 1.1:1 compared with a genome average of 1.3:1. This is consistent with high rates of transversions resulting from translesion polymerases acting to repair DNA lesions at or close to recombination tracts (75, 76). Both homologous recombination and nonhomologous end-joining are considered to be error-prone mechanisms that can introduce indels close to the site of a DNA break (77). Our data now reveal that approximately one-third of all GOH indels and base substitutions in C. albicans arise in regions flanking LOH events and that this is likely due to mutagenic DNA repair mechanisms.
Large-Scale Chromosomal Changes Acquired During Microevolution.
Sequence read depth across the microevolved genomes revealed that a subset of isolates underwent changes at the chromosomal level. Aneuploidy was observed in 3 of 28 evolved isolates and in each case involved Chr 7 trisomies (SI Appendix, Fig. S5 A and B). These aneuploids emerged in three different strain backgrounds passaged in the GI SD model, suggesting a fitness benefit may be associated with Chr 7 trisomy in this niche. One of the aneuploid isolates also became monosomic for two terminal regions involving the right arm of Chr 2 (1.15-Mbp region) and the left arm of Chr 3 (0.27-Mbp region; P76055 GI SD isolate C) (SI Appendix, Fig. S4B). Previous studies have similarly observed aneuploid forms among natural isolates and that chromosome-level changes can arise during passaging or in response to antifungal treatment (24, 27, 29, 31, 32, 70, 78). An analysis of copy number variation across smaller genomic regions (100- to 1,000-bp windows) was also performed (SI Appendix, Figs. S6 and S7).
Chr 7 Trisomy Promotes Increased Fitness in the Mouse GI Tract.
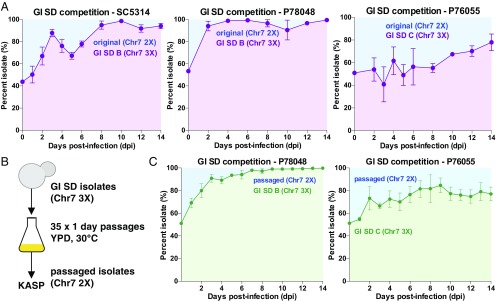
To determine whether the emergence of Chr 7 trisomies provides a fitness advantage in the GI, we performed direct competition experiments between strains that were disomic (2X) or trisomic (3X) for this chromosome. Three parental isolates (SC5314, P78048, and P76055) were genetically marked with the SAT1 nourseothricin resistance gene integrated at the NEUT5L locus (79), as this does not impact strain fitness per se (80) but allows for selection. Microevolved isolates harboring the Chr 7 trisomy (SC5314 GI SD isolate B, P78048 GI SD isolate B, and P76055 GI SD isolate C) and the corresponding SAT1+ parental strains were coinoculated at a 1:1 ratio into the GI and their relative proportions measured in mouse fecal pellets over 14 d by quantifying the ratio of SAT+/SAT− cells. In all cases, the strains that were trisomic for Chr 7 outcompeted the corresponding parental strain, with two of the evolved isolates representing >90% of the recovered population 8 d postinoculation (Fig. 6A). This is consistent with Chr 7 trisomies increasing the fitness of C. albicans isolates in the murine GI tract.
Fig. 6.
Chr 7 trisomy is associated with a fitness advantage for colonization of the GI tract. (A) Competition experiments comparing the relative fitness of evolved (Chr 7 3X) and parental isolates (Chr 7 2X) in the GI SD colonization model. Evolved isolates represent strains that had become trisomic for Chr 7 following passage in the GI SD model. C. albicans cells were recovered from mouse fecal pellets and the relative levels of evolved and parental isolates determined. (B) Trisomic isolates were passaged in vitro to induce loss of the trisomic Chr 7 and resulting isolates were genotyped using KASP. (C) Competition experiments comparing the fitness of GI SD evolved isolates (Chr 7 3X) and in vitro passaged derivatives that were disomic for Chr 7 (2X). Strains were competed in the GI SD colonization model, cells recovered from mouse fecal pellets, and the relative levels of evolved and passaged isolates determined. Error bars represent ±SD; four mice were used for each competition.
