Significance
Whereas crystals and other geometrical structures can be self-assembled under equilibrium conditions from a few particles with a few, often simple, identical interactions, functional materials require a multitude of specific interactions between many different types of particles and nonequilibrium, often hierarchical assembly. Here, we introduce a set of building blocks on the colloidal micrometer scale with designed valence and specificity that readily make linear, branched, or other programmed connected networks, yet maintain the flexibility needed to fold, unfold, or restructure from the original backbone. We demonstrate the self-assembly of alternating copolymers, hetero chains, trivalent clusters, and a dynamic programmability by opening and closing chain ends.
Keywords: patchy particle, self-assembly, DNA origami
Abstract
Nature self-assembles functional materials by programming flexible linear arrangements of molecules and then folding them to make 2D and 3D objects. To understand and emulate this process, we have made emulsion droplets with specific recognition and controlled valence. Uniquely monovalent droplets form dimers: divalent lead to polymer-like chains, trivalent allow for branching, and programmed mixtures of different valences enable a variety of designed architectures and the ability to subsequently close and open structures. Our functional building blocks are a hybrid of micrometer-scale emulsion droplets and nanoscale DNA origami technologies. Functional DNA origami rafts are first added to droplets and then herded into a patch using specifically designated “shepherding” rafts. Additional patches with the same or different specificities can be formed on the same droplet, programming multiflavored, multivalence droplets. The mobile patch can bind to a patch on another droplet containing complementary functional rafts, leading to primary structure formation. Further binding of nonneighbor droplets can produce secondary structures, a third step in hierarchical self-assembly. The use of mobile patches rather than uniform DNA coverage has the advantage of valence control at the expense of slow kinetics. Droplets with controlled flavors and valences enable a host of different material and device architectures.
Structures arising from colloidal self-assembly hold great promise for new functional materials and serve as model systems for other physical, chemical, and biological phenomena (1–3). The evolution of building blocks, from colloids with isotropic interactions to those with altered shapes and surface chemistries, contribute to many beautiful examples of self-assembled crystalline and quasi-crystalline lattices (4–11). In the past decades, patchy particles (12), as a powerful tool, gave access to numerous new microscopic architectures, such as colloidal micelles (13) and molecules (14). However, many sophisticated structures that nature builds, e.g., proteins and organelles, are still far from realization in colloidal systems, owing to the limited programmability. From a materials perspective, such systems require the building blocks to have diverse functionalities, hierarchical interactions, structural flexibility, and dynamic programmability.
Here we focus on a particular functionality, the ability of a droplet to bind to a specific number and type of particle. Flexible bonds between particles have been achieved previously (15, 16) but the valence, number of neighbors for each specific particle, was not controlled. We overcome this problem by preassembling a fixed number of mobile DNA patches on each droplet. For instance, we make a droplet with one α-"flavored" patch and two β-flavored patches. Placed in a bath with complementary droplets it will bind with valence three to one droplets with an α′ patch and two droplets each with a β′ patch. Such valence control allows formation of dimers, trimers, chains, and branched and folded structures. It adds a tool to the self-assembly tool box.
However, we pay a price in the reaction rates. In refs. 15 and 16 the particles have a uniform DNA coating and the reactions can be diffusion limited; particles bind when they touch. The use of patches typically requires many attempts since patches on the two particles must be aligned to bind. The rate reduction in our case is approximately 1/400 (SI Appendix). It may be worth paying this price for specific designs which here can be assembled in bulk without the use of additional separation processes or for example in the formation of a simple chain of droplets identical except for their colored order, e.g., red–orange–green–blue. Without valence control these chains would branch and aggregate and the desired structure would be difficult to isolate by a separation process other than by individual observation.
DNA nanotechnology (17) has provided us with a wide variety of structures, far exceeding what nature has built with DNA, ranging from arbitrary architectures made of DNA motifs (18–20), DNA origami (21–24), and DNA bricks (25, 26) to logic circuits (27), reaction networks, (28) and self-organization on lipid surfaces (29–31), duplicating some of the functional complexity of biological systems. The integration of DNA constructs with nanoparticles has been extensively explored (32–35). The present work is a hybrid which attempts to use DNA nanotechnology to control the association of larger emulsion structures.
