ABSTRACT
The release of soluble ligands of activating Natural Killer (NK) cell receptors may represent a regulatory mechanism of NK cell function both in physiologic and in pathologic conditions. Here, we identified the extracellular matrix protein Nidogen-1 (NID1) as a ligand of NKp44, an important activating receptor expressed by activated NK cells. When released as soluble molecule, NID1 regulates NK cell function by modulating NKp44-induced IFN-γ production or cytotoxicity. In particular, it also modulates IFN-γ production induced by Platelet-Derived Growth Factor (PDGF)-DD following NKp44 engagement. We also show that NID1 may be present at the cell surface. In this form or when bound to a solid support (bNID1), NID1 fails to induce NK cell cytotoxicity or cytokine release. However, analysis by mass spectrometry revealed that exposure to bNID1 can induce in human NK cells relevant changes in the proteomic profiles suggesting an effect on different biological processes.
Introduction
NK cells are Innate Lymphoid Cells (ILCs) involved in various immune processes ranging from the direct elimination of pathogens or tumor cells to the release of cytokines and chemokines and to regulatory interactions with different immune cells.1-9 In order to fulfill this variety of functions, NK cells use an array of receptors which sense microenvironmental stimuli and mediate appropriate responses.3,10,11 Several NK receptors are capable of regulating different NK cell functions. For example, the Natural Cytotoxicity Receptors (NCRs) NKp46, NKp30, and NKp44, play an important role in human NK cell-mediated recognition and killing of virally infected and tumor cells, and also induce the release of a number of cytokines and chemotactic factors.10,12 In addition, NKp30 and NKp46 mediate regulatory interactions occurring between NK and different leukocytes, including dendritic cells (DCs), neutrophils, eosinophils, macrophages, and T cells.5-9 NCRs were identified and molecularly characterized in ‘90s.13-15 Since then, numerous studies attempted to identify their ligands. This information is crucial for a better exploitation of the NK cell potential in the therapy of tumors, infections, or immune-mediated diseases.16-21 In spite of many efforts, so far, the panel of the NCR ligands has been only partially defined.10,12,22 A reason of these difficulties is related to the fact that study models of receptor-ligand interaction for the NCR are rather limited, as only NKp46 is expressed also on NK cells of rat and mouse. Moreover, although the NCRs belong to the Ig superfamily, they greatly differ in their molecular structure, implying that their ligands may be rather heterogeneous. In addition, each NCR may interact with different ligands, not necessarily displaying similar molecular features. Thus, for example, NKp46 and NKp44 have been shown to bind viral hemagglutinins.23 On the other hand, molecularly unrelated cellular ligands have been shown to bind NKp30 (B7H6 and BAT3/BAG6) and NKp44 (21spe-MLL5).24-26
To further complicate this issue, NCRs may interact with ligands by different modalities and even opposite functional outcomes. For example, NKp44 appears to differently modulate NK cell function by cis- or trans-interactions with heparan-sulphate proteoglycans (present at the NK or target cell surface).27,28 In addition, this receptor has been reported to transduce inhibitory signals upon interaction with Proliferating Cell Nuclear Antigen (PCNA).29,30 NKp30 triggers cytotoxicity and cytokine release upon binding B7H6 or BAT3 at the cell surface.24,25,31 Besides NCRs, other activating receptors, including NKG2D and DNAM-1, contribute to trigger the NK cell-mediated cytotoxicity.32-34 Ligands specific for these receptors have been extensively characterized: MICA/B and ULBPs molecules are recognized by NKG2D, while PVR and Nectin-2 (belonging to the Nectin family), are ligands for DNAM-1.32,33,35,36
Binding to soluble ligands may in some instances inhibit the triggering capability of activating NK receptors. In these cases, the release of such ligands in the extracellular environment may represent an effective mechanism to dampen NK cell function, not only in physiologic immune interactions but also in tumor-driven escape strategies.37-40 In this context, soluble ligands, endowed with suppressive capability, have been described for NKG2D and DNAM-1 activating receptors (sMICA, sULBPs, and sPVR) and for NKp30 (sB7H6 and sBAT3/BAG6).32,33,35,41-50 On the other hand, extracellular ligands for NKp46 and NKp44 have been recently identified and demonstrated to have a positive effect on the NK cell function. NKp46 has been shown to bind an extracellular molecule: the Complement Factor P (CFP or properdin).51 Recognition of CFP has been indicated as an important tool for innate responses to pathogens. Regarding NKp44, this receptor was recently shown to recognize a soluble factor, namely PDGF-DD. NKp44 engagement by PDGF-DD resulted in NK cell-mediated release of IFN-γ, TNF-α, and other proinflammatory cytokines and chemokines.52
In this study, we provide evidence that NKp44 recognizes a novel extracellular ligand, namely the Nidogen-1 (NID1) protein (also known as Entactin). We show that the NKp44/NID1 interaction results in a reduced NKp44-mediated induction of cytokine release by NK cells. Further analysis of the proteomic changes induced in NK cells by the exposure to NID1 revealed a substantial modulation of different molecules and pathways involved in important biological processes.
