ABSTRACT
Immunotherapy can revolutionize anti-cancer therapy if specific targets are available. Immunogenic peptides encoded by cancer-specific genes (CSGs) may enable targeted immunotherapy, even of oligo-mutated cancers, which lack neo-antigens generated by protein-coding missense mutations. Here, we describe an algorithm and user-friendly software named RAVEN (Rich Analysis of Variable gene Expressions in Numerous tissues) that automatizes the systematic and fast identification of CSG-encoded peptides highly affine to Major Histocompatibility Complexes (MHC) starting from transcriptome data. We applied RAVEN to a dataset assembled from 2,678 simultaneously normalized gene expression microarrays comprising 50 tumor entities, with a focus on oligo-mutated pediatric cancers, and 71 normal tissue types. RAVEN performed a transcriptome-wide scan in each cancer entity for gender-specific CSGs, and identified several established CSGs, but also many novel candidates potentially suitable for targeting multiple cancer types. The specific expression of the most promising CSGs was validated in cancer cell lines and in a comprehensive tissue-microarray. Subsequently, RAVEN identified likely immunogenic CSG-encoded peptides by predicting their affinity to MHCs and excluded sequence identity to abundantly expressed proteins by interrogating the UniProt protein-database. The predicted affinity of selected peptides was validated in T2-cell peptide-binding assays in which many showed binding-kinetics like a very immunogenic influenza control peptide. Collectively, we provide an exquisitely curated catalogue of cancer-specific and highly MHC-affine peptides across 50 cancer types, and a freely available software (https://github.com/JSGerke/RAVENsoftware) to easily apply our algorithm to any gene expression dataset. We anticipate that our peptide libraries and software constitute a rich resource to advance anti-cancer immunotherapy.
KEYWORDS: Immunotherapy, bioinformatics, microarray, cancer-specific genes
Introduction
Immunotherapy is currently transforming clinical oncology and holds promise for cure even for patients with metastatic disease.1 The success of many immunotherapeutic approaches, e.g. adoptive T cell therapy, largely depends on the availability of specific immunogenic target structures presented via Major Histocompatibility Complexes (MHCs) on the surface of cancer cells, but not on that of normal tissues.2 Genetically instable and hyper-mutated cancer entities such as malignant melanoma and lung carcinoma offer such highly specific target structures through missense mutations in the protein coding genome that generate ‘neo-antigens’.3
However, many cancer types such as pediatric cancers are characterized by a remarkably stable and oligo-mutated genome.4 In addition, the few recurrent somatic mutations found in pediatric cancers are hardly immunogenic.5 Thus, specific immunotherapy of oligo-mutated cancers is challenging, but may be enabled by the expression of non-mutated cancer-specific genes (CSGs).2
Many CSGs are only expressed during early embryogenesis or in immune-privileged germline tissues such as testis.6,7 This restricted expression pattern increases the likelihood of circulating lymphocytes directed against immunogenic peptides encoded by these CSGs,7 which can be exploited clinically. In neuroblastoma and Ewing sarcoma, which are aggressive and oligo-mutated pediatric cancers,8,9 adoptive T cell therapy targeting CSGs has been successfully applied in humanized mouse models10–13 and patients.14 Screening for additional CSGs could be enabled by comprehensive and already available transcriptome datasets of cancer and normal tissues,15 However, due to the lack of specific algorithms and user-friendly tools, the identification of CSGs and derivative peptides with high affinity to MHCs continues to be laborious and slow.16
To accelerate this process and to identify CSGs suitable for targeting various oligo-mutated cancer entities, we developed an algorithm and provide an intuitive software termed RAVEN (Rich Analysis of Variable gene Expressions in Numerous tissues), which automatizes the systematic and fast identification of cancer-specific peptides with high affinity to MHCs starting from gene expression data. By applying RAVEN to a dataset of 2,678 gene expression microarrays comprising 50 tumor entities and 71 normal tissue types, we identified a library of peptides suitable for targeting multiple cancers. Our datasets and software represent a rich resource for the development of immunotherapies.
Results
Dataset assembly, workflow, and basic concepts of RAVEN
In order to automatize the systematic and fast identification of CSGs as well as the prediction of corresponding highly affine peptides for any given MHC, we developed a user-friendly software named RAVEN (Rich Analysis of Variable gene Expressions in Numerous tissues). An overview on the workflow conducted by RAVEN is given in Figure 1. The software, a detailed user manual enabling researchers to easily use the software and our gene expression datasets are freely available under https://github.com/JSGerke/RAVENsoftware.
Figure 1.

Schematic illustration of the assembly, quality-check, and normalization of gene expression data as well as tasks executed by RAVEN.
Transcriptome-wide detection of CSGs overexpressed in multiple cancer entities with RAVEN
Previous studies have shown that many established CSGs are only expressed in subsets of specific cancer entities, which is often referred to as ‘outlier’ expression.17,18 Indeed, many CSGs are either virtually not expressed in somatic normal tissues or exclusively expressed in specific lineages such as embryonal and germline tissues.6,7 This outlier expression discriminates cancer cells from normal somatic cells and may offer a therapeutic window for preferentially targeting cancer cells, e.g. by adoptive T cell therapy.2 Also, it may increase the likelihood that lymphocytes responsive to the proteins encoded by CSGs are preserved in the mature lymphocyte repertoire,7 because they are not counter-selected during lymphocyte development. However, an outlier expression profile implies that conventional statistical tests, which either simply aim at identifying generally upregulated CSGs across many cancer samples (e.g. student’s t-test) or ignore the strength of overexpression in a small subset of patients (e.g. rank-based nonparametric tests), would fail to detect such clinically relevant CSGs.
Therefore, we developed a scoring algorithm to scan transcriptome-wide for CSGs by assigning an ‘outlier score’ (OS) to each gene for high expression in a given cancer entity, which is penalized by a ‘penalty score’ (PS) if high expression in any normal tissue type is present.
