Significance
Microbial arsenate respiration enhances the mobility of arsenic and contributes to the poisoning of tens of millions of people worldwide. Our ability to quantitatively predict how microbial activities shape arsenic geochemistry depends on a detailed understanding of how the enzymes that catalyze arsenate reduction work under environmentally relevant conditions. The structural and kinetic findings of the Arr enzyme complex reported here both help rationalize its extracytoplasmic localization and allow us to predict that the rate of arsenate release from minerals likely constrains its activity in sedimentary environments. Moreover, this work illustrates that engineering environmental bacteria to overexpress their native proteins can be straightforward, a strategy that may advance the study of enzymes that are challenging to express in traditional hosts.
Keywords: bacterial arsenate respiration, ArrAB, structure, enzymology, biogeochemistry
Abstract
Arsenate respiration by bacteria was discovered over two decades ago and is catalyzed by diverse organisms using the well-conserved Arr enzyme complex. Until now, the mechanisms underpinning this metabolism have been relatively opaque. Here, we report the structure of an Arr complex (solved by X-ray crystallography to 1.6-Å resolution), which was enabled by an improved Arr expression method in the genetically tractable arsenate respirer Shewanella sp. ANA-3. We also obtained structures bound with the substrate arsenate (1.8 Å), the product arsenite (1.8 Å), and the natural inhibitor phosphate (1.7 Å). The structures reveal a conserved active-site motif that distinguishes Arr [(R/K)GRY] from the closely related arsenite respiratory oxidase (Arx) complex (XGRGWG). Arr activity assays using methyl viologen as the electron donor and arsenate as the electron acceptor display two-site ping-pong kinetics. A Mo(V) species was detected with EPR spectroscopy, which is typical for proteins with a pyranopterin guanine dinucleotide cofactor. Arr is an extraordinarily fast enzyme that approaches the diffusion limit (Km = 44.6 ± 1.6 μM, kcat = 9,810 ± 220 seconds−1), and phosphate is a competitive inhibitor of arsenate reduction (Ki = 325 ± 12 μM). These observations, combined with knowledge of typical sedimentary arsenate and phosphate concentrations and known rates of arsenate desorption from minerals in the presence of phosphate, suggest that (i) arsenate desorption limits microbiologically induced arsenate reductive mobilization and (ii) phosphate enhances arsenic mobility by stimulating arsenate desorption rather than by inhibiting it at the enzymatic level.
Arsenic mobilization into drinking water is a health threat affecting tens of millions of people worldwide (1). This threat often originates from the underlying mineralogy and microbiology of water reservoirs (2), resulting in arsenic concentrations that exceed 10 μg/L, the World Health Organization’s guideline limit for arsenic in potable water (3). For example, as of 2009, over 5 million people in Bangladesh were exposed to arsenic concentrations >200 μg/L in drinking water, motivating mitigation efforts (4). In oxidizing environments, the predominant water-soluble form of arsenic is arsenate (found as H2AsO4− and HAsO42− with a pKa of 6.94; we use arsenate to reference both species interchangeably), which adsorbs onto minerals and does not rapidly contaminate water reservoirs (2). However, if the water becomes polluted with organic material, microbes consume the dissolved oxygen and turn to alternative electron acceptors to power metabolism. Many sediments contain bacteria and/or archaea with the capacity to generate energy by respiring arsenate, reducing it to arsenite (found as H3AsO3) (5–9), and thereby stimulate the release of arsenic into the aqueous phase because arsenite more readily leeches from mineral surfaces into water (10–12). Consequently, understanding the microbiology and biochemistry of arsenate respiration is of great interest to public health.
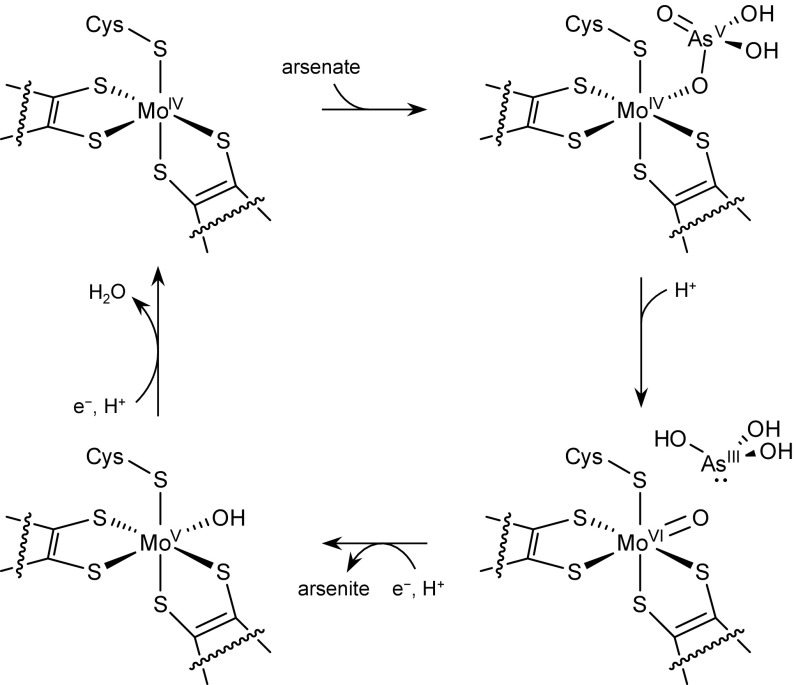
A microbe’s capacity for arsenate respiration corresponds with the genes encoding the arsenate respiratory reductase (Arr) complex, present in phylogenetically diverse organisms, including Gram-negative and Gram-positive bacteria (13). The Arr complex is a member of the DMSO reductase family of molybdoenzymes and comprises two proteins, a large subunit ArrA and a smaller subunit ArrB (13, 14). The ArrA subunit is predicted to contain a Mo atom coordinated by a pyranopterin guanine dinucleotide cofactor (Mo-bisPGD), as well as a single [4Fe–4S] cluster (14, 15). The ArrB subunit copurifies with ArrA (14, 16, 17) and is predicted to contain several [4Fe–4S] and/or [3Fe–4S] clusters (13, 15). The ArrA subunit contains a predicted N-terminal twin-arginine translocation (TAT) signal (13, 15), suggesting the assembled ArrAB complex is transferred posttranslationally across the cytoplasmic membrane, and its periplasmic localization has been confirmed experimentally in Gram-negative bacteria (16, 17). By analogy to the well-studied DMSO and nitrate reductase complexes (18, 19), the reaction mechanism likely involves oxygen atom transfer from arsenate to the Mo atom followed by two one-electron reductions of the Mo atom (Scheme 1). In some organisms, ArrAB likely associates with a third subunit ArrC, a heme-containing quinol oxidase that is presumably the electron donor for the complex (18, 20). In species lacking ArrC, such as Shewanella sp. ANA-3 (21), ArrAB instead acquires electrons from the tetraheme cytochrome CymA (20).
Scheme 1.
Proposed reaction cycle for Arr based on the mechanism of other Mo-bisPGD proteins. The reduced form of Arr contains Mo(IV) coordinated by two PGD cofactors and a cysteine side chain (Top Left). Upon binding to the protein, arsenate coordinates to the Mo atom (Top Right). An oxygen atom from arsenate is transferred to the Mo atom to form Mo(VI) and arsenite (Bottom Right). The cofactor is regenerated by sequential one-electron reductions, first to the EPR active Mo(V) species (Bottom Left) and then to Mo(IV) (Top Left). Although we have illustrated it here as H2AsO4−, the protonation state of bound arsenate is unknown and might instead be HAsO42−. Also unknown is whether the final water dissociates as illustrated (Bottom Left to Top Left) or if is displaced when arsenate binds (Top Left to Top Right).
Although arsenate respiration is a specialized microbial metabolism, small cytosolic arsenate reductases (ArsC and Acr2p) are present in numerous microorganisms (22). In contrast to Arr, which enables arsenate respiration using electrons from the quinone pool, ArsC and Acr2p likely evolved to detoxify cytoplasmic arsenate (23) and work through a disulfide redox cascade with glutaredoxin or thioredoxin (23–25). Emphasizing their separate physiological roles, Shewanella sp. ANA-3 does not require the Ars detoxification system for arsenate respiration (21) and the arr genes are induced by arsenite at concentrations 1,000 lower than those that induce ars (26). The Arr respiration system, located extracytoplasmically, has a greater affinity (Km = 44.6 μM, kcat/Km = 2.2 × 108 M−1⋅s−1, determined in this work) for dissolved arsenate than Ars [Km = 68 μM, kcat/Km = 5.2 × 104 M−1⋅s−1 (25)] and is able to reduce low levels of arsenate in equilibrium with iron minerals (11, 27). However, both arr and ars transcripts have been detected in arsenic-contaminated soils (6) and organisms expressing either enzyme can contribute to solubilized arsenate reduction (21, 28, 29).