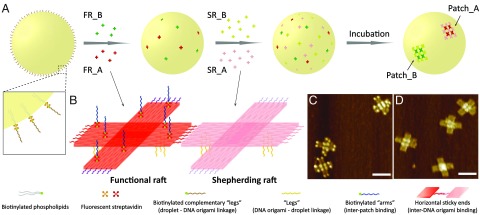
Our design involves assembling one, two, or three distinct “sticky” patches to the surfaces of micrometer-sized oil-in-water droplets (36, 37). Each patch is designed to bind specifically to a complementary patch on another droplet. A “sticky end” is a single strand of DNA, sticky because it can hybridize with a complementary sticky end with Watson–Crick pairing. A sticky patch is an organized array of sticky ends. To implement this design experimentally, we took advantage of DNA origami technology and generated a pair of rafts: functional rafts and shepherding rafts (Fig. 1B), by modifying a cross-shaped DNA origami (38). We decorated both origami with sticky ends on their bottom surfaces, “legs” to facilitate anchoring to the droplet surface. The two sets of horizontal sticky ends encoded into the edges of the functional rafts and shepherding rafts are complementary, allowing these rafts to self-organize into a 2D array, a patch. We can easily control the valence number by adding another pair of functional and shepherding rafts with different horizontal sticky ends for each additional patch. There is a specific type of shepherding raft for each type of functional raft. Introducing a pair of rafts, instead of self-complementary rafts, and adding the functional rafts and shepherding rafts separately, prevents them from forming arrays in free solution before attaching to the droplet surface.
Fig. 1.
Patch generation and valency control. (A) A droplet coated with complementary leg sticky ends is fabricated first. (Inset) The DNA strands are coated on the droplet surface through biotin–streptavidin–biotin linkages. The functional DNA origami rafts (FR_A in red and FR_B in green) carrying multiple single-stranded legs are then constructed, and mixed with the droplets at a ratio of 400:1. The functional rafts are anchored to the droplet surface via leg sticky-end hybridization. In the next step, an excess quantity of the shepherding rafts (SR_A in pink and SR_B in light green) carrying the same leg sticky ends are introduced and hybridized to the droplet surface gradually. Finally, the self-assembly of functional rafts and shepherding rafts via horizontal sticky-end binding leads to the patch formation (Patch_A and Patch_B). (Our patches use functional and complementary shepherding rafts added sequentially, rather than self-complementary functional rafts alone, to avoid patch formation in the solution.) (B) Schematics of functional and shepherding DNA origami rafts. Both rafts are decorated with six single-stranded extensions (leg sticky ends, yellow curls) on the bottom face. The two horizontal sticky-end sets (red and purple curls) on the functional raft are complementary to the two sets (pink and light purple curls) on the shepherding raft. The eight biotinylated staples (green dots) extruding from the top of the functional raft bind to the fluorescent streptavidin (red quatrefoil, for patch imaging) conjugated with arm sticky ends (blue curls, for patch functionalization). (C) AFM image of functional DNA origami rafts. (D) AFM image of shepherding DNA origami rafts. (Scale bars, 100 nm.)
To functionalize the patches for interdroplet assembly, we modified each functional raft with eight biotinylated staple strands extending from its top surface and then attached sticky ends, “arms,” to it via a biotin–streptavidin–biotin linkage (SI Appendix, Fig. S1). The streptavidin molecules on the functional rafts are dye-labeled, providing fluorescent signals for patch identification. Atomic force microscopy (AFM) images in Fig. 1 C and D show that functional and shepherding rafts are successfully constructed (see SI Appendix, Fig. S2 for low-magnification AFM images).
Fig. 1A illustrates a schematic drawing of our assembly procedure for creating a divalent droplet––a droplet with two distinct DNA origami-assembled patches. To begin, we coated the droplets with sticky ends complementary to the legs on DNA origami (Fig. 1A, Inset), then introduced two types of functional rafts (FR_A and FR_B) carrying orthogonal horizontal and arm sticky ends to the droplet solution, and allowed these rafts to anchor on the surface through leg sticky-ended hybridization. Next we added the shepherding rafts (SR_A and SR_B) three times so that the final amount is in excess, three SR for each FR. Upon attachment, the shepherding rafts can diffuse freely on the surface and gradually recruit all corresponding functional rafts (SR_A with FR_A and SR_B with FR_B) into two distinct patches via specific horizontal DNA interactions. This multistep addition protocol (see Methods and SI Appendix, Fig. S3) also avoids the formation of multiple fractional patches on the droplet surfaces.