Results
Identification of Nidogen-1 as an extracellular ligand for NKp44 activating NK receptor
Aim of this study was the identification of putative extracellular ligands for NKp44. To this end, we analyzed the HEK293T cells as possible source of such ligands since these cells were found to bind NKp44Fc chimeric receptor at their cell surface. This data suggested that HEK293T cells could synthesize a putative NKp44-ligand. Importantly, these cells could be cultured extensively in protein-free medium, thus facilitating the analysis of proteins released in the culture supernatant.
Thus, HEK293T cells were cultured in protein-free medium and the supernatants (HEK293T-SN) were collected, concentrated, and coated on ELISA plates. Direct ELISA was performed using NKp44Fc, NKp30Fc, NKp46Fc, and DNAM-1Fc soluble chimeric receptors (Fc molecules). As shown in Fig. S1A, NKp44Fc strongly bound to HEK293T-SN-coated wells. Regarding the other Fc molecules tested, only NKp46Fc showed some reactivity (although significantly weaker than NKp44Fc), while NKp30Fc and DNAM-1Fc displayed no binding. These data suggested that NKp44 could recognize ligand(s) released by HEK293T cells.
To gain further information on such putative NKp44 soluble ligand(s), a metabolic labeling of HEK293T cells was performed in the presence of azido-sugars. As a result of this procedure, cells synthesize glycoproteins characterized by modified glycosylated residues, capable of linking covalently to biotin, enabling the detection of glycosylated proteins. The supernatant of labeled HEK293T cells (HEK293T-SN-biot) was analyzed by ELISA on plates coated with NKp44Fc or NKp30Fc molecules. As shown in Fig. S1B, HEK293T-SN-biot reacted with NKp44Fc- (and not with NKp30Fc-) coated wells, indicating that NKp44Fc can bind glycosylated protein(s) secreted by HEK293T cells.
The NKp44Fc-reactive glycoprotein(s) were further analyzed by Western blot. Proteins from concentrated HEK293T-SN were separated by SDS-PAGE and immunoblotted with NKp44Fc. As shown in Fig. S2, NKp44Fc, but not other Fc molecules analyzed (NKp30Fc, NKp46Fc, DNAM-1Fc), recognized a band of approximately 180 KDa under non-reducing conditions.