To induce loss of the trisomic Chr 7, aneuploid strains were serially passaged in vitro for 35 d (∼238 generations) and nine isolates evaluated for Chr 7 copy number by KASP genotyping (Fig. 6B). We found that two of the nine in vitro-passaged isolates had lost the trisomic Chr 7, and these strains were competed against the corresponding Chr 7 3X isolates in the GI SD infection model. In both strain backgrounds (P78048 and P76055), Chr 7 3X strains displayed a significant fitness advantage relative to their Chr 7 2X derivatives (Fig. 6C) (P < 0.001). This provides further evidence that an extra copy of Chr 7 contributes to increased fitness of C. albicans cells in the GI infection model.
Discussion
This study defines the spectrum of mutations that emerge in heterozygous diploid genomes of C. albicans during microevolution, including a comparison of mutational patterns during in vitro culture with those that emerge during infection of the host. Numerous studies have established that C. albicans exhibits extensive genomic plasticity, from variation at the level of SNPs to changes at the whole-chromosome level (24, 25, 30, 32, 56, 81). However, this work provides a comprehensive picture of the genetic changes accompanying microevolution in this important pathogen.
Global Patterns of Mutation in C. albicans.
Microevolution resulted in similar mutational patterns regardless of strain background or environment, and was driven almost exclusively by changes in heterozygous polymorphisms. These included either new polymorphisms due to de novo base substitutions or LOH positions via gene-conversion events, as well as a relatively small number of indels. This establishes that “microscale” changes are the most frequent events arising in the C. albicans genome. We reveal that C. albicans displays an average de novo base-substitution rate of 1.17 × 10−10 per base pair per generation during in vitro passaging. This genome-wide estimate of C. albicans mutation rates is close to those reported for mitotically dividing cells of S. cerevisiae and S. pombe (8, 39, 40, 44), indicating that C. albicans exhibits a de novo substitution rate similar to that of haploid or homozygous diploid yeast genomes. Notably, microvariation is equally driven by LOH, as gene-conversion events occur at frequencies (1.61 × 10−10 per base pair per generation) that are close to those of de novo substitution rates and impact a similar number of nucleotide positions.
Genome Architecture and Environmental Pressures Impact C. albicans Microevolution.
While overall mutational patterns were similar between microevolved lineages, de novo substitution and LOH frequencies varied between different strain backgrounds and environments. Mutation frequencies varied by sixfold between lineages, although no obvious genetic differences were found (such as “mutator” genotypes due to disruptions in DNA repair genes) that could account for these differences (SI Appendix). The niche in which strains were passaged also impacted mutation frequencies; strains passaged in vivo showed up to 11-fold higher mutation frequencies than those passaged in vitro (frequencies calculated relative to the total time of passage). C. albicans may therefore experience environment- or stress-induced mutagenesis, as demonstrated for a number of bacterial, fungal, plant, and human studies (82–84). In support of this, C. albicans was previously shown to undergo stress-induced LOH events in vitro (22), and LOH events were more frequent during both bloodstream passage and infection of the oral cavity than in vitro (31, 85).
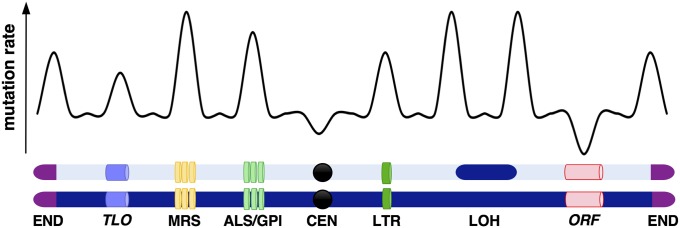
Our studies also establish that a number of chromosomal features impact C. albicans mutation rates. Mutation rates were higher in repeat regions, telomeric regions, and in genes encoding GPI-linked cell wall proteins (including ALS family genes) than in the rest of the genome (Fig. 7). These results are consistent with multiple reports linking higher mutation rates with repetitive and telomere-proximal regions of the C. albicans genome (55, 56, 60, 64, 66, 86).
Fig. 7.
Schematic illustrating the pattern of mutational events across the C. albicans genome. Figure highlights how certain chromosomal features are associated with elevated mutation rates.
Evidence That Purifying Selection Acts on Emerging Mutations.