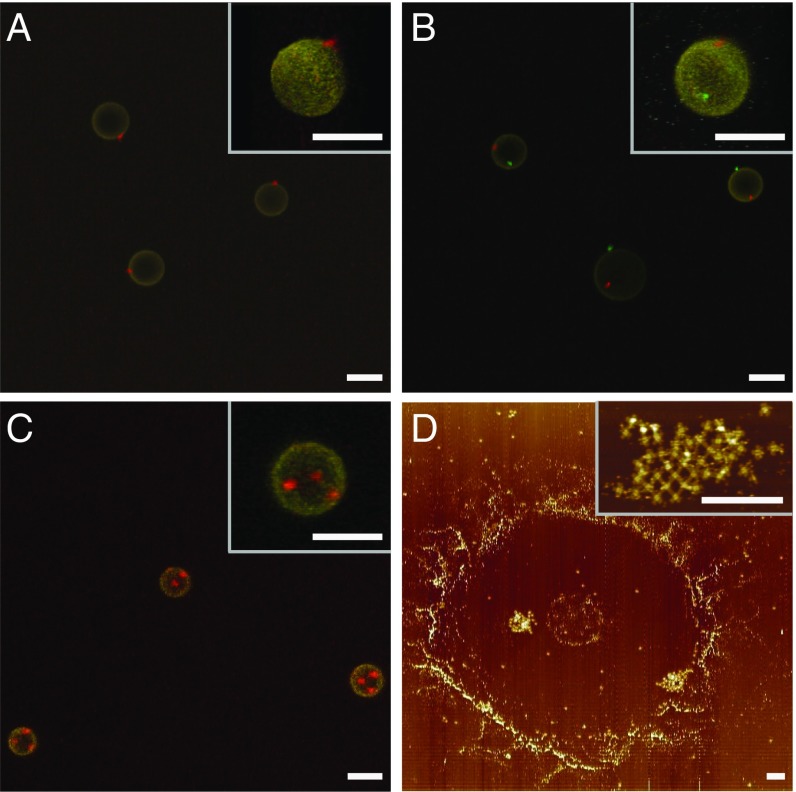
We examined patch formation on monovalent droplets using confocal fluorescent microscopy. Movie S1 shows that all of the droplets in solution have a single red patch. To make sure that we did not miss the presence of multiple fragmentary patches, we “froze” the patchy droplets in a 10% polyacrylamide gel and performed a 3D confocal scan. The z projection (Fig. 2A) and 3D reconstruction (Fig. 2A, Inset), confirms that there is one patch per droplet. The same verification procedure was conducted on the divalent and trivalent droplets, which shows the formation of two and three distinct patches per droplet in those cases (Fig. 2 B and C and Movies S2 and S3). To determine the fine structure of the patches, we probed the divalent droplets using AFM. The droplets were deposited onto a mica surface and subsequently collapsed when rinsed with water. Fig. 2D shows two patches located in a ring-shaped droplet stain, with a zoomed image demonstrating that the patch is a 2D array of DNA origami rafts as expected. The number of rafts in a patch ranges from 25 to 44 (each raft’s area is 0.01 μm2). Here we simply use 0.25 μm2 as an estimation of the patch size for further calculation. This patch size variation will also affect the binding dynamics, which has been observed in experiments as well.
Fig. 2.
Patch characterization. (A–C) Projections of confocal Z-stack images of monovalent, divalent, and trivalent droplets frozen in gels. (Insets) Zoomed images of 3D reconstructions. Emulsion droplets in yellow are labeled with SA-AF 546. The red and green DNA origami patches are labeled with SA-AF 647 and 488, respectively. (Scale bars, 5 μm.) (D) AFM image of a divalent droplet deposited on mica surface. (Inset) High-magnification AFM image of a DNA origami patch. (Scale bars, 500 nm.)