Both scores are calculated for each gene separately as the mean expression level of the 95th and 75th percentile. Then, we calculated an overall score for each gene named ‘CSG-score’, which is built by subtracting the gene-specific PS from the OS. This function highlights all genes overexpressed in only a subset of cancer samples, while avoiding the misrepresentation caused by extremely high outlier expression signals in single samples.
In addition, our algorithm takes into account gender-specific normal tissue types such as uterus and prostate. Specifically, our algorithm calculated gender-specific CSG-scores for each gene excluding normal tissues of sexual organs specific for the other gender (see Materials and Methods).
To analyze the expression profiles of human genes in normal and cancer tissues we compiled 83 Affymetrix HG-U133-Plus2.0 microarray datasets for 71 normal tissues and 50 cancer types with a focus on oligo-mutated pediatric cancers and sarcomas, totaling to 2,678 high-quality and simultaneously normalized samples (Supplementary Table 1). In prospect of a future exploitation of our CSGs as clinical immunotargets, we included graft versus host disease (GvHD)-sensitive normal tissue types such as retina and colonic mucosa as well as normal B and T cells to obviate fratricide effects, which can compromise adoptive T cell therapies.19,20
Applying our scoring algorithm to this well-curated gene expression dataset, RAVEN identified 806 non-redundant CSGs (defined by a CSG-score above the 99.9th percentile of all scores across 50 cancer entities) (Figure 2, Supplementary Table 5). Among them we found not only many established CSGs such as LIPI for Ewing sarcoma,21 PRAME for neuroblastoma22,23 and members of the MAGE-family for germinoma,24 neuroblastoma,25 synovial sarcoma,26 multiple myeloma,27 diffuse large B cell lymphoma (DLBCL),28 and osteosarcoma,29 but also many novel candidates of which some appear to be suitable for targeting multiple cancer entities (Figure 2, Supplementary Table 5, Supplementary Figure 1).
Figure 2.
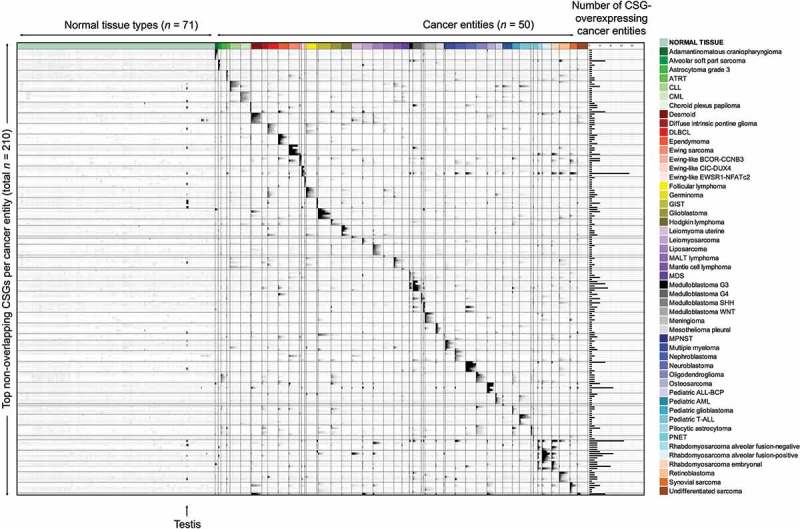
Overexpressed CSGs in multiple cancer entities identified with RAVEN. Relative gene expression intensities of the top-5 CSGs for each cancer entity (excluding overlapping CSGs with other tumor entities) indicated in greyscale with black color representing high and white color low expression. Each line represents an individual CSG (for a complete list see Supplementary Table 5); each column represents one primary tumor/leukemia/normal tissue sample. The bar graph on the right displays the number of different cancer entities in which the corresponding CSG reached a CSG-score above the 99.9th percentile of all CSG-scores. ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ATRT, atypical teratoid/rhabdoid tumor; CLL, chronic lymphatic leukemia; CML, chronic myeloid leukemia; DLBCL, diffuse large B cell lymphoma; GIST, gastrointestinal stromal tumor; MALT, mucosa associated lymphatic tissue; MPNST, malignant peripheral nerve sheath tumor; PNET, primitive neuroectodermal tumor.
The specific expression of nine selected CSGs was confirmed by qRT-PCR in a panel of cancer cell lines from six different tumor entities. As shown in Figure 3A, there was a high concordance of calculated CSG-scores and expression intensities measured by microarrays in primary tumors with relative mRNA expression levels measured by qRT-PCR in corresponding cancer-derived cell lines.
Figure 3.