Given the comparatively limited biochemical characterizations of Arr, it is difficult to disentangle the relative contributions of the Arr and Ars/Acr systems to arsenic mobilization. A major challenge for mechanistic enzymological studies of Arr has been obtaining enough protein for spectroscopic studies. The Arr complex expresses poorly in the heterologous host Escherichia coli (17), and ordinarily native induction and purification generate large volumes of arsenic-laden waste (14, 16). In this work, we successfully overcame both obstacles by overexpressing the Arr in its native host, Shewanella sp. ANA-3. With an abundance of protein, we optimized Arr crystal growth and used X-ray crystallography to solve the structure of an Arr complex. We also used enzyme kinetics and EPR spectroscopy to validate the presumed overall reaction cycle. Together, these mechanistic studies advance our ability to predict the extent to which microbial arsenate respiration will mobilize arsenic in the environment.
Results
Overview of the ArrAB Complex.
We obtained initial Arr crystal hits using protein from the heterologous host E. coli (17). To purify more protein for crystal optimization, we developed an overexpression vector for the native host Shewanella sp. ANA-3 (SI Appendix, SI Materials and Methods), providing ∼100-fold greater yields compared with E. coli (up to 5 mg of protein per L of culture; SI Appendix, Fig. S1 A and B). Subsequent crystal optimization (SI Appendix, Fig. S1C) enabled us to solve the X-ray crystal structure of ArrAB, bound to the substrate arsenate and product arsenite, and to the inhibitor phosphate. Data collection and refinement statistics are shown in Table 1. Purified Arr crystallized as the ArrAB heterodimer with two noncrystallographic symmetry (NCS) copies of ArrAB per asymmetric unit. The single ArrAB heterodimer is consistent with the size of the complex observed during size-exclusion chromatography (14, 16, 17), and so it likely represents the physiological complex rather than the crystallographic dimer of heterodimers. The structures do not include the N-terminal TAT sequence as it was removed to facilitate expression and purification. For consistency, the residue numbers refer to positions in the full ORF including the TAT sequence with the N-terminal methionine starting at position 1.
Table 1.
Crystallography data collection and refinement statistics
| Parameters | Arr (PDB ID 6CZ7) | Arr with arsenate (PDB ID 6CZ8) | Arr with arsenite (PDB ID 6CZ9) | Arr with phosphate (PDB ID 6CZA) |
| Cryoprotection | ||||
| Buffer | Hepes, pH 7.5 | MES, pH 6.0 | Hepes, pH 7.5 | MES, pH 6.0 |
| Data collection | ||||
| Space group | C2 | C2 | C2 | C2 |
| Unit cell | ||||
| a, b, c, Å | 231.26, 85.64, 146.68 | 233.162, 86.344, 148.221 | 232.512, 86.306, 147.367 | 232.397, 86.366, 147.429 |
| α, β, γ, ° | 90, 127.84, 90 | 90, 127.675, 90 | 90, 127.72, 90 | 90, 127.608, 90 |
| Resolution range, Å | 46.91–1.62 (1.678–1.62) | 39.1–1.78 (1.844–1.78) | 39.37–1.80 (1.864–1.80) | 37.70–1.71 (1.771–1.71) |
| Unique reflections | 286,352 (28,526) | 222,359 (22,113) | 209,823 (20,962) | 245,023 (23,784) |
| Rpim, % | 3.1 (32.0) | 3.4 (30.3) | 3.2 (31.3) | 3.3 (31.1) |
| <I/σ(I)> | 17.77 (2.40) | 15.94 (2.35) | 17.49 (2.27) | 15.71 (2.17) |
| Completeness, % | 99.93 (99.99) | 99.85 (99.84) | 98.35 (98.89) | 98.33 (95.72) |
| Multiplicity | 6.8 (6.6) | 6.8 (7.0) | 6.9 (7.0) | 6.9 (6.6) |
| Wilson B factor, Å2 | 18.10 | 23.40 | 23.77 | 20.40 |
| Refinement | ||||
| Rwork/Rfree | 0.1374/0.1547 | 0.1372/0.1681 | 0.1422/0.1694 | 0.1317/0.1535 |
| No. of non-H atoms | ||||
| Protein | 16,452 | 16,429 | 16,320 | 16,448 |
| Water | 2,435 | 2,241 | 2,288 | 2,378 |
| RMSD | ||||
| Bond lengths, Å | 0.007 | 0.010 | 0.006 | 0.009 |
| Bond angles, ° | 1.27 | 1.23 | 1.16 | 1.29 |
| Average B factors, Å2 | ||||
| Protein | 25.85 | 30.22 | 31.21 | 26.76 |
| Ligands | 23.04 | 27.30 | 28.64 | 23.78 |
| Water | 35.34 | 38.37 | 39.37 | 36.77 |
| Ramachandran, % | ||||
| Favored | 96.88 | 96.73 | 96.78 | 96.92 |
| Allowed | 2.88 | 2.98 | 2.98 | 2.79 |
| Outliers | 0.24 | 0.29 | 0.24 | 0.29 |
Values in parentheses are for the highest-resolution shell.
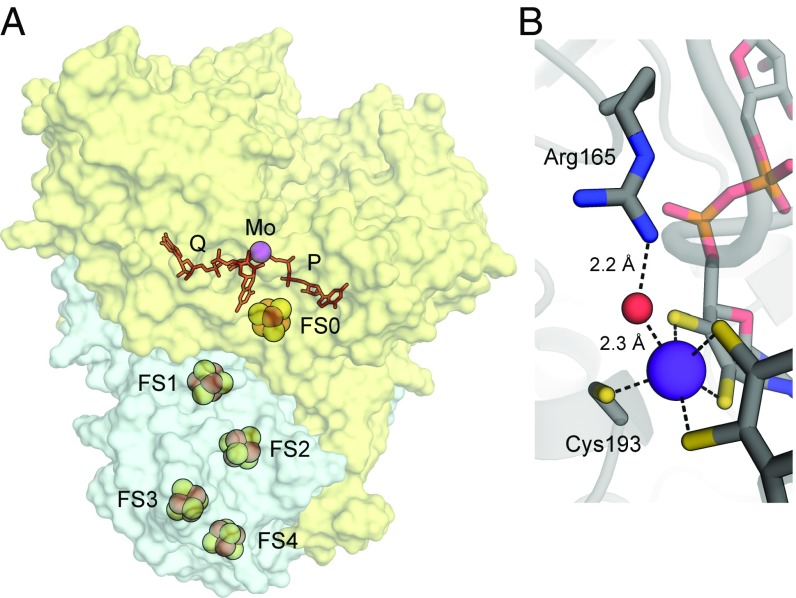
The overall structure and arrangement of the complex (Fig. 1A) resembles that of other Mo-bisPGD protein complexes including nitrate reductase (NarGH) (30), formate dehydrogenase-N (FdnGHI) (31), polysulfide reductase (PsrABC) (32), and perchlorate reductase (PcrAB) (33). All five Fe–S metal clusters are [4Fe−4S] and all 20 iron atoms are coordinated by cysteine side chains. The ArrA and ArrB subunits are bound together by extensive surface contact covering ∼3,400 Å2. Reflecting their close phylogeny (13, 34), the backbones of ArrAB and PsrAB are almost completely superimposable including their [4Fe–4S] clusters and Mo-bisPGD cofactors (SI Appendix, Fig. S2A). The active sites of ArrA and PsrA are distinguished by a unique set of residues that occupy distinct positions in the protein fold (SI Appendix, Fig. S2 B and C).
Fig. 1.
Overview of the ArrAB heterodimer from Shewanella sp. ANA-3. (A) View of the ArrA (yellow) and ArrB (blue) subunits with cofactors highlighted. The [4Fe–4S] clusters (FS0–FS4) and Mo atom are shown as spheres (Fe in orange, S in yellow, and Mo in purple). The PGD cofactors are shown in red labeled Q and P according to the DMSO reductase nomenclature. (B) Close-up view of the Mo-binding site in ArrA, focusing on the coordination of Mo (purple sphere) by water (red sphere), Cys193, and two PGD cofactors (C in gray, N in blue, O in red, S in yellow, and P in orange). Dashed lines illustrate coordination or hydrogen bonding.
ArrA and the Arsenate Binding Site.