Building structures with patchy droplets rely not only on how well the patch forms but also its binding ability and specificity. We first formed dimers using two species of monovalent droplets with complementary arm sticky ends (SI Appendix, Fig. S4). The assembly, however, proceeded unusually slowly in a conventional 2D reaction–diffusion system, as predicted by a kinetic model (39). The average binding time τb can be estimated from the following equation:
| [1] |
where τd is the 2D diffusion-limited aggregation (DLA) time; τr is the conditional reaction time given two patchy droplets that are held in contact with each other; R, L, and C0 are the average droplet radius (∼2.4 μm), thickness of the contact region (∼20 nm), and droplet number density, respectively. In a dilute sample (C0 ∼ 1,500/mm2), τd is on the order of a few minutes. τr, studied previously (40), is merely dependent on the patch’s diffusion (41) and surface area. In our case, τr ∼ 3 h, so it would take several months to form dimers. To accelerate this process, we simply tilt the sample so that all droplets are densely packed at an upper corner of the sample cell (∼2–3 layers). This suppresses the translational diffusion of droplets, resulting in τb = τr, a 1,000-fold increase in the binding kinetics. In addition, we incorporate a sample-flipping step to our incubation protocol, which enables the rearrangement of droplets every 6 h.
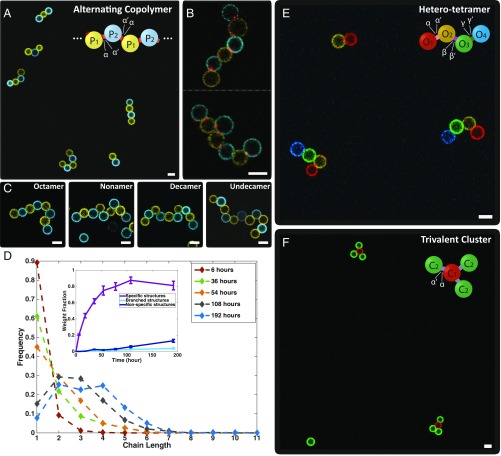
Following this procedure, dimers were formed via interpatch binding and the specificity was verified by confocal fluorescent microscopy. We have also self-assembled “alternating copolymers” using two species of divalent droplets: P1 and P2 (Fig. 3A). The two patches on P1 and P2 were functionalized with complementary arm sticky ends, α and α′, respectively, which “link” the droplets, mimicking step growth polymerization. A representative image taken at the final stage of incubation (192 h, equivalently 16 flips) shows that most of the monomers have self-assembled into oligomers, and close-up views (Fig. 3B) demonstrate the patch-mediated bonds at each joint. Furthermore, several long chains, e.g., linear octamers to undecamers, are displayed in Fig. 3C and Movie S4. To quantify the chain length distributions, we counted the numbers of all of the linear oligomers of various lengths at different time points (SI Appendix, Table S5). As plotted in Fig. 3D, the majority of the chains grew progressively from monomers to tetramers (SI Appendix, Figs. S5 and S6).
Fig. 3.
Self-assembly of patchy droplets. (A) A representative confocal image of assembled alternating copolymers after a 192-h incubation. Alternating copolymer: two types of divalent droplets (P1 and P2) with complementary patches (α on P1, α′ on P2) self-assemble into a linear chain. (B) High-magnification images of two chains with fluorescent patches (red, labeled with SA-AF 647) at each joint, confirming that the assembly is through interpatch binding. (C) A gallery of several long chains. (D) The chain length distributions at various time points (only unbranched oligomers are counted in the main panel). Each data point represents the frequency of the linear oligomer of a certain length in the sample. (Inset) Number fractions of droplets in linear alternating chains, branched structures (e.g., a structure containing one P1 binds to more than two P2), and structures containing nonspecific bindings (e.g., P1 binds to P1) versus time. (E) A representative confocal image of self-assembled hetero tetramers. “Hetero tetramer”: Two monovalent droplets (O1 and O4) and two divalent droplets (O2 and O3) with patches bearing preprogrammed binding specificity (α on O1, α′ and β on O2, β′ and γ on O3, γ′ on O4) self-assemble into a tetramer. (F) A representative confocal image of trivalent clusters. “Trivalent cluster”: One trivalent droplet (C1) and three monovalent droplets (C2) with patches bearing preprogrammed binding specificity (three α on C1 and one α′ on C2) self assemble into a trivalent cluster. (Scale bars, 5 μm.)