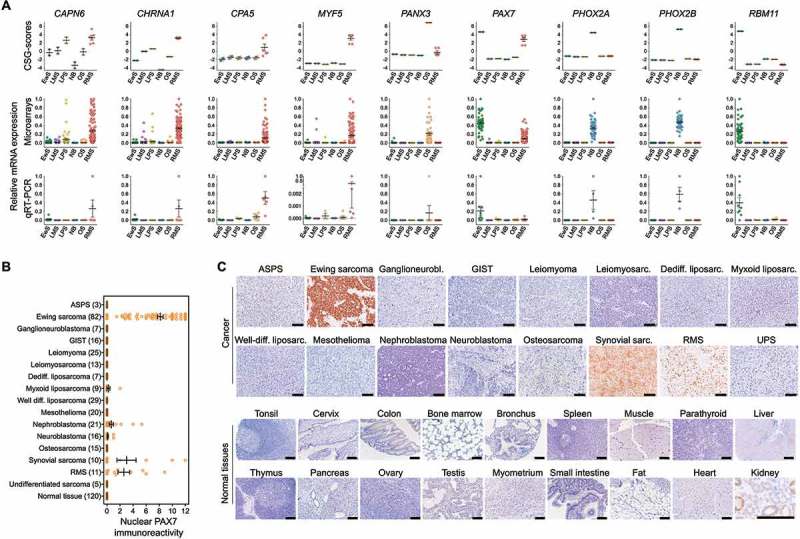
Validation of the expression pattern of selected CSGs by qRT-PCR and IHC. A) Upper and middle panel: CSG-scores and corresponding expression intensities (natural scale) of selected genes in primary Ewing sarcoma (EwS, n = 50), neuroblastoma (NB; n = 49), rhabdomyosarcoma (RMS; n = 101), liposarcoma (LPS; n = 50), leiomyosarcoma (LMS, n = 50) and osteosarcoma tumors (OS, n = 40). Lower panel: Relative expression levels of the same genes as determined by qRT-PCR in EwS (n = 9), NB (n = 4), RMS (n = 5) and LPS (n = 3), LMS (n = 3) and OS (n = 6) cell lines. B) Analysis of nuclear PAX7 immunoreactivity by IHC in indicated primary tumors and normal tissues. ASPS, alveolar soft part sarcoma; GIST, gastrointestinal stromal tumor. Numbers of analyzed samples are given in parentheses. C) Representative images of nuclear PAX7 IHC staining in cancer and selected normal tissues. Scale bar = 300 µm. UPS, undifferentiated pleomorphic sarcoma. Note: In renal proximal tubules non-specific cytoplasmic staining for PAX7 was observed, while all nuclei showed no PAX7 immunoreactivity. This non-specific cytoplasmic stain has been previously described for the employed anti-PAX7-antibody.56
In particular, the transcription factor PAX7 (paired box 7) showed a very high CSG-score (>4) in multiple cancer entities including oligo-mutated Ewing sarcoma. Therefore, we validated its strong overexpression on protein level in a subset of these cancer entities by immunohistochemistry in a comprehensive tissue microarray (TMA, n = 409 samples) also containing somatic and germline normal tissue types. As shown in Figure 3B,C, PAX7 was exclusively expressed in cell nuclei of cancer entities with high CSG-scores, while being virtually not expressed in normal tissues. Collectively, these data demonstrate that RAVEN can reliably identify CSGs with specific overexpression in multiple cancers as compared to normal tissues.
Prediction of non-redundant CSG-encoded peptides with high MHC-affinity by RAVEN
To identify peptides encoded by CSGs suitable for a targeted immunotherapy, we implemented the artificial neural network (ANN) algorithm30,31 provided by the immune epitope database IEDB 3.0.32 RAVEN can apply this ANN algorithm to predict peptide-affinities for different peptide lengths and the most common human and murine MHC-subtypes.
In our list of 806 CSGs, RAVEN predicted potential highly affine peptides for 9-mers, which usually show optimal binding to most MHC class I molecules,30,33 and for HLA-A02:01, which is the most common MHC-I in Caucasians34 with an allele frequency of 0.2755.35 RAVEN automatically crosschecked these peptides by a text search algorithm with ApacheLucene36,37 against the human reference-proteome (UniProt release 2015_06) to exclude sequence identity with non-specifically expressed proteins. In total, RAVEN predicted 7247 9-mer peptides with high MHC-I-affinity (defined as a dissociation constant Kd ≤ 150 nM) of which 6589 had no sequence identity with any other protein (Supplementary Table 6).
Predicted CSG-encoded peptides exhibit strong affinity to MHCs
We next sought to confirm the predicted affinity of peptides to human HLA-A02:01 proposed by RAVEN. Therefore, we selected among the unique 6589 peptides 79, which covered all analyzed tumor entities except of Pediatric ALL-BCP and AML and which had high to very high CSG-scores. For these 79 peptides, we designed a customized solid-phase synthesized peptide-library and assessed whether they can stabilize MHC-I on the surface of TAP2-deficient cells in T2-binding assays. As shown in Figure 4A, 38 of 79 tested peptides (48.1%) achieved at least 50% of the MHC-stabilizing effect of a highly immunogenic influenza control peptide (GILGFVFTL, Supplementary Table 6) at a saturation dose of 100 µM. For these CSG-peptides, we repeated the T2-assays with six different peptide concentrations (0.1 to 100 µM). Strikingly, some of them, including the one encoded by PAX7, showed MHC-stabilization kinetics similar to the influenza peptide (Figure 4B). Taken together, these experiments demonstrated that RAVEN can identify highly affine CSG-encoded peptides suitable for targeting multiple cancer types by leveraging publicly available gene expression data.
Figure 4.
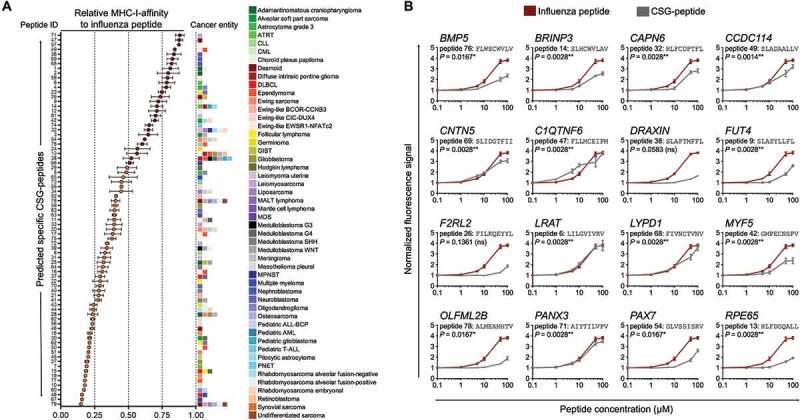
Validation of MHC-affinity of CSG-encoded peptides in a T2-binding assay. A) Relative MHC-I-affinity of 79 selected peptides at 100 µM in T2-binding assays as compared to a highly affine influenza peptide (peptide sequences are given in Supplementary Table 6). The colored boxes at the right side of the graph represent the number and type of cancer entities in which the corresponding CSG encoding the indicated peptide is overexpressed. Peptides with an MHC-affinity of ≥ 50% of the influenza peptide are highlighted in red color. Data are presented as mean and SEM of n ≥ 3 experiments. ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ATRT, atypical teratoid/rhabdoid tumor; CLL, chronic lymphatic leukemia; CML, chronic myeloid leukemia; DLBCL, diffuse large B cell lymphoma; GIST, gastrointestinal stromal tumor; MALT, mucosa associated lymphatic tissue; MPNST, malignant peripheral nerve sheath tumor; PNET, primitive neuroectodermal tumor. B) Normalized fluorescence signals of 16 selected peptides with high MHC-affinity as compared to that of a highly affine influenza peptide in T2-binding assays. Data are presented as mean and SEM of n ≥ 3 experiments. P values of a Spearman’s rank-order correlation are reported.