Mutations accumulated at significantly higher rates in intergenic regions than in coding regions, and the ratio of synonymous to nonsynonymous mutations was also greater than that expected by chance. Based on these observations, we estimate that 71–79% of nonsynonymous mutations were effectively removed from the population by purifying selection during microevolution. This implies that selection frequently removes fitness-reducing mutants from the population. Natural isolates of S. cerevisiae also show a significant bias toward intergenic over genic SNPs (87), and more than a third of nonsynonymous mutations were implicated as being deleterious in one study (88). The present study provides evidence for purifying selection acting broadly on the diploid C. albicans genome even over relatively short evolutionary periods.
Aneuploid Forms Frequently Arise During Commensal Colonization.
Aneuploid forms have frequently been observed in C. albicans (22, 24, 25, 89), and 3 of 28 lineages became aneuploid during microevolution. Whole-chromosome aneuploidies emerged in distinct strain backgrounds and in each case involved acquisition of a third copy of Chr 7 during passaging in the GI tract in mice on a SD with antibiotics. To determine if Chr 7 trisomy provides a fitness advantage under these conditions, direct competition experiments were performed between Chr 7 trisomic strains and euploid controls. In each case, the presence of the trisomy was associated with increased fitness in the GI. Interestingly, Chr 7 trisomies did not emerge in isolates passaged in an alternative GI model that used a purified mouse diet without antibiotics. This suggests that this trisomy may not enhance growth in the GI tract per se, but could provide a specific advantage to attributes of the GI environment when mice are on the SD.
mLOH Events Are a Major Driver of Genome Dynamics.
LOH has long been recognized as an important mechanism for introducing diversity into C. albicans populations (21, 22, 24, 25, 30), although a global analysis of dynamic events had not been performed. Previous studies have examined LOH in diploid S. cerevisiae strains and showed that mitotic tracts are generally longer than meiotic tracts, with the former averaging 2–12 kb (90, 91). A recent study also examined genome-wide mitotic events in S. cerevisiae and showed that LOH tracts ranged from <100 bp to >100 kb, with small LOH tracts (<1 kb) attributed to local gene conversions, although the smallest LOH events were excluded from analysis (8). The present study defines the complete spectrum of LOH events occurring during C. albicans microevolution and establishes in vitro LOH rates of 1.61 × 10−10 per base pair per generation (or 4.5 × 10−3 per cell division), which are slightly lower than previous estimates based on LOH events at three loci (63, 92). However, we note that LOH rates varied considerably between different genomic regions, with MRS, cell surface proteins, and telomeric regions representing relative hotspots for LOH.
We found that the majority of LOH events involved very short mLOH tracts (estimated Lavg size = 368 bp). Indeed, over half of all LOH events impacted only a single heterozygous position and, critically, a number of these events were validated by KASP genotyping. In fact, when examining all genetic changes accrued during microevolution, >30% of these events were due to LOH at single heterozygous positions, revealing that these represent a high-frequency event in the C. albicans genome. Consistent with our data, previous experiments studying the repair of DNA double-strand breaks in C. albicans revealed SLOH events due to gene conversion, and only rare LLOH events observed due to BIR or reciprocal recombination (71). Similarly, recent analysis of passaged lineages in the water flea Daphnia pulex found that SLOH events (median ∼221 bp) were prevalent (7). We therefore propose that SLOH events represent a common mutation in heterozygous genomes that will have been missed by studies that lack nucleotide-level resolution.
We note that global LOH frequencies were often balanced by equivalent frequencies of de novo GOH mutations during microevolution, regardless of genetic background or evolution niche. Because of this balance, overall heterozygosity levels were stably maintained (±2%) in the majority of passaged lineages, unless very large LOH events occurred (described below). The observation that de novo mutation frequencies often match LOH frequencies indicates that genomes can retain their heterozygosity levels even in the absence of outcrossing events over short time spans.
In contrast to frequent microscale LOH events, large LOH tracts were only observed in two isolates. These events involved very long LOH tracts (0.27–1.23 Mb) that extended to the ends of the chromosomes. Previous studies also detected large LOH events in C. albicans strains grown both in vitro and in vivo (21, 22, 24, 25, 71, 93). We note that while large-scale chromosomal changes are relatively rare, these impact numerous genes and are therefore the most likely to have phenotypic consequences. Overall, the three large LOH events identified here led to homozygosis of over 10,000 heterozygous positions, resulting in 2,135 nonsynonymous changes, 14 nonsense mutations, and 3 readthrough mutations. This reveals that large recombination tracts drive extensive genotypic changes but may also be heavily selected against given the large number of heterozygous positions impacted.