To demonstrate that patch interactions can be diversified on one droplet, we made a linear hetero tetramer with four different modules: monovalent droplets O1, O4 and divalent droplets O2, O3, among which O2 and O3 are equipped with multiflavored patches. The prescribed structures were achieved after 144-h incubation (Fig. 3E and SI Appendix, Fig. S8). Moreover, we mixed trivalent droplets C1 (three α patches) and complementary monovalent droplets C2 (patch α′) to form a nonlinear structure, a trivalent cluster, after 72-h incubation (Fig. 3F). A hetero trivalent cluster (SI Appendix, Fig. S9) was also achieved by altering the binding interaction of one of the three patches on C1 (patch β), and introducing another type of monovalent droplet (patch β′).
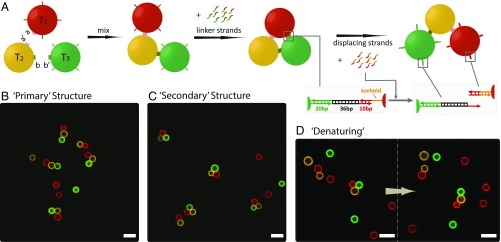
Finally, we demonstrate the versatility of the patchy droplets for dynamic hierarchical assembly by making reconfigurable trimers (Fig. 4A). To achieve this, another dimension of interaction and dynamic control are both needed. We therefore grafted the surface of the droplets with different single-stranded DNA handles. Initially, the “primary” structures, linear trimers (Fig. 4B and SI Appendix, Table S6), were assembled through interpatch bindings, similar to the formation of the hetero tetramer. Next, we added the linker strands, containing two segments that bridge the DNA handles on the “head” (T1) and the “tail” (T3) of the linear trimer, to fold it into a “triangle” (Fig. 4C and SI Appendix, Fig. S10). To “denature” the “secondary” structures, we then added an excess amount of displacing strands that have segments complementary to the toeholds on the DNA handles of T1. They triggered the dehybridization of the “bridges” between T1 and T3 via strand displacement (42, 43), leading to the unfolding of the triangles (Fig. 4D and SI Appendix, Fig. S11). The structure’s closing and opening occurred on the time scale of an hour, and were both captured in real time (Movies S5 and S6).
Fig. 4.
Reversible hierarchical assembly. (A) Two monovalent droplets (T1 and T3) and one divalent droplet (T2) with patches bearing preprogrammed binding specificity (α on T1, α′ and β on T2, β′ on T3) first assemble into a linear trimer. The linker strands are then added to bridge the handles on T1 (red) and T3 (green), resulting in the folding of the linear trimer into a triangle. The displacing strands are introduced to unfold the triangle via toehold strand displacement (see insets). Representative confocal images of (B) the linear trimers formed in the first step (trimer yield 59% after 142 h, SI Appendix, Table S6), (C) the triangles formed after adding the linkers with an ∼95% yield (SI Appendix, Fig. S9), (D) the unfolding of a triangle after adding the displacing strands with an ∼80% yield (SI Appendix, Fig. S10). (Scale bars, 10 μm.)
Although our technique enables a well-defined predetermination of particle valence and binding specificity it suffers from a great decrease in formation kinetics. In particular the use of patches is prohibitive for the conventional assembly of colloids in 3D suspensions controlled by diffusion (15). Compared with uniformly coated adhesive particles, the assembly of particles of radius R, each with a single patch of radius r, is reduced by a factor of α2, α = (πr2/4πR2). α ∼ 1/400 for our patchy droplets (R ∼ 2.5 μm, r ∼ 0.25 μm), i.e., the assembly time is increased by 160,000. This problem has been cleverly addressed by other researchers previously (44) by bringing the particles into surface contact using magnetic droplets and chaining them in a field. A detailed analysis of the resulting kinetics is found in ref. 40. The binding rate is proportional to the patch surface diffusion, D, and a single factor of α, 1/τ ∼ (D/R2)α. In our case the particles surfaces are held together by the buoyancy-driven concentration of droplets in the upper corner of a tilted sample container similar to the processing used in ref. 16 where uniformly coated droplets were used. Consequently our binding rate for dimers is ∼400 times slower, ∼50 h compared with the 10 min found in the previous study. If we used larger patches the time would decrease. The limiting patch size, still assuring that only one droplet binds to each patch, corresponds to α = 1/12 since only 12 monodispersed particles can pack on a like particle. This would allow a rate increase of 33 over our present droplets.