Discussion
High-throughput gene expression analyses of cancers and normal tissues generated comprehensive and freely available transcriptome datasets.15 However, identification of CSGs and derivative peptides with high affinity to MHCs continued to be laborious and slow.16
Here, we reported on the development and application of a mathematical scheme for transcriptome-wide detection of CSGs and their corresponding highly MHC-affine peptides as immunologic and clinical targets, and provide a use-friendly software (RAVEN) along with a detailed user manual, which automatizes this process. Applying RAVEN to a large gene expression dataset comprising multiple and often oligo-mutated pediatric cancer types as well as a broad spectrum of normal tissues revealed many CSGs with diagnostic and therapeutic potential. Moreover, we provide an analogous dataset including 19 of the most common carcinoma entities (1,462 samples; Supplementary Table 1, https://github.com/JSGerke/RAVENsoftware/relaeses), which can be used for identification CSG-encoded peptides in these tumor types. The CSG-scores for this ‘carcinoma’ dataset are given in Supplementary Table 7.
In both the pediatric and carcinoma datasets, we observed significant enrichments (P < 0.0001, two-tailed Chi2-test with Yates’ correction) of established cancer-testis antigens (Supplementary Figure 1, CTDatabase, www.cta.lncc.br38), but also identified many novel candidates including the pioneer transcription factor PAX7.39 PAX7 encodes a paired box transcription factor required for embryonal neural development40 and renewal of skeletal muscle stem cells.41 Translocations involving PAX7 and FKHR are found in the majority of alveolar rhabdomyosarcomas (ARMS), indicating a role of PAX7 in the pathogenesis of myogenic tumors.42 Using RAVEN, we identified PAX7 as a strong CSG in multiple oligo-mutated cancer entities such as Ewing sarcoma, Ewing-like sarcomas with a BCOR-CCNB3-translocation and embryonal as well as alveolar fusion-negative rhabdomyosarcoma. Its exclusive expression in these cancer entities was confirmed on protein level by IHC. Strikingly, PAX7 encodes a 9-mer peptide (GLVSSISRV) with very high affinity for the most frequent MHC-I subtype in Caucasians (HLA-A02:01),34 rendering PAX7 as an attractive target for immunotherapy for multiple oligo-mutated cancers. As we focused here on the validation of peptide affinities for HLA-A02:01, future experimental validation for predicted peptides for other HLAs is required.
The parameters of the analysis applied in RAVEN have been optimized to discover CSGs, which are virtually not expressed in most somatic tissues. Although some identified CSGs did not encode peptides suitable for immunotargets, a subset of them could constitute interesting targets for conventional pharmacotherapy. In fact, the CSGs FGFR4, CDK4, and several MMPs, which are specifically overexpressed in rhabdomyosarcoma (FGFR4), liposarcoma (CDK4), and desmoid tumors, leiomyoma, osteosarcoma and adamantinomatous craniopharyngioma (MMPs) (Supplementary Table 5), respectively, could be targeted by specific inhibitors currently in clinical trials.43–45
Besides their potential utility as (immune)-therapeutic targets, some CSGs may harbor the potential to serve as diagnostic markers: While CSGs expressed in multiple tumor entities could be utilized for cancer-screening, CSGs exclusively expressed in certain cancer types can be used to identify and differentiate specific tumor entities. This could be important for determining treatment options, which is often difficult in cancers of unknown primary.
As RAVEN can also be applied to datasets only containing cancer samples, RAVEN can easily identify potential diagnostic markers among several cancers in parallel. In principle, our work-flow embedded in RAVEN provides an unbiased approach for transcriptome-wide detection of CSGs, which can be adapted to many specific applications, such as the identification of autoantibody signatures, biomarkers, tumor vaccine targets, or membrane antigen targets. Its performance could be further enhanced by combining it with other datasets, on cancer plasma or membrane proteomics. Since our algorithm provides a quantitative and gender-specific value for each gene in each tumor entity (Supplementary Table 5), the preferential expression of each CSG in different cancers is apparent at a glance. With more and more deep transcriptome sequencing data available and the advent of digital gene expression technology, we expect that RAVEN will be a highly beneficial tool to maximize the identification of CSGs and, hence, new diagnostic markers and therapeutic targets based on these data.
Materials and methods
Microarray data
Publicly available gene expression data generated with Affymetrix HG-U133Plus2.0 microarrays for 3,078 samples comprising 50 tumor entities and 71 normal tissue were retrieved from the Gene Expression Omnibus (GEO) or the Array Express database at the European Bioinformatics Institute (EBI). Accession codes are reported in Supplementary Table 1. Microarray quality checks were performed by analyzing the Relative Log Expression (RLE) and Normalized Unscaled Standard Error (NUSE) scores with the Bioconductor packages affyPLM46 and hgu133plus2hsentrezgcdf47 in the statistical language R.48 The cut-offs for defining high quality were set as (1st quartile – [1.5 × interquartile range]) and (3rd quartile + [1.5 × interquartile range]).