The ArrA subunit contains the four-domain structure of related Mo-bisPGD proteins (18, 19) with a funnel leading from solvent to the active site. The substrate funnel is lined mostly with basic and aromatic residues (SI Appendix, Fig. S3). The Mo atom coordination geometry is distorted octahedral, in contrast to the trigonal prismatic geometry in PsrA (32), and it includes the dithiolene of both PGD cofactors with the cysteine side chain of Cys193 in an apical position (Fig. 1B). The electron density map suggests an oxygen atom, observed in one NCS copy and refined to 0.55 occupancy, completes the coordination geometry in the equatorial plane. The weak density and Mo–O bond distance of 2.3 Å likely indicates reduction of an oxo group (Mo=O) in the synchrotron X-ray beam (19). As in PsrA (32), an arginine side chain (Arg165) provides a hydrogen bond to the MoO group, but it is present in a different loop in ArrA and donates with Nη2 instead of Nε (Fig. 1B and SI Appendix, Fig. S2B). The [4Fe–4S] cluster in ArrA (FS0) is coordinated by cysteines 61, 64, 68, and 96, in contrast to NarG and PcrA, which contain a coordinating histidine (30, 33).
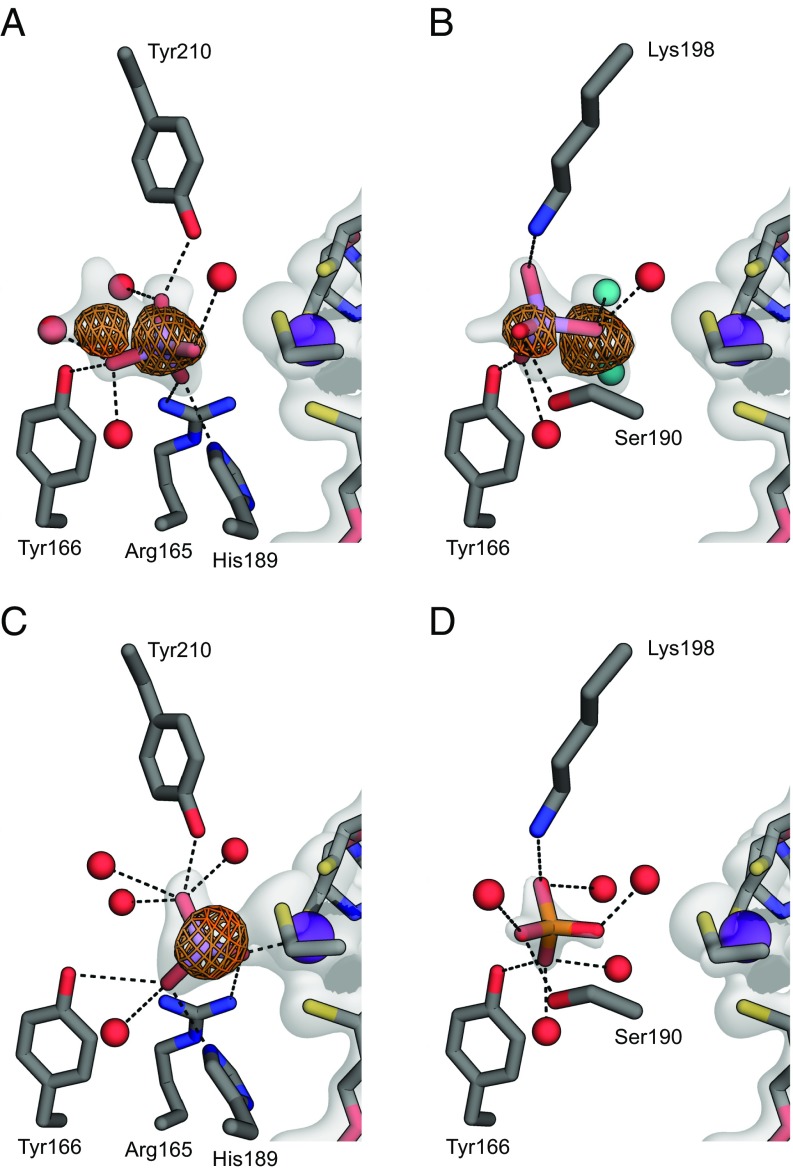
Cocrystallization yielded structures bound to arsenate (Fig. 2 A and B). Occupancy was highest at pH 6.0, lower at pH 7.5, and unobservable at pH 8.5, suggesting ArrA might bind the H2AsO4− state of arsenate. Alternatively, these binding differences might reflect changes in the protein’s protonation or subtle changes in structure. Unexpectedly, we observed arsenate in two alternate, mutually exclusive conformations, each with an occupancy of ∼0.5. This model is supported by two discrete features in the anomalous difference map collected at a wavelength of 1.0 Å, representing the locations of the central As atoms (Fig. 2 A and B). Both anomalous peaks disappeared at a wavelength of 1.1 Å, consistent with the As K-edge at 1.0448 Å. In the conformation closest to the Mo atom, arsenate forms hydrogen bonds with Arg165, His189, Tyr166, Tyr210, and several water molecules (Fig. 2A) with an As–Mo distance of 5.2 Å. In the second conformation, two waters substitute for arsenate oxygen atoms from the first conformation, and arsenate is hydrogen-bonded with Tyr166, Ser190, Lys198, and multiple water molecules (Fig. 2B) with an As–Mo distance of 7.3 Å. The alternative sites might reflect differences between the oxidized Mo(VI)O and the X-ray reduced Mo(V)OH [or Mo(IV) + H2O] forms of the protein. It is important to note that we are unable to resolve individual hydrogen atoms at the resolution of our data (∼1.8 Å), and the proposed hydrogen bonds are hypothesized based on bond distances and angles.
Fig. 2.
Close-up view of the arsenate-binding site in ArrA (C in gray, N in blue, O in red, S in yellow, P in orange, Mo in purple, and As in light purple). The 2mFo − Fc map around the relevant portions is shown as a white surface contoured to 1.5σ. For clarity, the map is shown only around ligands, the Mo-bisPGD cofactor, and the Mo-coordinating cysteine. The anomalous map collected at a wavelength of 1 Å is shown around arsenic atoms contoured to 6σ. Dashed lines indicate possible hydrogen bonds or coordination to the Mo atom. (A) Arsenate bound in the conformation nearest the Mo atom. (B) Arsenate bound in a second conformation farther from the Mo atom. The two water molecules in teal substitute for oxygen atoms from the first conformation shown in A. (C) Arsenite bound and coordinated to the Mo atom. (D) Phosphate bound to the active site in a conformation resembling the arsenate conformation from B.
We also obtained structures bound to arsenite, the product of arsenate respiration. Much like the first arsenate binding conformation, arsenite is coordinated by residues Arg165, Tyr166, His189, and Tyr210, but arsenite is shifted closer to the Mo atom with a Mo–As distance of 3.8 Å (Fig. 2C). In this position, arsenite forms a 2.3 Å Mo–O bond. While the proximity of arsenite to Cys193 resembles that observed in the ArsC enzymes, which form a covalent Cys-S–As intermediate with a bond distance of 2.2 Å (35), the crystallographic Arr Cys-S–As distance is too long to represent a covalent bond (3.0 Å as refined, or 2.8 Å to the center of the anomalous As peak). This structure might represent a nonphysiological form of the enzyme, as it likely illustrates arsenite binding to Mo(IV) or Mo(V) rather than to Mo(VI) as expected in the overall reaction cycle (Scheme 1). Similar structures have been observed in arsenite-inhibited aldehyde reductase (36, 37) and proposed as a dead-end pathway for the arsenite oxidase Aio (38).
Cocrystallization with phosphate also yielded structures bound to phosphate at pH 6.0, but not at pH 7.5. The structure resembles that of arsenate in the second conformation, with coordination by Tyr166, Ser190, Lys198, and several water molecules (Fig. 2D). We did not observe phosphate binding at the other arsenate site, suggesting the two sites have different arsenate specificities.
We found no differences in the protein side chains between arsenate and phosphate binding. However, we did observe a subtle change in dithiolene bond angles of the P-PGD cofactor upon arsenite binding. In the substrate-free and arsenate-bound structures, the S–C–C–S group is nearly planar (SI Appendix, Fig. S4), consistent with sp2 hybridization of the dithiolene carbon atoms. In the arsenite-bound structure, there is a small but notable deviation from planarity (SI Appendix, Fig. S4) that suggests partial reduction of the dithiolene carbons. As Arr is bidirectional in vitro (39), arsenite might be the electron donor for this reduction, and the shift might indicate that the dithiolene is electronically coupled to the Mo atom.
ArrB and the Electron Transfer Pathway.