In order to characterize the high molecular weight glycoprotein(s) recognized by NKp44Fc, concentrated HEK293T-SN was resolved by two-dimensional electrophoresis (2-DE) and analyzed by Western blot with NKp44Fc, NKp46Fc, or NKp30Fc (Figure 1A–C). Concentrated HEK293T-SN-biot was subjected to the same procedure and immunoblotted with Neutravidin-HRP (for the detection of glycoproteins) (Figure 1D). In parallel, a preparative 2-D gel was stained with Blue Coomassie to visualize all proteins and excise the spots of interest (Figure 1E). Following 2-DE and immunoblotting, NKp44Fc was found to recognize different proteins present in HEK293T-SN (Figure 1A). Thus, 27 spots, visualized in NKp44Fc blot, were excised, digested, and analyzed by high resolution mass spectrometry (Table S1). Thanks to the comparison with blots stained with NKp46Fc and NKp30Fc soluble receptors (Figure 1B, 1C) it was possible to subtract the background and further restrict the analysis to 17 spots. Finally, by the comparative analysis with the blot stained with Neutravidin (Figure 1D), we could further select 7 spots recognized by NKp44Fc and corresponding to glycosylated proteins. We focused on spot n. 26 because it corresponded to high MW proteins. Among the proteins identified in this spot, Nidogen-1 (NID1) displayed a predicted MW compatible with the electrophoretic mobility of the glycoprotein(s) that had been detected by NKp44Fc in mono-dimensional SDS-PAGE (i.e. approximately 180 kDa) (see Fig. S2).
Figure 1.
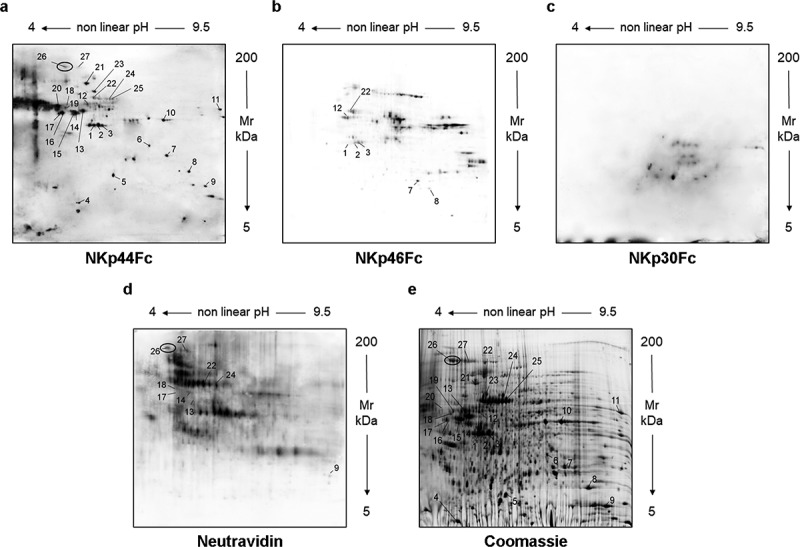
Analysis of culture supernatant from HEK293T cells by two-dimensional electrophoresis (2-DE). Concentrated HEK293T-SN was analyzed by two-dimensional electrophoresis (2-DE). After blotting, membranes were probed with NKp44Fc (A), NKp46Fc (B), or NKp30Fc (C) followed by HRP-conjugated anti-human IgG mAb. HEK293T-SN-biot was subjected to the same procedure and the membrane was incubated with HRP-conjugated Neutravidin (D). In parallel, HEK293T-SN (600 μg) was separated by 2-DE and proteins were stained with “blue silver” colloidal Coomassie (E). Numbers indicate the spots selected for mass spectrometry analysis; in panels (A), (D), and (E) spot 26 is highlighted with a black circle.
The direct and specific binding of NKp44 to NID1 was confirmed by different experimental evidences. Thus, immunoprecipitation experiments showed that NKp44Fc (but not NKp30Fc nor DNAM-1Fc used as controls) was able to immunoprecipitate NID1 protein from concentrated HEK293T-SN (Figure 2A). In addition, the ability of NKp44Fc to bind purified NID1 was investigated by Western blot. Recombinant human Nidogen-1 (rNID1) and HEK293T-SN were run in SDS-PAGE in parallel and immunoblotted with NKp44Fc (and NKp30Fc or DNAM1Fc as controls). Figure 2B shows that NKp44Fc was able to recognize both rNID1 and NID1 released from HEK293T-SN. In contrast, NKp30Fc and DNAM-1Fc did not display any reactivity. Finally, the interaction between NKp44 and rNID1 was assessed by ELISA. As shown in Figure 2C, different concentrations of NKp44Fc bound to rNID1-coated wells, while NKp30Fc, NKp46Fc, and DNAM-1Fc did not. This experiment shows that soluble NKp44Fc can specifically bind to NID1 also in its native conformation.