An Association Between Recombination Events and de Novo Mutations.
A striking correlation is reported here between de novo mutations in C. albicans and their proximity to recombination events. Specifically, GOH rates were elevated 840-fold within 500 bp of emergent LOH tracts relative to the genome average. Consequently, a third of all GOH events (substitutions and indels) were in regions flanking new LOH tracts, despite these tracts representing only ∼1.3% of the genome. This phenomenon was observed independent of strain background and was most evident during host passage where LOH frequencies were higher. The high rate of mutations adjacent to LOH tracts is likely due to gene conversion being mutagenic and introducing de novo mutations into neighboring regions of the genome. Increased rates of transversions in these mutated regions support the activity of translesion polymerases acting to repair DNA lesions at these sites. This is consistent with DNA double-strand break repair processes being both recombinogenic and mutagenic, as described in other species (50, 72, 94). Our data now indicate that recombination events in C. albicans also introduce de novo mutations during the repair process. Thus, LOH is a stress-inducible event in C. albicans (22) and this process introduces additional mutations into the genome, which will further accelerate adaptation.
Concluding Remarks.
This study provides a high-resolution analysis of the spectrum of mutations accumulating in a heterozygous diploid pathogen. Our data indicate that both cell-intrinsic properties (e.g., strain background, repetitive chromosomal features) and cell-extrinsic factors (e.g., host versus laboratory passage) impact the frequency and distribution of genetic mutations. Frequent microscale changes (predominantly de novo substitutions and short-tract gene-conversion events) and occasional larger-scale rearrangements (LLOH or chromosomal aneuploidies) determine genome dynamics. Furthermore, purifying selection plays a dominant role in dictating which genetic changes are retained during evolution. Our results provide a detailed picture against which genomic changes in other heterozygous diploid species can be compared, and establish the foundation for understanding how C. albicans adapts to diverse host niches.
Materials and Methods
A complete description of all materials and methods can be found in SI Appendix.
Strains and Growth Conditions.
C. albicans strains were grown at 30 °C in YPD medium (95). For in vitro evolution, cultures were serially diluted (1/100) daily for 80 d (bottlenecks every 6.8 generations) and cells collected once a week. In vitro isolates were collected as a pool and used to prepare genomic DNA.
Murine Experiments.
For animal infections, female BALB/c mice were housed together with free access to food and water. For systemic infection, C. albicans cells were grown overnight (YPD, 30 °C) and for each passage three mice were infected via the tail-vein with 6 × 105 cells. Three days postinfection, the mice were euthanized and fungal cells isolated from kidneys. Isolates for subsequent passages and for genomic DNA isolation were selected from the mouse showing the highest virulence outcome. For the GI SD model, mice were fed standard rodent chow and their water was supplemented with penicillin and streptomycin (96). For the GI PD model, mice were fed a purified diet (AIN-93G, Dyets) and their water was not supplemented with antibiotics (35). In both models, mice were orally gavaged with 108 cells and continued with their respective diet and water for 6 wk. At 42 d, isolates were collected from fecal pellets and used to prepare genomic DNA. For competition experiments using the GI SD model, C. albicans cells were grown overnight in YPD at 30 °C and washed with sterile water. Approximately 108 cells (containing a 1:1 ratio of each competing strain) were orally gavaged into the mouse GI tract. For each competition, one strain was nourseothricin-sensitive (SAT−) and one strain was nourseothricin-resistant (SAT1+). Fecal pellets were collected every other day for 14 d, after which mice (four per experiment) were euthanized. Abundance of each strain was quantified by plating both the inoculum and fecal pellets homogenates onto YPD and YPD with nourseothricin (200 µg/mL).
Ethics Statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (97) as defined by the National Institutes of Health (PHS Assurance #A3284-01). Animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Brown University. All animals were housed in a centralized and Association for Assessment and Accredidation of Laboratory Animal Care-accredited research animal facility that is fully staffed with trained husbandry, technical, and veterinary personnel.