All microarrays were pre-processed (normalized) simultaneously in R with the Robust Multi-chip Average (RMA) algorithm49 including background adjustment, quantile normalization and summarization using custom brainarray Chip Description Files (CDF; ENTREZG, v21) yielding one optimized probe-set per gene.47
Identification of CSG-scores from normalized expression intensities
To identify CSGs in any given gene expression dataset, we calculated the outlier expression of a gene x in a specific cancer c by considering the adjusted upper quartile mean of its expression signals, as such approach avoids bias through extreme outliers in a tiny subset of samples (above 95th quantile).18 The adjusted upper quartile mean, named ‘Outlier Score’ (OS), of gene x in cancer type c is given as
Next, a ‘Penalty Score’ (PS) for gene x was calculated on the basis of its adjusted upper quartile mean among different types of normal human tissues n as
The CSG-score of a gene x in a given cancer type c was then calculated as
Previously reported algorithms included weighting scores for each normal tissue type based on their possible degree of estimated ‘immuno-privilege’ or even excluded highly immune-privileged organs such as testis from calculation of a PS.18,50
In contrast, we considered each normal tissue type including testis as equally relevant for calculating the PS of a given gene, as otherwise our list of CSGs would be exceedingly enriched in established cancer-testis antigens. However, as gender-specific normal tissue types such as uterus/ovary or prostate/testis, respectively, are irrelevant to nominate CSGs for the respective other gender, we calculated gender-specific CSG-scores omitting gender-specific tissue types for calculation of the PS of a given gene for the respective other gender (Supplementary Table 1). A meaningful CSG-score was determined statistically as being equal or above the 99.9th percentile of all CSG-scores calculated across all cancer entities. Using this cut-off, the CSG-scores for CSGs potentially suitable for immunotherapeutic targets in a given cancer entity were usually greater than 2. CSG-scores greater than 3 were empirically considered as high and those greater than 4 as very high.
Development of RAVEN (rich analysis of variable gene expressions in numerous tissues)
We developed an application named RAVEN that incorporates several statistical methods to easily identify putative highly immunogenic peptides encoded by CSGs from any gene expression dataset including RNA sequencing data.
RAVEN and a detailed user manual as well as associated datasets can be downloaded free of charge and for academic use only under https://github.com/JSGerke/RAVENsoftware.
The graphical interface of RAVEN is simple and designed for scientists without bioinformatics background. The current program version developed with Java (for Windows, Linux and Mac) requires at least a Java 8 runtime environment.
RAVEN can interrogate gene expression datasets and compare expression levels of different genes in the same tissue or of the same gene in different tissues applying our algorithm as explained above. The statistical summary of such comparisons can be obtained in spreadsheet format and visualized by Java library JFreeChart. Additionally, the application enables users to retrieve either gene- or tissue-specific subsets of the interrogated gene expression dataset, which can then be further analyzed in RAVEN or other commonly used software such as Microsoft Excel or GraphPad Prism.
In addition, RAVEN includes a pipeline combining several bioinformatic services to offer a quick and simple way to obtain all peptide sequences of a pre-specified length (encoded by identified CSGs) and their affinity to different HLA-alleles. Furthermore, RAVEN nominates all MHCs that are predicted to present the identified peptides. To access the UniProtKB51 database via RAVEN, we used Protein API.52 RAVEN sends a query to match gene IDs with their corresponding protein IDs of different databases such as UniProt and NCBI as well as the proteins sequence. The implemented peptide-matching pipeline accesses the MHC-I binding prediction tool provided by the Immune Epitope Database (IEDB) Analysis Resource32 via a RESTful interface (IEDB-API). T Cell Epitope Prediction identifies peptides binding to MHC class I of a certain protein sequence. Therefore, RAVEN uses artificial neural networks (ANN) and a prediction algorithm developed by NetMHC.30,31 The peptide search service36 of UniProt is queried via a RESTful web service which API is provided and integrated by Protein Information Resource (PIR) using ApacheLucene for peptide text searches.36,37 In RAVEN, this approach is available for the most common alleles in human and mouse. In contrast to other methods provided by RAVEN, this pipeline is independent from the analyzed gene expression dataset but requires an internet connection.
Human cell lines and cell culture conditions
Cells were grown at 37°C in humidified 5% CO2 atmosphere in RPMI 1640 medium (Biochrom, Berlin, Germany) supplemented with 10% FCS (Biochrom) and 100 U/ml penicillin and 100 μg/ml streptomycin (Biochrom). TAP-deficient HLA*A02:01+ T2 cell line (somatic cell hybrid) was obtained from P. Cresswell (Yale University School of Medicine, New Haven, CT, USA). T2 cells were maintained in RPMI 1640 medium additionally supplemented with 1 mM sodium pyruvate and non-essential amino acids (both Biochrom). Cell line purity was confirmed by short tandem repeat profiling (latest profiling 15th December 2015) and cells were routinely examined by PCR for the absence of mycoplasma. A list of the used cell lines is provided in Supplementary Table 2.
RNA extraction, reverse transcription and qRT-PCR
RNA was extracted with the Nucleospin RNA kit (Macherey-Nagel, Düren, Germany) containing a 15 min on-column DNA digestion step to degrade genomic DNA. RNA was reverse-transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCRs were performed using SYBR Select Master Mix (Applied Biosystems). Oligonucleotides were purchased from MWG Eurofins Genomics (Ebersberg, Germany). Primer sequences are listed in Supplementary Table 3. Reactions were run in 10–20 µl final volume on a CFX Connect instrument and analyzed using the CFX Manager 3.1 (both Bio-Rad). Gene expression levels of specific genes were normalized to that of the housekeeping gene RPLP0.53
Human samples and ethics approval
Human tissue samples were collected at the Institute of Pathology of the LMU Munich (Germany) with approval of the corresponding institutional review boards. The ethics committee of the University Hospital of the LMU Munich approved the study (approval no. 307–16 UE).