ArrB shares the same overall fold of related electron transfer subunits in the Mo-bisPGD family (30–33). It contains four [4Fe–4S] clusters extending linearly from FS0 in ArrA to the distal FS4 (Fig. 1A). The coordinating cysteines differ in arrangement from the original prediction (13) and are as follows: (FS1) 12, 15, 18, 183; (FS2) 22, 164, 167, 179; (FS3) 57, 60, 65, 99; and (FS4) 69, 89, 92, 95. The last cluster (FS4) is separated from solvent by a single layer of the peptide backbone. By comparison with PsrABC, this region likely binds to the membrane protein CymA in vivo, and one NCS copy of ArrB forms extensive crystallographic contacts near FS4. The FS4 cluster is located near a protruding ridge distinguished by a series of positively charged residues including Lys67, Lys94, Lys93, Arg159, Lys158, and Arg113 (SI Appendix, Fig. S5), and it is tempting to speculate that these residues interact with CymA.
Arr and Arx Have Different Active Sites.
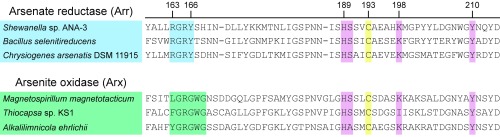
The Arx family of arsenite oxidases forms a clade closely related to Arr (40, 41). Both enzymes are bidirectional (39) but differ in the physiological direction of activity. An open question is whether Arr and Arx directionality is determined by the enzyme active site, redox potentials of the metal cofactors, the electron donor or acceptor in the electron transport chain, or a combination thereof (39). The structure of Arr reveals a conserved distinction between Arr and Arx in the active site, suggesting the two complexes are tuned for directionality. In Arr, the motif beginning at position 163 is (R/K)GRY(Fig. 3 and SI Appendix, Fig. S6) and is conserved with few exceptions across several hundred partial ArrA sequences analyzed previously (42). In Arx, the motif is XGRGWG (Fig. 3 and SI Appendix, Fig. S6), where X is F, L, T, or Y in the sequence diversity observed to date. It is unclear whether the Arx equivalent of Tyr166 is Gly or Trp. In Arr, Arg163 and Tyr166 form a π-stacking interaction that might affect the pKa of the tyrosine side chain. Accordingly, one possibility for Arx is that Trp substitutes for Tyr in the active site, forming a hydrogen bond with arsenite or stabilizing the transition state. Because tryptophan is a hydrogen bond donor, this model is difficult to reconcile with the high pKa of arsenite (9.2). Alternatively, this position might be occupied by Gly, allowing for hydrogen bonding with water or the protein backbone, or possibly disallowing hydrogen bonding to one –OH group of arsenite. Future structural and mutational studies of Arx will resolve the functions of these residues.
Fig. 3.
Alignment of representative ArrA and ArxA sequences highlighting the active-site residues. A more complete comparison that includes accession numbers is provided in SI Appendix, Fig. S6. Annotated ArrA enzymes contain a (R/K)GRY motif (blue), while annotated ArxA sequences contain a XGRGWG motif (green). Other active-site residues are generally conserved between ArrA and ArxA (pink), including the Mo-coordinating Cys residue (yellow). Sequence numbers are relative to the ArrA from Shewanella sp. ANA-3.
Arr Exhibits Two-Site Ping-Pong Kinetics.
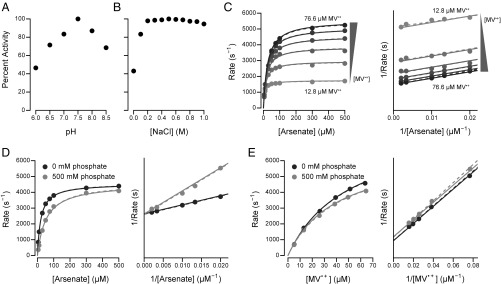
While confirming the activity of our enzyme preparations, we discovered some technical caveats that placed uncertainty on the originally reported arsenate Km of 5 μM and Vmax of 11,111 s−1 (17). First, we found that dithionite, used to generate the electron donor methyl viologen radical (MV•+) for the colorimetric assay, interfered with the assay at pH 7.0 and below, possibly due to contaminating sulfite in our stock of commercial dithionite (43). Second, we found that the affinity for MV•+ is surprisingly low compared with other Mo-bisPGD reductases, which was not accounted for previously. In light of these considerations, we performed kinetic experiments using Ti(III) citrate as a substitute for dithionite.
In our hands, the Shewanella sp. ANA-3 Arr complex has maximum activity at pH 7.5 (Fig. 4A) and above 200 mM NaCl (Fig. 4B) [as opposed to pH 7.0 and 150 mM NaCl as previously assayed (17)]. The comparatively low affinity for MV•+ enabled us to test for the ping-pong kinetics predicted by the proposed reaction cycle (Scheme 1). As expected, measurements of enzyme rate versus arsenate concentration, determined at multiple MV•+ concentrations, produce parallel lines on a double-reciprocal plot (Fig. 4C), illustrating the classical picture of ping-pong kinetics. A global nonlinear fit of a ping-pong kinetic model to the data yields the following parameters (±1 SE): arsenate Km = 44.6 ± 1.6 μM, MV•+ Km = 59.1 ± 2.4 μM, and kcat = 9,810 ± 220 s−1 (as arsenate reduced).
Fig. 4.
Kinetic characterization of Arr. Enzyme rates are shown as molecules of arsenate reduced per second per molecule of protein. Dashed lines illustrate a kinetic model fit for each curve individually. Solid lines illustrate a single kinetic model fit to all datasets together. For clarity, double-reciprocal plots omit the y-axis tick marks and begin at 0 s−1. For C–E, the assays were performed at pH 7.5 and 300 mM NaCl. (A) Relative activity of Arr as a function of pH. (B) Relative activity of Arr as a function of NaCl concentration. (C) Enzyme rates as a function of arsenate and MV•+ concentration. Parallel lines on the double-reciprocal plot (Right) indicate ping-pong kinetics. (D) Enzyme rates as a function of arsenate and phosphate concentration. The MV•+ concentration was 51.1 μM. Intersecting lines at the y axis on the double-reciprocal plot (Right) indicate competitive inhibition. (E) Enzyme rates as a function of MV•+ and phosphate concentration. The arsenate concentration was 250 μM. Parallel lines on the double-reciprocal plot (Right) indicate uncompetitive inhibition.
Based on the chemical similarity between arsenate and phosphate, and similar binding sites in our crystal structures, we expected phosphate to act as a competitive inhibitor for arsenate reduction. Rate measurements as a function of arsenate, with and without phosphate, confirmed competitive inhibition as illustrated by intersecting lines on a double-reciprocal plot (Fig. 4D). In contrast, similar rate measurements as a function of MV•+ yielded parallel lines on a double-reciprocal plot (Fig. 4E), indicative of uncompetitive inhibition. Using the Km and kcat values determined above, a global nonlinear fit to the inhibition data yields a phosphate Ki of 325 ± 12 μM. The different modes of phosphate inhibition indicate that the two substrates, MV•+ and arsenate, bind at distinct locations in the enzyme, consistent with arsenate reduction at the Mo atom in ArrA and protein reduction at FS4 in ArrB.
The Arr Reaction Cycle Produces Mo(V).
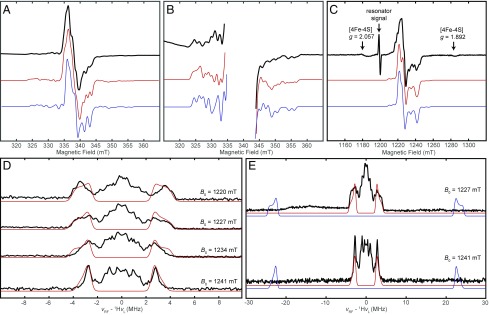
A feature common to Mo-bisPGD proteins is the generation of a discrete Mo(V) species in the reaction cycle (18, 19). A notable exception is the arsenite oxidase (Aio) complex for which no Mo(V) intermediate has been observed (44, 45). Because Arr and Aio catalyze the reverse reactions of one another, we wondered whether the Arr mechanism would more closely resemble that of Aio than the other characterized Mo-bisPGD proteins. In comparison with Mo(IV) and Mo(VI), which are invisible in EPR spectroscopy, Mo(V) produces a characteristic EPR signal. We therefore used EPR to confirm the presence of a Mo(V) species in a reaction mixture containing Arr, dithionite, arsenate, and varying concentrations of methyl viologen.
An X-band continuous-wave (CW) EPR spectrum collected on the reaction mixture produced an intense EPR signal at 120 K (Fig. 5A) attributable to Mo(V). This signal can be distinguished as arising from Mo(V) as opposed to the [4Fe–4S] clusters based on the acquisition temperature and its g values. Signals from at least one class of [4Fe–4S] cluster become evident only upon lowering temperature to 20 K (SI Appendix, Fig. S7). In control experiments, the reaction mixture or protein alone yielded no measurable signal. The Mo(V) EPR spectrum (Fig. 5A) exhibits a roughly axial symmetry (g = [1.990, 1.980, 1.957]), with clear low-intensity satellite peaks at the low and high field edges of the spectrum (Fig. 5B) due to hyperfine interactions between the unpaired electron and the two magnetic nuclei of molybdenum (95Mo and 97Mo, both I = 3/2; 15.92% and 9.55% natural abundance, respectively). Simulations including these interactions alone could not reproduce the complexity of the experimental spectrum (SI Appendix, Fig. S8), requiring the inclusion of an additional hyperfine coupling to produce the fine structure evident in the spectrum.