Our process of concentrating droplets in a corner of the sample container and then periodically flipping the container to allow new neighbors is clearly not an equilibrium process. In SI Appendix, we present an extremely simplified Monte Carlo calculation simulating the basic reactions involved in chain growth. Since the calculation uses phantom particles and chains, with no hydrodynamic interactions, excluded volume, or reduced mobilities for larger structures, we do not expect it to quantitatively compare with our results, as it does not, especially for long polymers. The sample flipping process, certainly not accounted for in the calculations, may further slow down the growth of long chains.
The fact that we are rate limited is evidenced in the data shown in SI Appendix for targeted growth of different structures. In SI Appendix, Table S6 we present the statistics for assembly of linear trimers, ABC. After 142 h the yield of ABC trimers is 59% (ABC trimers/initial B monomers). The fraction of unreacted Bs is 19%, the fraction of correctly formed dimers, AB and BC is 22%, and the fraction of mistakes (nonspecifically formed pairs) that will never form the target structure is 1%. In SI Appendix, Table S5 we present the statistics of divalent droplets in different polymerized structures. After 192 h 82% are in dimers or longer linear polymers, 16% are in “mistakes”––nonspecific or branched structures, and 2% remain as monomers. A decomposition in terms of valence shows 2% unbound, 52% bound to one neighbor (13% in dimers, 39% at the ends of longer chains), 43% bound to two neighbors, and 1.6% bound to three neighbors. The direct yield of divalently bound particles after 192 h is 43%, the yield of mistakes––particles which will never form specific divalent bonds––is 3%. This reinforces the idea that slow reaction kinetics is the greatest problem for this technique.
We have presented a bottom-up approach for fabricating a controllable number of multiflavored patches on liquid-based colloids by surface-mediated self-organization of DNA origami rafts. These patches were exploited to assemble a series of structures, including alternating polymers, finite chains, trivalent clusters, and reconfigurable triangles. This hybrid assembly, bridging the nanoscopic and the microscopic, can be extended in many directions. The valence number can be expanded easily, by applying more orthogonal pairs of rafts, to construct different structures, e.g., dendrimers or networks. Grafting different interactions directly on droplets would also facilitate multistep assembly, e.g., folding, ring formation. Our system, allowing for real-space, real-time observation of the nonequilibrium process, holds promise for serving as a model to study various biological processes, such as intercellular communication or protein folding. Rigid particles and structures with directional multiflavored bonds can be produced from our approach by polymerizing the droplet oil core (45) after the droplet chains fold into compact clusters. It should also be possible to assemble these specific adhesion patches on other systems with fluid phospholipid surfaces such as vesicles or cells.
Methods
DNA Sequence Generation and Purification.
DNA sequences of sticky ends were generated using the program Uniquimer (46). Single-stranded M13mp18 DNA genome was purchased from Bayou Biolabs. The staple sequences of the cross-shaped DNA origami rafts were adapted from Liu et al.’s paper (34). DNA strands were purchased from Integrated DNA Technology, Inc. (www.idtdna.com/pages). The biotinylated strands were HPLC-purified by Integrated DNA Technology, Inc. The other functional DNA sticky ends were purified via denaturing polyacrylamide gel electrophoresis.
AFM Imaging.
The AFM imaging was performed in tapping mode in air. A diluted sample (DNA origami rafts only or DNA origami-coated emulsion droplets) was deposited on freshly cleaved mica (Ted Pella, Inc.) for 5 min. The mica was washed with three drops of double-distilled water three times, and excess water was removed by blotting the mica with a filter paper. The mica was then blown dry using compressed air. All AFM imaging was performed on a NanoScope MultiMode 8-HR SPM (Bruker Corp.) with silicon tips (Bruker Corp.).