Immunohistochemistry (IHC) and evaluation of immunoreactivity
IHC analyses were performed on formalin-fixed, paraffin-embedded (FFPE) tissue samples. Paraffin blocks from several institutions were collected at the Institute of Pathology of the LMU Munich. From all blocks, we harvested 3 cores per sample with a core-diameter of 1 mm to assemble a tissue microarray (TMA). A list of the included tumor types and normal tissues is given in Supplementary Table 4. Of each TMA block 4 µm sections were cut and stained with an iView DAB detection kit (Ventana Medical System, Tucson, AZ) according to the company’s protocol. Subsequent antigen retrieval was carried out using TRIS-buffer and blockage of endogenous peroxidase with 7.5% aqueous H2O2. TMA sections were stained at a dilution of 1:180 for 60 min at room temperature with a monoclonal antibody against human PAX7 raised in mouse,40 which was purchased from the Developmental Studies Hybridoma Bank (Cat.No. PAX7-c; Iowa City, IA). Then slides were incubated with a secondary biotinylated anti-mouse IgG antibody (ImmPress Reagent Kit, Peroxidase-conjugated) followed by target detection using ABC-kit chromogen for 10 min (Dako, K3461).
At least three high-power fields (40x) of each core for every sample were assessed. Semi-quantitative evaluation of immunoreactivity was carried out by two independent physicians trained in histopathology. The percentage of cells with marker expression was scored and classified in five grades (grade 0 = 0–19%, grade 1 = 20–39%, grade 2 = 40–59%, grade 3 = 60–79% and grade 4 = 80–100%). In addition, the intensity of marker immunoreactivity was determined as grade 0 = none, grade 1 = faint, grade 2 = moderate and grade 3 = strong. For calculation of overall immunoreactivity for the given protein, we multiplied both grades in analogy to UICC guidelines for hormone receptor scoring in human breast cancers.54
Peptide binding assay using TAP deficient T2 cells
All peptides were solid-state synthesized with the highly-parallelized LIPS® technology (Elephants & Peptides, Potsdam, Germany). As a positive control, we used an established highly affine influenza matrix protein epitope (M158-66; sequence GILGFVFTL).55 T2 cells were washed twice with PBS and seeded in round-bottom 96-well plates (TPP, Trasadingen, Switzerland) at a concentration of 2 × 105 cells/well in a final volume of 200 µl. Cells were pulsed with increasing amounts of peptide to measure a concentration dependency of MHC-I binding. Unpulsed cells were used as a negative control. After incubation over-night, cells were washed twice with FACS-buffer consisting of PBS with 2% FCS and stained for HLA-A2 using a FITC mouse anti-human HLA-A2 antibody (BD Pharmingen™, Clone BB7) for 30 min at 4°C. For isotype control a BB515 mouse IgG2Ak antibody (BD Horizon™, Clone G155-178) was used. Then, cells were washed twice in FACS-buffer before being resuspended in PBS and analyzed using a FACSCalibur flow cytometer (Becton Dickinson). To determine the relative peptide binding, the fluorescence intensity of a peptide at a defined concentration was divided by the intensity of unpulsed T2 cells.
Funding Statement
The laboratory of TGPG is supported by grants from the ‘Verein zur Förderung von Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München (WiFoMed)’, the Daimler and Benz Foundation in cooperation with the Reinhard Frank Foundation, by LMU Munich’s Institutional Strategy LMUexcellent within the framework of the German Excellence Initiative, the ‘Mehr LEBEN für krebskranke Kinder – Bettina-Bräu-Stiftung’, the Walter Schulz Foundation, the Kind-Philipp Foundation, the Friedrich-Baur Foundation, the Fritz Thyssen Foundation (FTF-2015-01046), the Dr. Leopold and Carmen Ellinger Foundation, the Wilhelm Sander-Foundation (2016.167.1), the Matthias-Lackas Foundation, the Barbara und Hubertus Trettner Foundation, the Deutsche Forschungsgemeinschaft (DFG 391665916) and by the German Cancer Aid (DKH-111886 and DKH-70112257). Deutsche Krebshilfe [70112257].
Acknowledgments
We thank Mrs. Andrea Sendelhofert, Mrs. Anja Heier and Ms. Mona Melz for excellent technical assistance.
Author contributions
MCB, JSG and TGPG conceived the study, performed bioinformatic and wet-lab analyses, and drafted and wrote the paper. MFO helped with bioinformatic analyses and assembly of gene expression datasets. ME, KS, and DHB provided immunological guidance and helped in experiments. MMK, NA, ANA, FCR, ÖÖ, Ta.K, SS, TH, HS and DB contributed to the tissue microarray. JSG programmed and developed RAVEN. AK and UT performed T2-cell assays. FB and TF provided immunological guidance. MD, RAR, JM, AM, SO, MMLK, GS helped in wet-lab analyses. JL helped in IHC experiments. Th.K provided laboratory infrastructure, tissue samples and histological guidance. TGPG supervised the study and performed histological analyses. All authors read and approved the final manuscript.
Supplemental Material
Supplemental data for this article can be accessed here.