Figure 2.
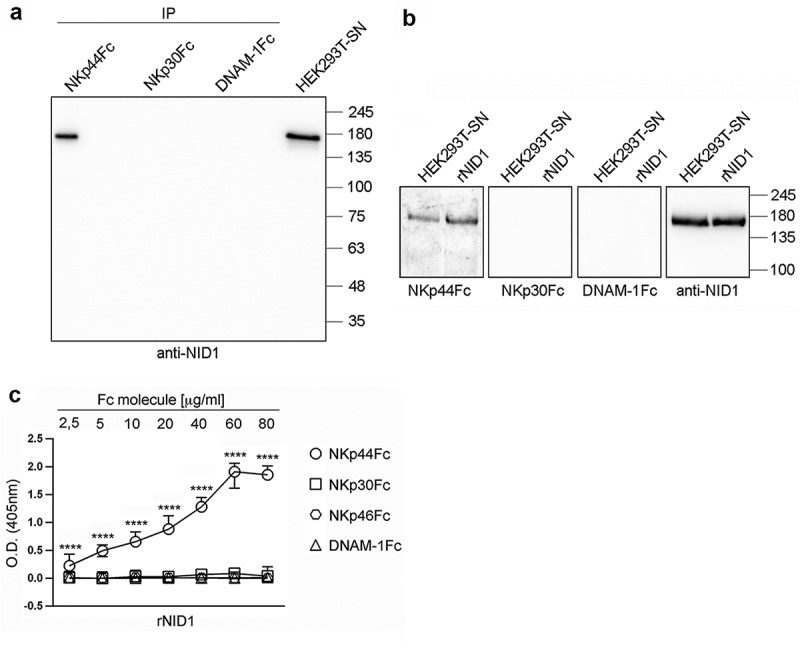
Specific recognition of NID1 by NKp44Fc chimeric receptor. (A) Concentrated HEK293T-SN was immunoprecipitated with the indicated Fc molecules and analyzed in 7.5% SDS-PAGE under non-reducing conditions; in parallel, concentrated HEK293T-SN was loaded as a positive control. After blotting, the membrane was probed with mouse anti-NID1 mAb followed by HRP-conjugated anti-mouse IgG mAb. One representative experiment of three is shown. (B) Concentrated HEK293T-SN and rNID1 were analyzed in SDS-PAGE on a 7.5% polyacrylamide gel under non-reducing conditions and, after blotting, probed with the indicated Fc molecules or with anti-NID1 mAb, followed by HRP-conjugated anti-human or anti-mouse IgG secondary reagent, respectively. Molecular weight (MW) markers (kDa) are indicated on the right. One representative experiment of five is shown. (C) ELISA plates were coated with rNID1 and incubated with different concentrations of the indicated Fc molecules followed by HRP-conjugated anti-human IgG mAb. Graph represents absorbance at 405 nm after normalization to background (nonspecific binding of the secondary reagent). Data are medians of triplicates ± interquartile range and are the pooled results of three independent experiments. ****p < 0.0001 by two-tailed Mann-Whitney test.