Fig. 5.
EPR characterization of the Mo(V) species generated in Arr. Samples contained 50 µM protein, 50 mM Hepes buffer (pH 7.5), 10 mM sodium dithionite, 1 µM methyl viologen, and 10 mM sodium arsenate. Simulation parameters are indicated in Table 2. (A) X-band CW EPR spectrum of Arr acquired at 120 K (black trace) with a simulation of the arsenite-bound Mo(V) species (red trace) compared with a simulation of a putative Mo(V)–OH species (blue trace) with the same simulation parameters except that the coupling to 75As has been replaced with a hyperfine coupling to 1H of A = [44, 49, 45] MHz. Acquisition parameters: microwave frequency, 9.390 GHz; microwave power, 1 mW; modulation amplitude, 0.4 mT. (B) Enlarged view of same X-band CW EPR spectrum (black trace) in A and simulations of the arsenite-bound Mo(V) species (red trace) and putative Mo(V)–OH species (blue trace) showing the low-intensity features from the hyperfine interaction with 95/97Mo. (C) Pseudomodulated Q-band ESE–EPR spectrum of Arr acquired at 12 K (black trace) with a simulation of the arsenite-bound Mo(V) species (red trace) compared with a simulation of a putative Mo(V)–OH species (blue trace) with the same simulation parameters except that the coupling to 75As has been replaced with a hyperfine coupling to 1H of A = [44, 49, 45] MHz. (D) Field-dependent Q-band 1H Davies ENDOR of the Mo(V) species in Arr (black trace) with a simulation of the largest observed 1H hyperfine coupling (red trace). (E) Wide Q-band 1H Davies ENDOR of the Mo(V) species in Arr (black trace) with a simulation of the largest observed 1H hyperfine coupling (red trace) compared with a simulation of the expected 1H ENDOR signals for the putative Mo(V)–OH species with 1H A = [44, 49, 45] MHz, which do not appear in the experimental spectrum to any significant degree.
Table 2.
EPR parameters for the Mo(V) signal observed in Arr under reaction conditions
| Axis | g | |95/97Mo A|, MHz | |1H A|, MHz | |75As A|, MHz | |75As P|, MHz |
| 1 | 1.990 | 122 | 8.5 | 23 | ≤12 |
| 2 | 1.980 | 68 | 5.2 | 23 | ≤24 |
| 3 | 1.957 | 143 | 5.2 | 32 | ≤24 |
The hyperfine and nuclear quadrupole coupling tensors are colinear with the g-tensor.
Simulations of the X-band CW EPR spectrum consistent with the observed data could be achieved with inclusion of either a large, relatively isotropic hyperfine coupling to a single proton (|1H A| = [44, 49, 45] MHz, 99.99% natural abundance, I = 1/2) or with a somewhat smaller hyperfine coupling to arsenic (|75As A| = [23, 23, 32]; 100% natural abundance, I = 3/2) in addition to a relatively rhombic hyperfine coupling to Mo (|95/97Mo A| = [122, 68, 143]). Because CW EPR is typically insensitive to the relative sign of each of the principal components of the hyperfine tensor, 75As A could be [23, 23, 32], [−23, −23, 32], [23, 23, −32], etc., and so we only note the absolute value here. In the case of the 75As simulation, addition of a nuclear quadrupole interaction was not necessary to simulate the CW spectrum, but an upward limit was able to be placed on this term of |75As P| ≤ [12, 12, 24]. Both simulation models are consistent with a Mo(V) species that could be generated in Arr, with a large isotropic 1H hyperfine coupling consistent with Mo(V)–OH as previously observed in xanthine oxidase (46, 47), or with a 75As hyperfine coupling consistent with an arsenite-bound Mo(V) species as has been observed in arsenite-inhibited xanthine oxidase (48) and aldehyde reductase (36, 37).
To interrogate the identity of the observed Mo(V) species, Q-band (34-GHz) pulse EPR and electron nuclear double-resonance (ENDOR) spectroscopies were used to further evaluate the potential magnetic interactions. The higher-frequency Q-band field-swept electron spin echo (ESE)–EPR spectrum provides a more rigorous constraint on the g values due to the increase in resolution of these field-dependent parameters in comparison with X-band (Fig. 5C). Field-dependent Q-band 1H Davies ENDOR (Fig. 5D) revealed the presence of only relatively small hyperfine couplings to protons, with the largest coupling corresponding to 1H A = [8.5, 5.2, 5.2] MHz. This hyperfine interaction is not large enough to account for the resolved splittings present in the X-band CW spectrum (SI Appendix, Fig. S8), and the lack of any features consistent with the large 1H hyperfine coupling necessary to reproduce the CW spectrum eliminates the possibility of any appreciable Mo(V)–OH being present in the experimental samples. The 1H hyperfine coupling observed in the ENDOR likely corresponds to the proximal β-proton of the Mo-ligating Cys193, which should be within ∼3.2 Å of the Mo(V) center. Attempts to detect signals associated with 75As via ENDOR were unsuccessful, likely owing to a combination of decreased sensitivity due to the lower gyromagnetic ratio of 75As in comparison with 1H (75As γ/1H γ = 0.172), as well as the potential for significant hyperfine anisotropy due to localization of electron density in As p-orbitals, and a large 75As nuclear quadrupole interaction that would cause substantial broadening of the ENDOR spectrum at any given field. These factors would not limit resolution of these interactions in the CW-EPR spectrum and would not be present for 1H, and so we assign the observed species to an arsenite-bound Mo(V).
We observed the Mo(V)–arsenite signal only with higher MV concentrations (1 nM to 1 µM) (SI Appendix, Fig. S9). Based on the rate of Arr (kcat = 9,810−1), these concentrations are likely to allow near completion of the reaction over the timescale of sample preparation (1–5 min). Lower MV concentrations (0.1 nM) produced no measurable signal, and we were unable to observe a signal consistent with Mo(V)–OH. We interpret this result to indicate that the putative Mo(V)–OH intermediate does not accumulate to measurable amounts during turnover conditions. By driving the reaction to completion, we instead captured an off-pathway Mo(V)–arsenite conjugate that formed after converting all arsenate into arsenite. Indeed, the Mo(V) concentration was estimated to be 43 µM for the 1 µM MV reaction sample by spin quantitation of the Mo(V) species performed via comparison of the doubly integrated intensity to that of a Cu(II) standard sample [200 µM CuSO4 in H2O with 30% glycerol (vol/vol)] acquired under identical acquisition conditions. Given the concentration of Arr (50 μM), this species represents the majority of Arr under these conditions. Using this same quantitation method, the Mo(V) concentration for the 1 nM MV reaction was estimated to be 30 μM. Also consistent with this interpretation, we observed no Mo(V) signal when the enzyme was incubated with arsenite and MV (SI Appendix, Fig. S10), indicating that generating the signal requires reduction (e.g., by dithionite) as observed for xanthine oxidase (36, 37) or possibly enzyme turnover. Nonetheless, our detection of the Mo(V)–arsenite conjugate establishes the ability of Arr to adopt a Mo(V) redox state in contrast to Aio, suggesting Arr behaves similarly to other well-characterized Mo-bisPGD enzymes.
Discussion
Microbial arsenate reduction is a key driver of arsenic mobilization in carbon-rich anoxic environments, where arsenate is commonly found adsorbed to minerals. To better understand this biogeochemical phenomenon, we determined the X-ray crystal structure of the Arr complex and performed a detailed characterization of its kinetics. Our results help clarify what may limit arsenate respiration in the environment and provide insight into the cell biological strategy taken to achieve it. The resulting kinetic picture of arsenate respiration is summarized in Fig. 6 and described below.