References
- 1.Mellman I, Coukos G, Dranoff G.. 2011. Cancer immunotherapy comes of age. Nature 480(7378):480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. 2014. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 14(2):135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher TN, Schreiber RD. 2015. Neoantigens in cancer immunotherapy. Science 348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orentas RJ, Lee DW, Mackall C. 2012. Immunotherapy targets in pediatric cancer. Front Oncol 2. doi: 10.3389/fonc.2012.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monk M, Holding C. 2001. Human embryonic genes re-expressed in cancer cells. Oncogene 20(56):8085–8091. doi: 10.1038/sj.onc.1205088. [DOI] [PubMed] [Google Scholar]
- 7.Simpson AJG, Caballero OL, Jungbluth A, Chen Y-T, Old LJ. 2005. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 8.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, et al. 2013. The genetic landscape of high-risk neuroblastoma. Nat Genet 45(3):279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, Zhang Z, Lapouble E, Grossetête-Lalami S, Rusch M, et al. 2014. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 4(11):1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaeschke F, Thiel U, Kirschner A, Thiede M, Rubio RA, Schirmer D, Kirchner T, Richter GHS, Mall S, Klar R, et al. 2016. Human HLA-A*02:01/CHM1+ allo-restricted T cell receptor transgenic CD8+ T cells specifically inhibit Ewing sarcoma growth in vitro and in vivo. Oncotarget 7(28):43267–43280. doi: 10.18632/oncotarget.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirschner A, Thiede M, Tgp G, Rubio RA, Richter GHS, Kirchner T, Busch DH, Burdach S, Thiel U. 2017. Pappalysin-1 T cell receptor transgenic allo-restricted t cells kill ewing sarcoma in vitro and in vivo. OncoImmunology 6(ja):e1273301. doi: 10.1080/2162402X.2016.1273301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schirmer D, Grünewald TGP, Klar R, Schmidt O, Wohlleber D, Rubío RA, Uckert W, Thiel U, Bohne F, Busch DH, et al. 2016. Transgenic antigen-specific, HLA-A*02:01-allo-restricted cytotoxic T cells recognize tumor-associated target antigen STEAP1 with high specificity. Oncoimmunology 5(6):e1175795. doi: 10.1080/2162402X.2016.1175795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh N, Kulikovskaya I, Barrett DM, Binder-Scholl G, Jakobsen B, Martinez D, Pawel B, June CH, Kalos MD, Grupp SA. 2016. T cells targeting NY-ESO-1 demonstrate efficacy against disseminated neuroblastoma. Oncoimmunology 5(1):e1040216. doi: 10.1080/2162402X.2015.1040216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiel U, Schober SJ, Einspieler I, Kirschner A, Thiede M, Schirmer D, Gall K, Blaeschke F, Schmidt O, Jabar S, et al. 2017. Ewing sarcoma partial regression without GvHD by chondromodulin-I/HLA-A*02:01-specific allorestricted T cell receptor transgenic T cells. OncoImmunology 6(5):e1312239. doi: 10.1080/2162402X.2017.1312239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rung J, Brazma A. 2013. Reuse of public genome-wide gene expression data. Nat Rev Genet 14(2):89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]
- 16.Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. 2015. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest 125(9):3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu B. 2007. Cancer outlier differential gene expression detection. Biostatistics 8(3):566–575. doi: 10.1093/biostatistics/kxl029. [DOI] [PubMed] [Google Scholar]
- 18.Xu Q-W, Zhao W, Wang Y, Sartor MA, Han D-M, Deng J, Ponnala R, Yang J-Y, Zhang Q-Y, Liao G-Q, et al. 2012. An integrated genome-wide approach to discover tumor-specific antigens as potential immunologic and clinical targets in cancer. Cancer Res 72(24):6351–6361. doi: 10.1158/0008-5472.CAN-12-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirschner A, Thiede M, Blaeschke F, Richter GHS, Gerke JS, Baldauf MC, Grünewald TGP, Busch DH, Burdach S, Thiel U. 2016. Lysosome-associated membrane glycoprotein 1 predicts fratricide amongst T cell receptor transgenic CD8+ T cells directed against tumor-associated antigens. Oncotarget 7(35):56584–56597. doi: 10.18632/oncotarget.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leisegang M, Wilde S, Spranger S, Milosevic S, Frankenberger B, Uckert W, Schendel DJ. 2010. MHC-restricted fratricide of human lymphocytes expressing survivin-specific transgenic T cell receptors. J Clin Invest 120(11):3869–3877. doi: 10.1172/JCI43437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foell JL, Hesse M, Volkmer I, Schmiedel BJ, Neumann I, Staege MS. 2008. Membrane-associated phospholipase A1 beta (LIPI) Is an Ewing tumour-associated cancer/testis antigen. Pediatr Blood Cancer 51(2):228–234. doi: 10.1002/pbc.21602. [DOI] [PubMed] [Google Scholar]
- 22.Oberthuer A, Hero B, Spitz R, Berthold F, Fischer M. 2004. The tumor-associated antigen PRAME is universally expressed in high-stage neuroblastoma and associated with poor outcome. Clin Cancer Res Off J Am Assoc Cancer Res 10(13):4307–4313. doi: 10.1158/1078-0432.CCR-03-0813. [DOI] [PubMed] [Google Scholar]
- 23.Spel L, Boelens -J-J, van der Steen DM, Blokland NJG, van Noesel MM, Molenaar JJ, Heemskerk MHM, Boes M, Nierkens S. 2015. Natural killer cells facilitate PRAME-specific T-cell reactivity against neuroblastoma. Oncotarget 6(34):35770–35781. doi: 10.18632/oncotarget.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara I, Hara S, Miyake H, Yamanaka K, Nagai H, Gohji K, Arakawa S, Kamidono S. 1999. Expression of MAGE genes in testicular germ cell tumors. Urology 53(4):843–847. doi: 10.1016/S0090-4295(98)00618-9. [DOI] [PubMed] [Google Scholar]
- 25.Söling A, Schurr P, Berthold F. 1999. Expression and clinical relevance of NY-ESO-1, MAGE-1 and MAGE-3 in neuroblastoma. Anticancer Res 19(3B):2205–2209. [PubMed] [Google Scholar]
- 26.Iura K, Maekawa A, Kohashi K, Ishii T, Bekki H, Otsuka H, Yamada Y, Yamamoto H, Harimaya K, Iwamoto Y, et al. 2017. Cancer-testis antigen expression in synovial sarcoma: NY-ESO-1, PRAME, MAGEA4, and MAGEA1. Hum Pathol 61:130–139. doi: 10.1016/j.humpath.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Condomines M, Hose D, Raynaud P, Hundemer M, De Vos J, Baudard M, Moehler T, Pantesco V, Moos M, Schved J-F, et al. 2007. Cancer/testis genes in multiple myeloma: expression patterns and prognosis value determined by microarray analysis. J Immunol Baltim Md 1950 178(5):3307–3315. [DOI] [PubMed] [Google Scholar]