Effect of soluble NID1 on NKp44-mediated cell activation
In order to investigate the potential effect of NID1-NKp44 interaction, we took advantage of a model available in our lab based on the use of murine Bw5147 (Bw) cell transfectants expressing either the NKp44/DAP12 receptor complex (Bw-NKp44) or the chimeric receptor NKp30-CD3ζ (Bw-NKp30). In this model, cell activation induced by mAb-mediated cross-linking of NKp44 or NKp30 results in the release in culture SN of IL-2, which can be measured by ELISA. Since soluble ligands of different activating receptors have been shown to interfere with receptor function, we asked whether rNID1 pretreatment of Bw-NKp44 cells had a similar inhibitory effect. As shown in Figure 3, different rNID1 concentrations inhibited the NKp44-induced IL-2 production. As control, Bw-NKp30 cells were also analyzed. A minor inhibition of NKp30-induced IL-2 production could be detected only at the highest rNID1 concentration. Since NID1 has been reported to interact with α3β1 (CD49a/CD29) and αvβ3 (CD51/CD61) integrins,53,54 we analyzed the expression of CD29, CD49a, and CD61 in Bw-NKp44 and Bw-NKp30 cells, to rule out possible biases in the functional data on NID1. As shown in Fig. S3, expression levels of these integrins on the two Bw cell transfectants were comparable, indicating that the effects induced by rNID1 pretreatment were not reasonably influenced by integrin expression.
Figure 3.
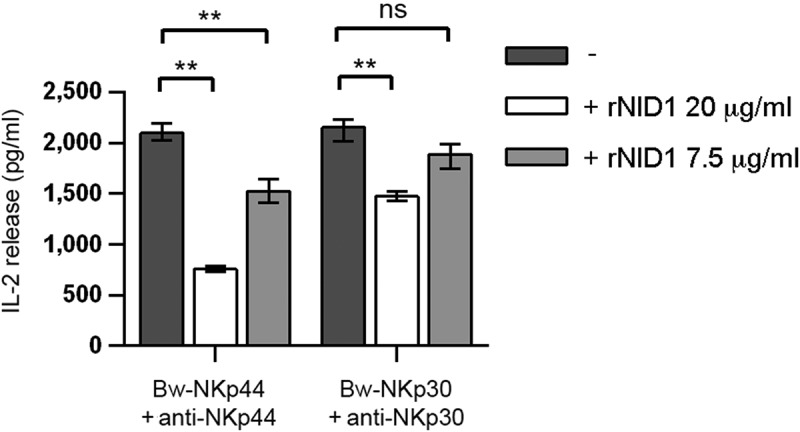
Effect of rNID1 pretreatment on NKp44-induced IL-2 production in Bw-NKp44 cells. Bw-NKp44 and Bw-NKp30 cells were left untreated or were pre-treated with rNID1 at the indicated concentrations for 1 h at 37°C and subsequently incubated on anti-NKp44 or anti-NKp30 mAb-coated plates, respectively, for 20 h. IL-2 release in the SN was evaluated by ELISA. The background (from GAM-stimulated cells) was subtracted for each value. Data are medians of duplicates ± interquartile range and are the pooled results of three independent experiments. **p = 0.0022, ns (not significant) = 0.584, by two-tailed Mann-Whitney test.
Next, we asked whether also NID1 released from cells could inhibit NKp44-mediated cell activation. To this end, the NID1 construct was transfected in NID1-negative cells. The culture supernatant of such transfected cells was assessed in functional assays. The human K562 cell line was used as recipient, since it does not express NID1 mRNA (Figure 4A). Stable transfection with the NID1 construct resulted in NID1 transcript expression and protein secretion in cell culture supernatants (Figure 4A, B). In ELISA, NKp44Fc reacted with K562-NID1-SN and not with K562-SN (Figure 4C) and in Western blot experiments it specifically reacted with the band corresponding to NID1 protein, while it didn’t bind to any band of K562-SN (Figure 4D). In order to investigate the potential functional consequences of NID1-NKp44 interaction we used again Bw-NKp44 and Bw-NKp30. Bw-NKp44 cells were stimulated with anti-NKp44 mAb in the absence or in the presence of K562-SN or K562-NID1-SN. As shown in Figure 5A, K562-SN had no effect, while K562-NID1-SN inhibited NKp44-induced IL-2 release. In contrast, K562-NID1-SN didn’t induce any inhibitory effect in Bw-NKp30 cells stimulated via NKp30, as compared to K562-SN. A similar inhibitory effect on NKp44-induced IL-2 production was also detected with culture SN from NID1-transfected Bw cells (Figs. S4-S5).