Supplementary Material
Acknowledgments
Y.Z. acknowledges support from US Department of Energy (DOE), Center for Bio-Inspired Energy Science for initiation, design, sample preparation, confocal microscopy, and data analysis. This research has been primarily supported by DOE DE-SC0007991 (to P.M.C., N.C.S., R.S., R.Z., and X.H.) for initiation, design, analysis, and imaging. We acknowledge partial support of Award GBMF3849 from the Gordon and Betty Moore Foundation (to P.M.C., N.C.S., R.S., and X.H.) for DNA sequence design, preparation, and characterization; partial support from National Science Foundation Awards EFRI-1332411 and CCF-1526650 (to R.S. and N.C.S.), and to X.H. for laboratory supplies. X.H. acknowledges partial support from the Materials Research Science & Engineering Centers (MRSEC) program of the National Science Foundation under Award DMR-1420073 for synthesis and characterization of the DNA origami. R.S. and N.C.S. acknowledge Department of Defense Multidisciplinary Research Program of the University Research Initiative (MURI) Award W911NF-11-1-0024 from the Army Research Office, MURI Award N000140911118 from the Office of Naval Research for partial salary support. R.S. and N.C.S. acknowledge partial support from DOE DE-SC0007991 for DNA synthesis and partial salary support. J.B. acknowledges partial support from National Science Foundation under Award DMR-1710163. The authors are grateful for shared facilities provided through the MRSEC program of the National Science Foundation under Award DMR-1420073.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. V.N.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718511115/-/DCSupplemental.
References
- 1.Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 2.Whitesides GM, Boncheva M. Beyond molecules: Self-assembly of mesoscopic and macroscopic components. Proc Natl Acad Sci USA. 2002;99:4769–4774. doi: 10.1073/pnas.082065899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frenkel D. Colloidal systems. Playing tricks with designer “atoms”. Science. 2002;296:65–66. doi: 10.1126/science.1070865. [DOI] [PubMed] [Google Scholar]
- 4.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 5.Manoharan VN, Elsesser MT, Pine DJ. Dense packing and symmetry in small clusters of microspheres. Science. 2003;301:483–487. doi: 10.1126/science.1086189. [DOI] [PubMed] [Google Scholar]
- 6.Leunissen ME, et al. Ionic colloidal crystals of oppositely charged particles. Nature. 2005;437:235–240. doi: 10.1038/nature03946. [DOI] [PubMed] [Google Scholar]
- 7.Talapin DV, et al. Quasicrystalline order in self-assembled binary nanoparticle superlattices. Nature. 2009;461:964–967. doi: 10.1038/nature08439. [DOI] [PubMed] [Google Scholar]
- 8.Sacanna S, Irvine WTM, Chaikin PM, Pine DJ. Lock and key colloids. Nature. 2010;464:575–578. doi: 10.1038/nature08906. [DOI] [PubMed] [Google Scholar]
- 9.Chen Q, Bae SC, Granick S. Directed self-assembly of a colloidal kagome lattice. Nature. 2011;469:381–384. doi: 10.1038/nature09713. [DOI] [PubMed] [Google Scholar]
- 10.Macfarlane RJ, et al. Nanoparticle superlattice engineering with DNA. Science. 2011;334:204–208. doi: 10.1126/science.1210493. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Lu F, Yager KG, van der Lelie D, Gang O. A general strategy for the DNA-mediated self-assembly of functional nanoparticles into heterogeneous systems. Nat Nanotechnol. 2013;8:865–872. doi: 10.1038/nnano.2013.209. [DOI] [PubMed] [Google Scholar]
- 12.Glotzer SC, Solomon MJ. Anisotropy of building blocks and their assembly into complex structures. Nat Mater. 2007;6:557–562. doi: 10.1038/nmat1949. [DOI] [PubMed] [Google Scholar]
- 13.Kraft DJ, et al. Surface roughness directed self-assembly of patchy particles into colloidal micelles. Proc Natl Acad Sci USA. 2012;109:10787–10792. doi: 10.1073/pnas.1116820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Colloids with valence and specific directional bonding. Nature. 2012;491:51–55. doi: 10.1038/nature11564. [DOI] [PubMed] [Google Scholar]
- 15.Chakraborty I, Meester V, van der Wel C, Kraft DJ. Colloidal joints with designed motion range and tunable joint flexibility. Nanoscale. 2017;9:7814–7821. doi: 10.1039/c6nr08069c. [DOI] [PubMed] [Google Scholar]
- 16.Feng L, Pontani LL, Dreyfus R, Chaikin PM, Brujic J. Specificity, flexibility and valence of DNA bonds guide emulsion architecture. Soft Matter. 2013;9:9816–9823. [Google Scholar]