- 28.Hudolin T, Kastelan Z, Ilic I, Levarda-Hudolin K, Basic-Jukic N, Rieken M, Spagnoli GC, Juretic A, Mengus C. 2013. Immunohistochemical analysis of the expression of MAGE-A and NY-ESO-1 cancer/testis antigens in diffuse large B-cell testicular lymphoma. J Transl Med 11:123. doi: 10.1186/1479-5876-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudo T, Kuramoto T, Komiya S, Inoue A, Itoh K. 1997. Expression of MAGE genes in osteosarcoma. J Orthop Res Off Publ Orthop Res Soc 15(1):128–132. doi: 10.1002/jor.1100150119. [DOI] [PubMed] [Google Scholar]
- 30.Andreatta M, Nielsen M. 2016. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinforma Oxf Engl 32(4):511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, Brunak S, Lund O. 2003. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci Publ Protein Soc 12(5):1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, et al. 2015. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43(Database issue):D405–412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichmann M, de Ru A, van Veelen PA, Peakman M, Kronenberg-Versteeg D. 2014. Identification and characterisation of peptide binding motifs of six autoimmune disease-associated human leukocyte antigen-class I molecules including HLA-B*39:06. Tissue Antigens 84(4):378–388. doi: 10.1111/tan.12413. [DOI] [PubMed] [Google Scholar]
- 34.González-Galarza FF, Takeshita LYC, Santos EJM, Kempson F, Maia MHT, Silva ALS, Da Silva ALT, Ghattaoraya GS, Alfirevic A, Jones AR, et al. 2015. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res 43(D1):D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gragert L, Madbouly A, Freeman J, Maiers M. 2013. Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum Immunol 74(10):1313–1320. doi: 10.1016/j.humimm.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Li Z, Huang H, Suzek BE, Wu CH, UniProt Consortium . 2013. A fast peptide match service for UniProt knowledgebase. Bioinforma Oxf Engl 29(21):2808–2809. doi: 10.1093/bioinformatics/btt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CH, Yeh L-SL, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, et al. 2003. The protein information resource. Nucleic Acids Res 31(1):345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almeida LG, Sakabe NJ, deOliveira AR, Silva MCC, Mundstein AS, Cohen T, Chen Y-T, Chua R, Gurung S, Gnjatic S, et al. 2009. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res 37(Databaseissue):D816–819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayran A, Khetchoumian K, Hariri F, Pastinen T, Gauthier Y, Balsalobre A, Drouin J. 2018. Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat Genet 50(2):259–269. doi: 10.1038/s41588-017-0035-2. [DOI] [PubMed] [Google Scholar]
- 40.Kawakami A, Kimura-Kawakami M, Nomura T, Fujisawa H. 1997. Distributions of PAX6 and PAX7 proteins suggest their involvement in both early and late phases of chick brain development. Mech Dev 66(1–2):119–130. doi: 10.1016/S0925-4773(97)00097-X. [DOI] [PubMed] [Google Scholar]
- 41.Oustanina S, Hause G, Braun T. 2004. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J 23(16):3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barr FG. 1999. The role of chimeric paired box transcription factors in the pathogenesis of pediatric rhabdomysarcoma. Cancer Res 59(7 Suppl):1711s–1715s. [PubMed] [Google Scholar]
- 43.Chen F, Zhang J, Xu W, Zhang Y. 2017. April 12 Progress of CDK4/6 inhibitor palbociclib in the treatment of cancer. Anticancer Agents Med Chem. doi: 10.2174/1871521409666170412123500. [DOI] [PubMed] [Google Scholar]
- 44.Hagel M, Miduturu C, Sheets M, Rubin N, Weng W, Stransky N, Bifulco N, Kim JL, Hodous B, Brooijmans N, et al. 2015. First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov 5(4):424–437. doi: 10.1158/2159-8290.CD-14-1029. [DOI] [PubMed] [Google Scholar]
- 45.Matziari M, Dive V, Yiotakis A. 2007. Matrix metalloproteinase 11 (MMP-11; stromelysin-3) and synthetic inhibitors. Med Res Rev 27(4):528–552. doi: 10.1002/med.20066. [DOI] [PubMed] [Google Scholar]
- 46.Brettschneider J, Collin F, Bolstad BM, Speed TP. 2008. Quality assessment for short oligonucleotide microarray data. Technometrics 50(3):241–264. doi: 10.1198/004017008000000334. [DOI] [Google Scholar]
- 47.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. 2005. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.R Development Core Team 2014. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing: http://www.R-project.org/ [Google Scholar]
- 49.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostat Oxf Engl 4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 50.Kadota K, Ye J, Nakai Y, Terada T, Shimizu K. 2006. ROKU: a novel method for identification of tissue-specific genes. BMC Bioinformatics 7:294. doi: 10.1186/1471-2105-7-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.UniProt Consortium 2015. UniProt: a hub for protein information. Nucleic Acids Res 43(Database issue):D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nightingale A, Antunes R, Alpi E, Bursteinas B, Gonzales L, Liu W, Luo J, Qi G, Turner E, Martin M. The Proteins API: accessing key integrated protein and genome information. Nucleic Acids Res. 2017;45(W1):W539–W544. doi: 10.1093/nar/gkx237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin JL. 2016. Validation of reference genes for oral cancer detection panels in a prospective blinded cohort. PloS One 11(7):e0158462. doi: 10.1371/journal.pone.0158462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Remmele W, Stegner HE. 1987. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathology 8(3):138–140. [PubMed] [Google Scholar]
- 55.Gotch F, Rothbard J, Howland K, Townsend A, McMichael A. 1987. Cytotoxic T lymphocytes recognize a fragment of influenza virus matrix protein in association with HLA-A2. Nature 326(6116):881–882. doi: 10.1038/326881a0. [DOI] [PubMed] [Google Scholar]
- 56.Charville GW, Varma S, Forgó E, Dumont SN, Zambrano E, Trent JC, Lazar AJ, van de Rijn M. 2016. PAX7 expression in rhabdomyosarcoma, related soft tissue tumors, and small round blue cell neoplasms. Am J Surg Pathol 40(10):1305–1315. doi: 10.1097/PAS.0000000000000717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