Whole-Genome Sequencing and Variant Identification.
For genomic DNA, C. albicans cells were grown in YPD at 30 °C and DNA isolated using a Qiagen Genomic Buffer Set, a Qiagen Genomic-tip 100/G or the MasterPure Yeast DNA Purification kit (Epicentre). DNA was sequenced using Illumina HiSeq 2000 generating 101-bp paired reads. Reads were aligned to the SC5314 reference genome using Burrows–Wheeler Aligner (98), and converted to sorted BAM format using Samtools (99). Picard tools (picard.sourceforge.net/) were used to preprocess the alignments. The Genome Analysis Toolkit (100) and Pilon (37) were used to call variant and reference bases from the alignments. Mutations were inspected in IGV (Broad Institute) and annotated using the VCFannotator (Broad Institute). Ploidy, copy number variation, and LOH analyses were performed as described in SI Appendix, Materials and Methods.
KASP Genotyping.
Genomic DNA was subjected to allele specific PCR (KASP, LGC), a fluorescent technique which enables testing of SNPs and indels at specific loci. Primers were designed for testing of 80 unique sites representing mutations from diverse genomic locations.
Supplementary Material
Acknowledgments
We thank Adrian Vladu for help with computational analyses, and Daniel Weinreich and members of the R.J.B. laboratory for feedback on the manuscript. This work was supported by NIH Grants AI081704, AI122011, and AI112363 (to R.J.B.), AI139592 (to I.V.E.), and F31DE023726 (to M.P.H.); a PATH award from the Burroughs Wellcome Fund (to R.J.B.); a Vessa Notchev fellowship from Sigma Delta Epsilon-Graduate Women in Science (to I.V.E.); a Wellcome Trust/Massachusetts Institute of Technology postdoctoral fellowship (to R.A.F.); and funds from the National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services, under Contract HHSN272200900018C and Grant U19AI110818 to the Broad Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/bioproject (BioProject ID PRJNA345600).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806002115/-/DCSupplemental.
References
- 1.Byrnes EJ, 3rd, et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6:e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gire SK, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jerison ER, Desai MM. Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments. Curr Opin Genet Dev. 2015;35:33–39. doi: 10.1016/j.gde.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang S, et al. Parent-progeny sequencing indicates higher mutation rates in heterozygotes. Nature. 2015;523:463–467. doi: 10.1038/nature14649. [DOI] [PubMed] [Google Scholar]
- 5.McDonald MJ, Rice DP, Desai MM. Sex speeds adaptation by altering the dynamics of molecular evolution. Nature. 2016;531:233–236. doi: 10.1038/nature17143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sellis D, Kvitek DJ, Dunn B, Sherlock G, Petrov DA. Heterozygote advantage is a common outcome of adaptation in Saccharomyces cerevisiae. Genetics. 2016;203:1401–1413. doi: 10.1534/genetics.115.185165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keith N, et al. High mutational rates of large-scale duplication and deletion in Daphnia pulex. Genome Res. 2016;26:60–69. doi: 10.1101/gr.191338.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutta A, et al. Genome dynamics of hybrid Saccharomyces cerevisiae during vegetative and meiotic divisions. G3 (Bethesda) 2017;7:3669–3679. doi: 10.1534/g3.117.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrer RA, et al. Microevolutionary traits and comparative population genomics of the emerging pathogenic fungus Cryptococcus gattii. Philos Trans R Soc Lond B Biol Sci. 2016;371:20160021. doi: 10.1098/rstb.2016.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goddard MR, Godfray HC, Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005;434:636–640. doi: 10.1038/nature03405. [DOI] [PubMed] [Google Scholar]
- 11.Heitman J. Sexual reproduction and the evolution of microbial pathogens. Curr Biol. 2006;16:R711–R725. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- 12.Ryland GL, et al. Australian Ovarian Cancer Study Group Loss of heterozygosity: What is it good for? BMC Med Genomics. 2015;8:45. doi: 10.1186/s12920-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell CD, et al. Estimating the human mutation rate using autozygosity in a founder population. Nat Genet. 2012;44:1277–1281. doi: 10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown GD, et al. Hidden killers: Human fungal infections. Sci Transl Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 15.Kullberg BJ, Arendrup MC. Invasive candidiasis. N Engl J Med. 2015;373:1445–1456. doi: 10.1056/NEJMra1315399. [DOI] [PubMed] [Google Scholar]