- 17.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 18.Aldaye FA, Sleiman HF. Guest-mediated access to a single DNA nanostructure from a library of multiple assemblies. J Am Chem Soc. 2007;129:10070–10071. doi: 10.1021/ja073305n. [DOI] [PubMed] [Google Scholar]
- 19.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 20.Zheng J, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 22.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han D, et al. DNA gridiron nanostructures based on four-arm junctions. Science. 2013;339:1412–1415. doi: 10.1126/science.1232252. [DOI] [PubMed] [Google Scholar]
- 24.Gerling T, Wagenbauer KF, Neuner AM, Dietz H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science. 2015;347:1446–1452. doi: 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- 25.Wei B, Dai M, Yin P. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012;485:623–626. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke Y, Ong LL, Shih WM, Yin P. Three-dimensional structures self-assembled from DNA bricks. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 28.Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 29.Langecker M, et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012;338:932–936. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kocabey S, et al. Membrane-assisted growth of DNA origami nanostructure arrays. ACS Nano. 2015;9:3530–3539. doi: 10.1021/acsnano.5b00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki Y, Endo M, Sugiyama H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nat Commun. 2015;6:8052. doi: 10.1038/ncomms9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzyk A, et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012;483:311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- 33.Schreiber R, et al. Hierarchical assembly of metal nanoparticles, quantum dots and organic dyes using DNA origami scaffolds. Nat Nanotechnol. 2014;9:74–78. doi: 10.1038/nnano.2013.253. [DOI] [PubMed] [Google Scholar]
- 34.Liu W, et al. Diamond family of nanoparticle superlattices. Science. 2016;351:582–586. doi: 10.1126/science.aad2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat Chem. 2016;8:476–483. doi: 10.1038/nchem.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pontani LL, Jorjadze I, Viasnoff V, Brujic J. Biomimetic emulsions reveal the effect of mechanical forces on cell-cell adhesion. Proc Natl Acad Sci USA. 2012;109:9839–9844. doi: 10.1073/pnas.1201499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Sequential self-assembly of DNA functionalized droplets. Nat Commun. 2017;8:21. doi: 10.1038/s41467-017-00070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu W, Zhong H, Wang R, Seeman NC. Crystalline two-dimensional DNA-origami arrays. Angew Chem Int Ed Engl. 2011;50:264–267. doi: 10.1002/anie.201005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu KT, et al. Kinetics of DNA-coated sticky particles. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;88:022304. doi: 10.1103/PhysRevE.88.022304. [DOI] [PubMed] [Google Scholar]
- 40.Lee NK, et al. Ligand-receptor interactions in chains of colloids: When reactions are limited by rotational diffusion. Langmuir. 2008;24:1296–1307. doi: 10.1021/la701639n. [DOI] [PubMed] [Google Scholar]
- 41.Joshi D, et al. Kinetic control of the coverage of oil droplets by DNA-functionalized colloids. Sci Adv. 2016;2:e1600881. doi: 10.1126/sciadv.1600881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 43.Rogers WB, Manoharan VN. DNA nanotechnology. Programming colloidal phase transitions with DNA strand displacement. Science. 2015;347:639–642. doi: 10.1126/science.1259762. [DOI] [PubMed] [Google Scholar]
- 44.Baudry J, et al. Acceleration of the recognition rate between grafted ligands and receptors with magnetic forces. Proc Natl Acad Sci USA. 2006;103:16076–16078. doi: 10.1073/pnas.0607991103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ugelstad J, El-Aasser MS, Vanderhoff JW. Emulsion polymerization: Initiation of polymerization in monomer droplets. J Polym Sci C. 1973;11:503–513. [Google Scholar]
- 46.Wei B, Wang Z, Mi Y. Uniquimer: A de novo DNA sequence generation computer software for DNA self-assembly. J Comput Theor Nanosci. 2007;4:133–141. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.