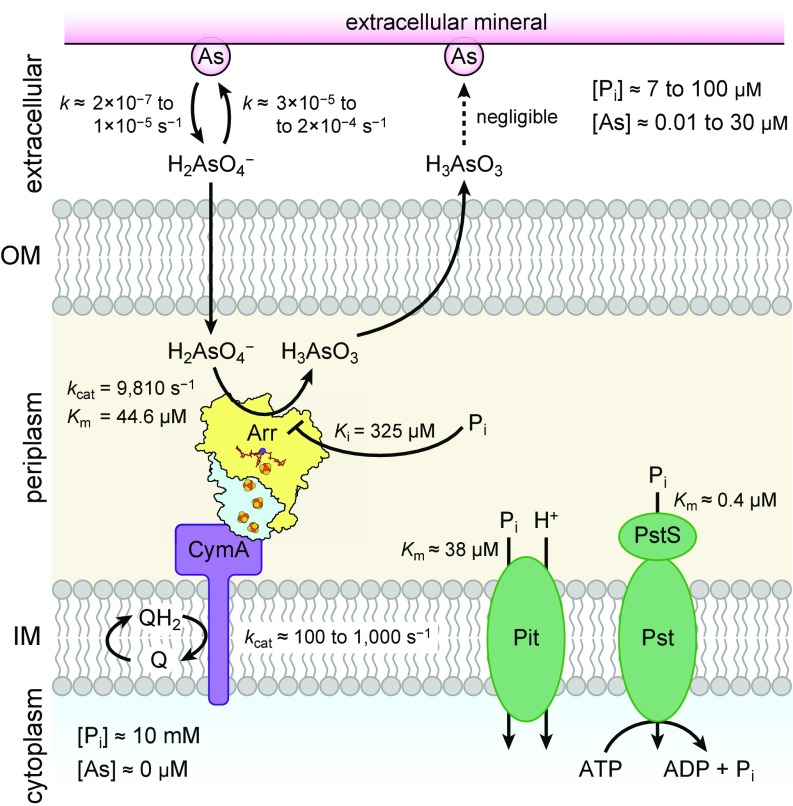
Fig. 6.
A summary of the localization and kinetics of key processes involved in arsenate respiration in Gram-negative bacteria. Arsenate is illustrated as H2AsO4−, which is in equilibrium with HAsO42− with a pKa of 6.94. References for literature values are provided in the main text. IM, inner membrane; OM, outer membrane.
Arsenate adsorption and desorption have been modeled as multistep processes (49). In a fast phase (minutes to hours), arsenate rapidly equilibrates with surface-accessible binding sites. In a subsequent slow phase (hours to weeks), arsenate equilibrates with less accessible sites, possibly representing slow diffusion through the insoluble material, and it is this slower phase that likely controls arsenic mobility (50). The slow phase in model soils proceeds with first-order rate constants of 0.1–0.8 h−1 (3 × 10−5 to 2 × 10−4 s−1) for adsorption and 0.007–0.035 h−1 (2 × 10−7 to 1 × 10−5 s−1) for desorption (49). In comparison, our kinetic data reveal that Arr is an exceptionally fast enzyme that approaches the diffusion limit. A similar rate has been reported for the Arr complex from Chrysiogenes arsenatis, a member of a deeply branching bacterial lineage (16) (kcat = 14,000 s−1). A slower rate has been reported for the Gram-positive bacterium Bacillus selenitireducens (kcat = 5 s−1), possibly owing to metal lability during purification (14). Given the fast rate of Arr, the steady-state rate of arsenate respiration is likely to be limited by other features of the electron transport chain and/or the environmental arsenate concentration. The use of quinones as electron donors (through CymA or ArrC) for the sequential one-electron reductions of Arr necessitates formation of the semiquinone radical, and this process is known to be rate-limiting in the cytochrome bc1 complex (with a rate of <1,000 s−1) (51). Similarly, Aio is rate limited by the reduction of cytochrome c (kcat = 427 s−1) rather than by arsenite oxidation (kcat > 4,000 s−1) (52). Therefore, the steady-state rate of Arr coupled to its electron donor is expected to be on the order of 100–1,000 s−1 in vivo. Moreover, arsenic concentrations in contaminated groundwaters are generally below the Km of Arr, ranging from 1 μg/L (0.013 μM) to 2,500 μg/L (33 μM) (53). At the recommended limit of 10 μg/L (0.13 μM), Michaelis–Menten kinetics predicts that Arr would have a maximum rate of 29 s−1. Nonetheless, these lower expected rates are still orders of magnitude faster than the rates of arsenate adsorption onto minerals, and so Arr is poised to easily outcompete mineral surfaces for arsenate. Together, this kinetic picture is consistent with observations that arsenate respiration proceeds markedly slower with solid-phase arsenate (11, 29), suggesting that desorption is the rate-limiting step of arsenate mobilization in the environment.
In terms of arsenate acquisition, the similarity between phosphate and arsenate ions poses challenges for arsenate respirers. As a natural phosphate analog, arsenate is improperly incorporated by phosphorylases into arsenate esters with aqueous half-lives up to 100,000-fold shorter than the corresponding phosphate esters (54), leading to an uncoupling of energy generation through ADP-arsenate hydrolysis (55). Arsenate enters bacterial cells through the low-affinity Pit phosphate transporter (56). Using Pit with a Vmax of 55 nmol Pi (mg cell dry weight)−1⋅min−1 (56) devoted solely to arsenate transport, we estimate (using a cell weight of 1 pg) a maximal uptake rate of ∼5 × 105 cell−1⋅s−1. Although this rate is comparable to the arsenate reduction rate observed in cultures (∼1 × 105 cell−1⋅s−1) (11) and this method of arsenic acquisition would not be rate limiting per se, it would flood the cytoplasm with toxic arsenic species. Moreover, we found phosphate to be an effective inhibitor of Arr (Ki = 325 µM) at levels far below the typical intracellular phosphate concentration of 10 mM (57). By localizing Arr extracytoplasmically, cells solve both problems simultaneously by providing a segregated environment for arsenate respiration. Compared with the cytosolic environment, the average phosphate concentration in arsenic-rich groundwaters is much lower and ranges from 7 to 100 µM (53). Assuming the extracytoplasmic phosphate concentration reflects that of the extracellular milieu, these concentrations would not be expected to significantly inhibit arsenate respiration. Additionally, in arsenic-rich environments, bacteria (including Shewanella sp. ANA-3; National Center for Biotechnology Information genome accession number NC_008577.1) often contain the high-affinity Pst phosphate transporter (56) that can maintain up to a 4,500-fold selectivity for phosphate over arsenate (58). With a Km of 0.4 µM (56), the Pst system might lower the extracytoplasmic phosphate concentration further below that which would inhibit arsenate respiration. Moreover, even in artificial medium with up to 5 mM phosphate, arrA gene expression is unaffected in Shewanella sp. ANA-3 (26), and higher phosphate concentrations actually increase growth rates from arsenate respiration, presumably by protecting against the toxicity of arsenic (26). Together, these considerations suggest that environmental phosphate concentrations do not deter microbial arsenate reduction. On the contrary, by competing with arsenate for mineral sorption sites (59), phosphate likely enhances arsenate reduction by making it more bioavailable.
Consistent with our kinetic results, our crystal structures illustrate that Arr struggles to distinguish between phosphate and arsenate with both ions binding at the same site (Fig. 2 B and D). Phosphate binding at this site sterically blocks arsenate from accessing the catalytic Mo atom. Interestingly, we also observed arsenate, but not phosphate, at a second overlapping site (Fig. 2A) that might be more selective for arsenate. Binding at this site might reflect changes in the protein structure owing to Mo reduction in the synchrotron X-ray beam, or it might compete with the nonspecific site. Phosphate selectivity has been studied in detail using subangstrom resolution crystal structures of PstS (58). This protein achieves selectivity through a single rigid hydrogen bond that is positioned to interact with the specific bond distances and angles of phosphate (58). While the resolution of our data (1.8 Å) and diffuse substrate electron density (Fig. 2) preclude a similar analysis here, we note that arsenate in the specific site forms hydrogen bonds ∼2.6 Å to Tyr210 and Arg165 (Fig. 2A), similar to the 2.5-Å distance observed in PstS phosphate selectivity. Both residues are universally conserved in all known ArrA homologs (SI Appendix, Fig. S6), suggesting that, like PstS, Arr might use a hydrogen-bonding mechanism to enforce arsenate selectivity near the catalytic Mo atom.
Regardless of their mechanistic roles, the active-site residues revealed by the Arr structure provide a powerful handle for the annotation of ambiguous sequences as Arr or Arx. The sequence similarity between these enzymes makes it difficult to distinguish them without a phylogenetic analysis (41, 42), but doing so is essential to understand the directionality of arsenic redox transformations based on metagenomic data. The (R/K)GRY motif in Arr and the XGRGWG motif in Arx provide a structural basis for this distinction. Although PCR amplification with arrA primers has been used to identify Arr-encoding organisms in the environment (27, 42), the similarity between ArrA and ArxA raises questions about the specificity of this detection. For example, the “cluster V” group of arrA sequences identified by PCR (42) contains the Arx motif, suggesting this group is Arx rather than Arr, even though the amplified sequences appeared more related to arrA than to arxA (42). Interestingly, two annotated ArxA sequences from the Methanoperedens genus of archaea (60) deviate from the ArxA motif and contain SGRN(R/Q)G (SI Appendix, Fig. S6). These sequences form a sister group to the other ArxA members, suggesting the full sequence diversity of Arr and Arx has yet to be explored, and it is presently ambiguous whether these Methanoperedens enzymes operate as Arr or Arx.