- 16.Calderone RA, Clancy CJ. Candida and Candidiasis. ASM Press; Washington, DC: 2011. [Google Scholar]
- 17.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: A persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones T, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun BR, et al. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005;1:36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muzzey D, Schwartz K, Weissman JS, Sherlock G. Assembly of a phased diploid Candida albicans genome facilitates allele-specific measurements and provides a simple model for repeat and indel structure. Genome Biol. 2013;14:R97. doi: 10.1186/gb-2013-14-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diogo D, Bouchier C, d’Enfert C, Bougnoux ME. Loss of heterozygosity in commensal isolates of the asexual diploid yeast Candida albicans. Fungal Genet Biol. 2009;46:159–168. doi: 10.1016/j.fgb.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Forche A, et al. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio. 2011;2:e00129-11. doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenberg SM. Stress-induced loss of heterozygosity in Candida: A possible missing link in the ability to evolve. mBio. 2011;2:e00200-11. doi: 10.1128/mBio.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirakawa MP, et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 2015;25:413–425. doi: 10.1101/gr.174623.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forche A. Large-scale chromosomal changes and associated fitness consequences in pathogenic fungi. Curr Fungal Infect Rep. 2014;8:163–170. doi: 10.1007/s12281-014-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrow CA, Fraser JA. Ploidy variation as an adaptive mechanism in human pathogenic fungi. Semin Cell Dev Biol. 2013;24:339–346. doi: 10.1016/j.semcdb.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F, et al. High-frequency genetic contents variations in clinical Candida albicans isolates. Biol Pharm Bull. 2011;34:624–631. doi: 10.1248/bpb.34.624. [DOI] [PubMed] [Google Scholar]
- 29.Rustchenko E. Chromosome instability in Candida albicans. FEMS Yeast Res. 2007;7:2–11. doi: 10.1111/j.1567-1364.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 30.Bennett RJ, Forche A, Berman J. Rapid mechanisms for generating genome diversity: Whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb Perspect Med. 2014;4:a019604. doi: 10.1101/cshperspect.a019604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forche A, Magee PT, Selmecki A, Berman J, May G. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics. 2009;182:799–811. doi: 10.1534/genetics.109.103325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ford CB, et al. The evolution of drug resistance in clinical isolates of Candida albicans. eLife. 2015;4:e00662. doi: 10.7554/eLife.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W, Lockhart SR, Pujol C, Srikantha T, Soll DR. Heterozygosity of genes on the sex chromosome regulates Candida albicans virulence. Mol Microbiol. 2007;64:1587–1604. doi: 10.1111/j.1365-2958.2007.05759.x. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Pande K, French SD, Tuch BB, Noble SM. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe. 2011;10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi N, et al. Gastric colonization of Candida albicans differs in mice fed commercial and purified diets. J Nutr. 2005;135:109–115. doi: 10.1093/jn/135.1.109. [DOI] [PubMed] [Google Scholar]
- 36.MacCallum DM, Odds FC. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses. 2005;48:151–161. doi: 10.1111/j.1439-0507.2005.01121.x. [DOI] [PubMed] [Google Scholar]
- 37.Walker BJ, et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu YO, Siegal ML, Hall DW, Petrov DA. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci USA. 2014;111:E2310–E2318. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishant KT, et al. The baker’s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. 2010;6:e1001109. doi: 10.1371/journal.pgen.1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keightley PD, et al. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19:1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graur D. Encyclopedia of Life Sciences. John Wiley and Sons; Chichester, UK: 2008. Single-base mutation. [Google Scholar]