Figure 4.
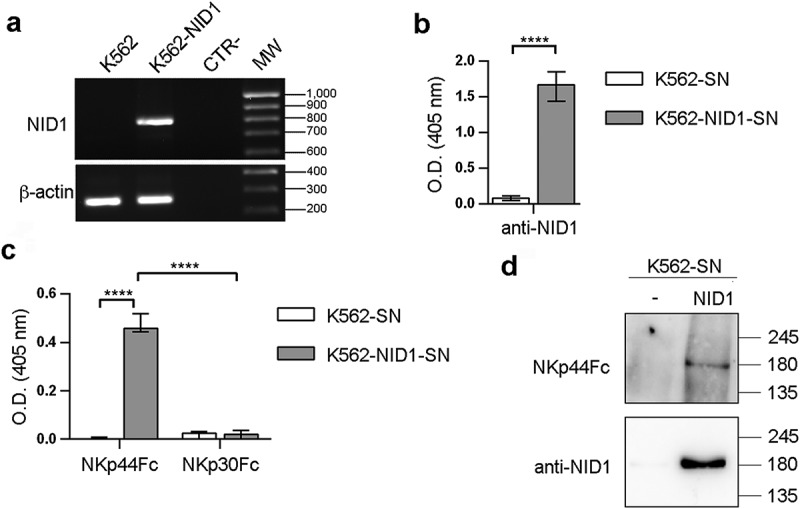
NKp44Fc recognizes NID1 released by NID1-transfected K562 cells. (A) NID1 mRNA expression was assessed by RT-PCR in wild type or NID1-transfected K562 cells. Primers specific for β-actin were utilized as positive control. PCR products were run on 1.5% agarose gel and visualized by ethidium bromide staining. One representative experiment of three is shown. (B, C) ELISA plates were coated with mouse anti-NID1 mAb (B) or with the indicated Fc molecules (C), followed by incubation with concentrated SN obtained from untransfected or NID1-transfected K562 cells cultured in protein-free medium. NID1 was detected using a goat anti-NID1 Ab followed by a HRP-conjugated anti-goat IgG Ab. Graphs represent absorbance at 405 nm after normalization to background (nonspecific binding of goat anti-NID1 + secondary reagent). Data are medians of triplicates ± interquartile range and are the pooled results of three independent experiments. ****p < 0.0001 by two-tailed Mann-Whitney test. (D) Concentrated SN (30 μl for each sample) derived from K562 cells and the corresponding NID1 transfectants cultured in protein-free medium were analyzed in SDS-PAGE on a 7.5% polyacrylamide gel; membrane was probed with NKp44Fc molecule or with mouse anti-NID1 mAb followed by the appropriate HRP-conjugated secondary mAb. MW markers (kDa) are indicated on the right. One representative experiment of two is shown.
Figure 5.