Both the crystal structure (Fig. 2C) and the EPR spectrum of Arr (Fig. 5) suggest that the Arr Mo atom forms stable complexes with arsenite. Similar structures have been observed in xanthine oxidase (36, 37) for which arsenite is a potent inhibitor (Kd < 0.1 µM for the reduced protein) (61). However, unlike the case of xanthine oxidase, arsenite is a relevant product necessarily encountered by Arr as part of its function. Interestingly, although we readily observed a Mo(V)-arsenite EPR signal in Arr (Fig. 5), a similar signal has never been observed for the arsenite oxidase Aio (44, 45), a distantly related molybdoenzyme to Arr and Arx (41, 62). Compared with Arr and Arx, the Aio active site contains primarily basic residues (45, 63), and, unique among all known Mo-bisPGD proteins, it does not coordinate its Mo atom with the polypeptide chain (19, 45, 63). Moreover, although both Arr and Arx are bidirectional enzymes (39), Aio has only been observed to operate unidirectionally (62), a possible phenotypic manifestation of these biochemical differences.
In conclusion, this study has clarified important aspects of microbial arsenate respiration that allow its environmental activity to be better predicted. In addition, the native host overexpression system described here will enable the nature of the relationship between Arr, Arx, and Aio to be probed mechanistically. Future studies of the Arr mechanism will benefit from a side-by-side comparison with Arx, especially as it pertains to the functions of the distinct active-site residues, and we anticipate that expression in Shewanella or a similarly facile native host will provide ample amounts of Arx. By establishing the mechanisms used to control the directionality of arsenic redox transformations, such comparisons will provide further insight into arsenic biogeochemical cycling.
Methods
Please see SI Appendix, SI Materials and Methods for information regarding cell culturing, protein expression and purification, crystallography, enzyme activity assays, and EPR spectroscopy.
Supplementary Material
Acknowledgments
This research is rooted in work performed in the laboratory of Francois M. M. Morel (Massachusetts Institute of Technology/Princeton). We thank Shu-ou Shan and Lisa Racki for assistance with enzyme kinetics, Barbara Schoepp-Cothenet for correspondence about the phylogeny of Arr and Arx, and Jonas Peters for helpful discussions. The Caltech Molecular Observatory provided essential training, knowledge, and equipment for the crystallography in this work. NIH Grant 1R01AI127850-01A1 (to D.K.N.) supported this research. The Caltech EPR Facility was supported by National Science Foundation Grant 1531940 and the Dow Next Generation Educator Fund. This research used resources of the Advanced Light Source, which is a US Department of Energy (DOE) Office of Science User Facility under Contract DE-AC02-05CH11231. The Molecular Observatory is supported by the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US DOE Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-76SF00515. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the US DOE Office of Biological and Environmental Research, and by the NIH–National Institute of General Medical Sciences (including Grant P41GM103393). Funding to J.M.S. was provided by Biotechnology and Biological Sciences Research Council Grant BB/N012674/1. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of National Institute of General Medical Sciences or NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6CZ7–6CZ9 and 6CZA).
See Commentary on page 9051.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807984115/-/DCSupplemental.
References
- 1.Naujokas MF, et al. The broad scope of health effects from chronic arsenic exposure: Update on a worldwide public health problem. Environ Health Perspect. 2013;121:295–302. doi: 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oremland RS, Stolz JF. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 2005;13:45–49. doi: 10.1016/j.tim.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization 2017. Guidelines for Drinking-Water Quality, Fourth Edition, Incorporating the First Addendum (World Health Organization, Geneva)
- 4.UNICEF . Bangladesh National Drinking Water Quality Survey of 2009. UNICEF; New York: 2011. [Google Scholar]
- 5.Kulp TR, et al. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl Environ Microbiol. 2006;72:6514–6526. doi: 10.1128/AEM.01066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo J, Bai Y, Liang J, Qu J. Metagenomic approach reveals variation of microbes with arsenic and antimony metabolism genes from highly contaminated soil. PLoS One. 2014;9:e108185. doi: 10.1371/journal.pone.0108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song B, Chyun E, Jaffé PR, Ward BB. Molecular methods to detect and monitor dissimilatory arsenate-respiring bacteria (DARB) in sediments. FEMS Microbiol Ecol. 2009;68:108–117. doi: 10.1111/j.1574-6941.2009.00657.x. [DOI] [PubMed] [Google Scholar]
- 8.Osborne TH, McArthur JM, Sikdar PK, Santini JM. Isolation of an arsenate-respiring bacterium from a redox front in an arsenic-polluted aquifer in West Bengal, Bengal Basin. Environ Sci Technol. 2015;49:4193–4199. doi: 10.1021/es504707x. [DOI] [PubMed] [Google Scholar]
- 9.Hassan Z, Sultana M, van Breukelen BM, Khan SI, Röling WF. Diverse arsenic- and iron-cycling microbial communities in arsenic-contaminated aquifers used for drinking water in Bangladesh. FEMS Microbiol Ecol. 2015;91:fiv026. doi: 10.1093/femsec/fiv026. [DOI] [PubMed] [Google Scholar]
- 10.Appelo CAJ, Van Der Weiden MJJ, Tournassat C, Charlet L. Surface complexation of ferrous iron and carbonate on ferrihydrite and the mobilization of arsenic. Environ Sci Technol. 2002;36:3096–3103. doi: 10.1021/es010130n. [DOI] [PubMed] [Google Scholar]
- 11.Zobrist J, Dowdle PR, Davis JA, Oremland RS. Mobilization of arsenite by dissimilatory reduction of adsorbed arsenate. Environ Sci Technol. 2000;34:4747–4753. [Google Scholar]
- 12.Dhar RK, et al. Microbes enhance mobility of arsenic in pleistocene aquifer sand from Bangladesh. Environ Sci Technol. 2011;45:2648–2654. doi: 10.1021/es1022015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA. 2003;100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afkar E, et al. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol Lett. 2003;226:107–112. doi: 10.1016/S0378-1097(03)00609-8. [DOI] [PubMed] [Google Scholar]
- 15.Stolz JF, Basu P, Santini JM, Oremland RS. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol. 2006;60:107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- 16.Krafft T, Macy JM. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur J Biochem. 1998;255:647–653. doi: 10.1046/j.1432-1327.1998.2550647.x. [DOI] [PubMed] [Google Scholar]
- 17.Malasarn D, Keeffe JR, Newman DK. Characterization of the arsenate respiratory reductase from Shewanella sp. strain ANA-3. J Bacteriol. 2008;190:135–142. doi: 10.1128/JB.01110-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimaldi S, Schoepp-Cothenet B, Ceccaldi P, Guigliarelli B, Magalon A. The prokaryotic Mo/W-bisPGD enzymes family: A catalytic workhorse in bioenergetic. Biochim Biophys Acta. 2013;1827:1048–1085. doi: 10.1016/j.bbabio.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Hille R, Hall J, Basu P. The mononuclear molybdenum enzymes. Chem Rev. 2014;114:3963–4038. doi: 10.1021/cr400443z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy JN, Saltikov CW. The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Shewanella species. J Bacteriol. 2007;189:2283–2290. doi: 10.1128/JB.01698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saltikov CW, Cifuentes A, Venkateswaran K, Newman DK. The ars detoxification system is advantageous but not required for As(V) respiration by the genetically tractable Shewanella species strain ANA-3. Appl Environ Microbiol. 2003;69:2800–2809. doi: 10.1128/AEM.69.5.2800-2809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y-G, Yoshinaga M, Zhao F-J, Rosen BP. Earth abides arsenic biotransformations. Annu Rev Earth Planet Sci. 2014;42:443–467. doi: 10.1146/annurev-earth-060313-054942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oden KL, Gladysheva TB, Rosen BP. Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol Microbiol. 1994;12:301–306. doi: 10.1111/j.1365-2958.1994.tb01018.x. [DOI] [PubMed] [Google Scholar]