- 44.Farlow A, et al. The spontaneous mutation rate in the fission yeast Schizosaccharomyces pombe. Genetics. 2015;201:737–744. doi: 10.1534/genetics.115.177329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prieto D, Pla J. Distinct stages during colonization of the mouse gastrointestinal tract by Candida albicans. Front Microbiol. 2015;6:792. doi: 10.3389/fmicb.2015.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forche A, Schönian G, Gräser Y, Vilgalys R, Mitchell TG. Genetic structure of typical and atypical populations of Candida albicans from Africa. Fungal Genet Biol. 1999;28:107–125. doi: 10.1006/fgbi.1999.1164. [DOI] [PubMed] [Google Scholar]
- 47.Gräser Y, et al. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balloux F, Lehmann L, de Meeûs T. The population genetics of clonal and partially clonal diploids. Genetics. 2003;164:1635–1644. doi: 10.1093/genetics/164.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arbeithuber B, Betancourt AJ, Ebner T, Tiemann-Boege I. Crossovers are associated with mutation and biased gene conversion at recombination hotspots. Proc Natl Acad Sci USA. 2015;112:2109–2114. doi: 10.1073/pnas.1416622112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–473. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 51.Amos W. Heterozygosity and mutation rate: Evidence for an interaction and its implications: The potential for meiotic gene conversions to influence both mutation rate and distribution. BioEssays. 2010;32:82–90. doi: 10.1002/bies.200900108. [DOI] [PubMed] [Google Scholar]
- 52.Hsueh YP, Idnurm A, Heitman J. Recombination hotspots flank the Cryptococcus mating-type locus: Implications for the evolution of a fungal sex chromosome. PLoS Genet. 2006;2:e184. doi: 10.1371/journal.pgen.0020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bensasson D. Evidence for a high mutation rate at rapidly evolving yeast centromeres. BMC Evol Biol. 2011;11:211. doi: 10.1186/1471-2148-11-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson MZ, Wigen LJ, Burrack LS, Berman J. Real-time evolution of a subtelomeric gene family in Candida albicans. Genetics. 2015;200:907–919. doi: 10.1534/genetics.115.177451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freire-Benéitez V, Price RJ, Tarrant D, Berman J, Buscaino A. Candida albicans repetitive elements display epigenetic diversity and plasticity. Sci Rep. 2016;6:22989. doi: 10.1038/srep22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanyal K, Baum M, Carbon J. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci USA. 2004;101:11374–11379. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genet Genomics. 2007;278:455–465. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- 59.Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 60.Chibana H, Magee PT. The enigma of the major repeat sequence of Candida albicans. Future Microbiol. 2009;4:171–179. doi: 10.2217/17460913.4.2.171. [DOI] [PubMed] [Google Scholar]
- 61.Chibana H, Beckerman JL, Magee PT. Fine-resolution physical mapping of genomic diversity in Candida albicans. Genome Res. 2000;10:1865–1877. doi: 10.1101/gr.148600. [DOI] [PubMed] [Google Scholar]
- 62.Lephart PR, Chibana H, Magee PT. Effect of the major repeat sequence on chromosome loss in Candida albicans. Eukaryot Cell. 2005;4:733–741. doi: 10.1128/EC.4.4.733-741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lephart PR, Magee PT. Effect of the major repeat sequence on mitotic recombination in Candida albicans. Genetics. 2006;174:1737–1744. doi: 10.1534/genetics.106.063271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoyer LL, Green CB, Oh SH, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—A sticky pursuit. Med Mycol. 2008;46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh SH, et al. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology. 2005;151:673–681. doi: 10.1099/mic.0.27680-0. [DOI] [PubMed] [Google Scholar]
- 66.Zhao X, et al. Analysis of ALS5 and ALS6 allelic variability in a geographically diverse collection of Candida albicans isolates. Fungal Genet Biol. 2007;44:1298–1309. doi: 10.1016/j.fgb.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao X, Pujol C, Soll DR, Hoyer LL. Allelic variation in the contiguous loci encoding Candida albicans ALS5, ALS1 and ALS9. Microbiology. 2003;149:2947–2960. doi: 10.1099/mic.0.26495-0. [DOI] [PubMed] [Google Scholar]
- 68.Dunkel N, Blass J, Rogers PD, Morschhäuser J. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol. 2008;69:827–840. doi: 10.1111/j.1365-2958.2008.06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coste A, et al. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics. 2006;172:2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 71.Feri A, et al. Analysis of repair mechanisms following an induced double-strand break uncovers recessive deleterious alleles in the Candida albicans diploid genome. mBio. 2016;7:e01109-16. doi: 10.1128/mBio.01109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hollister JD, Ross-Ibarra J, Gaut BS. Indel-associated mutation rate varies with mating system in flowering plants. Mol Biol Evol. 2010;27:409–416. doi: 10.1093/molbev/msp249. [DOI] [PubMed] [Google Scholar]