- 24.Messens J, et al. All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade. Proc Natl Acad Sci USA. 2002;99:8506–8511. doi: 10.1073/pnas.132142799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messens J, et al. Kinetics and active site dynamics of Staphylococcus aureus arsenate reductase. J Biol Inorg Chem. 2002;7:146–156. doi: 10.1007/s007750100282. [DOI] [PubMed] [Google Scholar]
- 26.Saltikov CW, Wildman RA, Jr, Newman DK. Expression dynamics of arsenic respiration and detoxification in Shewanella sp. strain ANA-3. J Bacteriol. 2005;187:7390–7396. doi: 10.1128/JB.187.21.7390-7396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malasarn D, et al. arrA is a reliable marker for As(V) respiration. Science. 2004;306:455. doi: 10.1126/science.1102374. [DOI] [PubMed] [Google Scholar]
- 28.Macur RE, Jackson CR, Botero LM, McDermott TR, Inskeep WP. Bacterial populations associated with the oxidation and reduction of arsenic in an unsaturated soil. Environ Sci Technol. 2004;38:104–111. doi: 10.1021/es034455a. [DOI] [PubMed] [Google Scholar]
- 29.Langner HW, Inskeep WP. Microbial reduction of arsenate in the presence of ferrihydrite. Environ Sci Technol. 2000;34:3131–3136. [Google Scholar]
- 30.Jormakka M, Richardson D, Byrne B, Iwata S. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure. 2004;12:95–104. doi: 10.1016/j.str.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 31.Jormakka M, Törnroth S, Byrne B, Iwata S. Molecular basis of proton motive force generation: Structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 32.Jormakka M, et al. Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol. 2008;15:730–737. doi: 10.1038/nsmb.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youngblut MD, et al. Perchlorate reductase is distinguished by active site aromatic gate residues. J Biol Chem. 2016;291:9190–9202. doi: 10.1074/jbc.M116.714618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duval S, Ducluzeau A-L, Nitschke W, Schoepp-Cothenet B. Enzyme phylogenies as markers for the oxidation state of the environment: The case of respiratory arsenate reductase and related enzymes. BMC Evol Biol. 2008;8:206. doi: 10.1186/1471-2148-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin P, et al. Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure. 2001;9:1071–1081. doi: 10.1016/s0969-2126(01)00672-4. [DOI] [PubMed] [Google Scholar]
- 36.Boer DR, Thapper A, Brondino CD, Romão MJ, Moura JJG. X-ray crystal structure and EPR spectra of “arsenite-inhibited” Desulfovibriogigas aldehyde dehydrogenase: A member of the xanthine oxidase family. J Am Chem Soc. 2004;126:8614–8615. doi: 10.1021/ja0490222. [DOI] [PubMed] [Google Scholar]
- 37.Thapper A, Boer DR, Brondino CD, Moura JJG, Romão MJ. Correlating EPR and X-ray structural analysis of arsenite-inhibited forms of aldehyde oxidoreductase. J Biol Inorg Chem. 2007;12:353–366. doi: 10.1007/s00775-006-0191-9. [DOI] [PubMed] [Google Scholar]
- 38.Warelow TP, Pushie MJ, Cotelesage JJH, Santini JM, George GN. The active site structure and catalytic mechanism of arsenite oxidase. Sci Rep. 2017;7:1757. doi: 10.1038/s41598-017-01840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richey C, et al. Respiratory arsenate reductase as a bidirectional enzyme. Biochem Biophys Res Commun. 2009;382:298–302. doi: 10.1016/j.bbrc.2009.03.045. [DOI] [PubMed] [Google Scholar]
- 40.Zargar K, Hoeft S, Oremland R, Saltikov CW. Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1. J Bacteriol. 2010;192:3755–3762. doi: 10.1128/JB.00244-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zargar K, et al. ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ Microbiol. 2012;14:1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x. [DOI] [PubMed] [Google Scholar]
- 42.Mirza BS, Sorensen DL, Dupont RR, McLean JE. New arsenate reductase gene (arrA) PCR primers for diversity assessment and quantification in environmental samples. Appl Environ Microbiol. 2017;83:e02725-16. doi: 10.1128/AEM.02725-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna CE, Gutheil WG, Song W. A method for preparing analytically pure sodium dithionite. Dithionite quality and observed nitrogenase-specific activities. Biochim Biophys Acta. 1991;1075:109–117. doi: 10.1016/0304-4165(91)90082-r. [DOI] [PubMed] [Google Scholar]
- 44.Anderson GL, Williams J, Hille R. The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J Biol Chem. 1992;267:23674–23682. [PubMed] [Google Scholar]
- 45.Warelow TP, et al. The respiratory arsenite oxidase: Structure and the role of residues surrounding the rieske cluster. PLoS One. 2013;8:e72535. doi: 10.1371/journal.pone.0072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bray RC, Knowles PF, Pick FM, Vänngård T. Electron-spin-resonance evidence for interaction of protons with Mo(V) in reduced forms of xanthine oxidase. Biochem J. 1968;107:601–602. doi: 10.1042/bj1070601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutteridge S, Tanner SJ, Bray RC. The molybdenum centre of native xanthine oxidase. Evidence for proton transfer from substrates to the centre and for existence of an anion-binding site. Biochem J. 1978;175:869–878. doi: 10.1042/bj1750869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.George GN, Bray RC. Reaction of arsenite ions with the molybdenum center of milk xanthine oxidase. Biochemistry. 1983;22:1013–1021. doi: 10.1021/bi00274a003. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H, Selim HM. Kinetics of arsenate adsorption-desorption in soils. Environ Sci Technol. 2005;39:6101–6108. doi: 10.1021/es050334u. [DOI] [PubMed] [Google Scholar]
- 50.Badruzzaman M, Westerhoff P, Knappe DRU. Intraparticle diffusion and adsorption of arsenate onto granular ferric hydroxide (GFH) Water Res. 2004;38:4002–4012. doi: 10.1016/j.watres.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Crofts AR, et al. The mechanism of ubihydroquinone oxidation at the Qo-site of the cytochrome bc1 complex. Biochim Biophys Acta. 2013;1827:1362–1377. doi: 10.1016/j.bbabio.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watson C, et al. Electron transfer through arsenite oxidase: Insights into Rieske interaction with cytochrome c. Biochim Biophys Acta. 2017;1858:865–872. doi: 10.1016/j.bbabio.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahin T, Ngai T, Murcott S, Mondal M. Importance of evaluating phosphate levels in tubewells in high arsenic areas of Asia. In: Jones H, editor. Access to Sanitation and Safe Water—Global Partnerships and Local Actions: Proceedings of the 33rd WEDC International Conference, Accra, Ghana, 7–11 April 2008. Water, Engineering and Development Centre, Loughborough University; Leicestershire, UK: 2008. pp. 467–473. [Google Scholar]
- 54.Long JW, Ray WJ., Jr Kinetics and thermodynamics of the formation of glucose arsenate. Reaction of glucose arsenate with phosphoglucomutase. Biochemistry. 1973;12:3932–3937. doi: 10.1021/bi00744a023. [DOI] [PubMed] [Google Scholar]
- 55.Moore SA, Moennich DM, Gresser MJ. Synthesis and hydrolysis of ADP-arsenate by beef heart submitochondrial particles. J Biol Chem. 1983;258:6266–6271. [PubMed] [Google Scholar]
- 56.Willsky GR, Malamy MH. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol. 1980;144:356–365. doi: 10.1128/jb.144.1.356-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosing J, Slater EC. The value of ΔG° for the hydrolysis of ATP. Biochim Biophys Acta. 1972;267:275–290. doi: 10.1016/0005-2728(72)90116-8. [DOI] [PubMed] [Google Scholar]
- 58.Elias M, et al. The molecular basis of phosphate discrimination in arsenate-rich environments. Nature. 2012;491:134–137. doi: 10.1038/nature11517. [DOI] [PubMed] [Google Scholar]
- 59.Hongshao Z, Stanforth R. Competitive adsorption of phosphate and arsenate on goethite. Environ Sci Technol. 2001;35:4753–4757. doi: 10.1021/es010890y. [DOI] [PubMed] [Google Scholar]
- 60.Oremland RS, Saltikov CW, Stolz JF, Hollibaugh JT. Autotrophic microbial arsenotrophy in arsenic-rich soda lakes. FEMS Microbiol Lett. 2017;364:fnx146. doi: 10.1093/femsle/fnx146. [DOI] [PubMed] [Google Scholar]
- 61.Stewart RC, Hille R, Massey V. Characterization of arsenite-complexed xanthine oxidase at room temperature. Spectral properties and pH-dependent redox behavior of the molybdenum-arsenite center. J Biol Chem. 1984;259:14426–14436. [PubMed] [Google Scholar]
- 62.van Lis R, Nitschke W, Duval S, Schoepp-Cothenet B. Arsenics as bioenergetic substrates. Biochim Biophys Acta. 2013;1827:176–188. doi: 10.1016/j.bbabio.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 63.Ellis PJ, Conrads T, Hille R, Kuhn P. Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 Å and 2.03 Å. Structure. 2001;9:125–132. doi: 10.1016/s0969-2126(01)00566-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.