- 74.Tian D, et al. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature. 2008;455:105–108. doi: 10.1038/nature07175. [DOI] [PubMed] [Google Scholar]
- 75.Mudrak SV, Welz-Voegele C, Jinks-Robertson S. The polymerase eta translesion synthesis DNA polymerase acts independently of the mismatch repair system to limit mutagenesis caused by 7,8-dihydro-8-oxoguanine in yeast. Mol Cell Biol. 2009;29:5316–5326. doi: 10.1128/MCB.00422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Livneh Z. DNA damage control by novel DNA polymerases: Translesion replication and mutagenesis. J Biol Chem. 2001;276:25639–25642. doi: 10.1074/jbc.R100019200. [DOI] [PubMed] [Google Scholar]
- 77.Rodgers K, McVey M. Error-prone repair of DNA double-strand breaks. J Cell Physiol. 2016;231:15–24. doi: 10.1002/jcp.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arbour M, et al. Widespread occurrence of chromosomal aneuploidy following the routine production of Candida albicans mutants. FEMS Yeast Res. 2009;9:1070–1077. doi: 10.1111/j.1567-1364.2009.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gerami-Nejad M, Zacchi LF, McClellan M, Matter K, Berman J. Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans. Microbiology. 2013;159:565–579. doi: 10.1099/mic.0.064097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seervai RN, Jones SK, Jr, Hirakawa MP, Porman AM, Bennett RJ. Parasexuality and ploidy change in Candida tropicalis. Eukaryot Cell. 2013;12:1629–1640. doi: 10.1128/EC.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.MacCallum DM, et al. Property differences among the four major Candida albicans strain clades. Eukaryot Cell. 2009;8:373–387. doi: 10.1128/EC.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shor E, Fox CA, Broach JR. The yeast environmental stress response regulates mutagenesis induced by proteotoxic stress. PLoS Genet. 2013;9:e1003680. doi: 10.1371/journal.pgen.1003680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang C, et al. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 2014;24:1821–1829. doi: 10.1101/gr.177659.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Forche A, et al. Rapid phenotypic and genotypic diversification after exposure to the oral host niche in Candida albicans. Genetics. 2018;209:725–741. doi: 10.1534/genetics.118.301019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clutterbuck AJ. Genomic evidence of repeat-induced point mutation (RIP) in filamentous ascomycetes. Fungal Genet Biol. 2011;48:306–326. doi: 10.1016/j.fgb.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 87.Wohlbach DJ, et al. Comparative genomics of Saccharomyces cerevisiae natural isolates for bioenergy production. Genome Biol Evol. 2014;6:2557–2566. doi: 10.1093/gbe/evu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Doniger SW, et al. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 2008;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weil T, et al. Adaptive mistranslation accelerates the evolution of fluconazole resistance and induces major genomic and gene expression alterations in Candida albicans. mSphere. 2017;2:e00167-17. doi: 10.1128/mSphere.00167-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Judd SR, Petes TD. Physical lengths of meiotic and mitotic gene conversion tracts in Saccharomyces cerevisiae. Genetics. 1988;118:401–410. doi: 10.1093/genetics/118.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol. 2000;182:5730–5736. doi: 10.1128/jb.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moorhouse AJ, Rennison C, Raza M, Lilic D, Gow NA. Clonal strain persistence of Candida albicans isolates from chronic mucocutaneous candidiasis patients. PLoS One. 2016;11:e0145888. doi: 10.1371/journal.pone.0145888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sakofsky CJ, Ayyar S, Malkova A. Break-induced replication and genome stability. Biomolecules. 2012;2:483–504. doi: 10.3390/biom2040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 96.Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45:1088–1091. doi: 10.1038/ng.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.National Research Council of the National Academies . Guide for the Care and Use of Laboratory Animals. 8th Ed The National Academies Press; Washington, DC: 2011. [Google Scholar]
- 98.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN]
- 99.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.