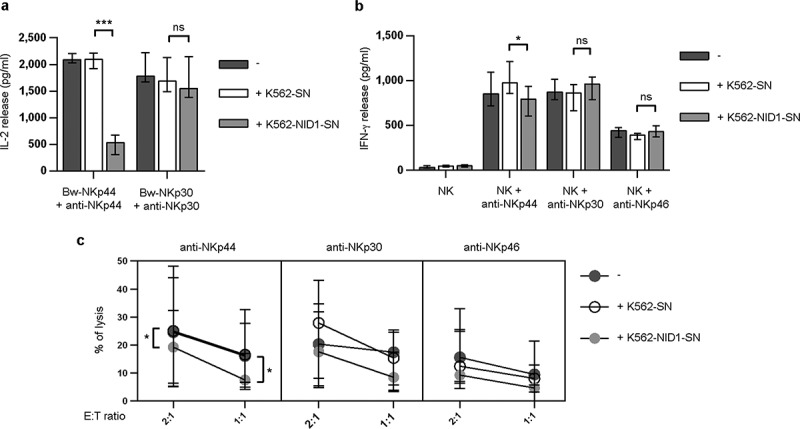
Effects of soluble NID1 on NKp44-induced function in Bw-NKp44 cells and human polyclonal NK cells. (A) Bw-NKp44 and Bw-NKp30 cells were incubated on anti-NKp44 or anti-NKp30 mAb-coated plates, respectively, for 20 h at 37°C in the absence or in the presence of SN derived from untransfected or NID1-transfected K562 cells. IL-2 release in the SN was evaluated by ELISA. The background (from GAM-stimulated cells) was subtracted for each value. Data are medians of duplicates ± interquartile range and are the pooled results of four independent experiments. ***p = 0.0002, ns = 0.1044, by two-tailed Mann Whitney test. (B) Polyclonal NK cell lines were incubated with SN derived from wild type or NID1-transfected K562 cells and cultured for 20 h at 37°C on plates coated with anti-NKp44, -NKp30, or -NKp46 mAbs. IFN-γ release in the SN was evaluated by ELISA. Data are medians of six independent experiments ± interquartile range performed with NK cells from three donors. *p = 0.0156, ns = 0.1094 (NK + anti-NKp30) and 0.0983 (NK + anti-NKp46) by one-tailed Wilcoxon test. (C) Polyclonal NK cell lines were incubated with medium or with SN derived from wild type or NID1-transfected K562 cells. After 20 h cells were utilized in a redirected killing assay against the FcγR+ P815 target cell line in the absence or in the presence of the indicated mAbs (E:T ratios 2:1 and 1:1). Data are medians of duplicates ± interquartile range and are the pooled results of six experiments performed with NK cells derived from three donors *p = 0.0313 (2:1 ratio) and 0.0156 (1:1 ratio) by one-tailed Wilcoxon test.
We next investigated whether NID1 released by NID1-expressing cells could exert an inhibitory effect on NKp44-mediated activation of normal, polyclonal NK cells. To this end, NK cells isolated from PB were cultured in the presence of IL-2 to induce the expression of NKp44 (absent on resting PB-NK cells). NK cells were then stimulated with anti-NKp44, anti-NKp30, or anti-NKp46 mAbs either in the absence or in the presence of K562-SN or K562-NID1-SN. After stimulation, SN of NK cells were collected and analyzed by ELISA for their IFN-γ content. As shown in Figure 5B, a significant reduction of NKp44-induced IFN-γ production was observed in the presence of K562-NID1-SN, as compared to K562-SN. On the other hand, neither K562-NID1-SN nor K562-SN inhibited IFN-γ secretion triggered via NKp46 or NKp30. Finally, we investigated whether NID1-containing SN could affect NKp44-induced NK cell cytotoxicity. To this end, NK cells were analyzed in a redirected killing assay either in the absence or in the presence of K562-SN or K562-NID1-SN. K562-NID1-SN (but not K562-SN) reduced anti-NKp44-triggered killing of P815 target cells, while it was ineffective on NKp30- and NKp46-mediated cytotoxic activity (Figure 5C).
Collectively, these results show that NID1 may interfere with target cell recognition via NKp44, exerting a regulatory effect on NKp44-induced NK cell activation.
In view of the recent identification of PDGF-DD as an extracellular NKp44 ligand, able to induce cytokine production by NK cells,52 we investigated whether sNID1 could inhibit NKp44 triggering induced by this novel ligand. We first tested this hypothesis in the Bw model, by stimulating Bw-NKp44 and Bw-NKp30 cells with recombinant (r)PDGF-DD. As shown in Figure 6A, recombinant PDGF-DD at 250 and 25 ng/ml was able to induce IL-2 production by Bw-NKp44 cells, while pretreatment of Bw-NKp44 cells with rNID1 resulted in a dose-dependent inhibition of PDGF-DD-induced IL-2 release. A similar inhibitory effect was induced by K562-NID1-SN. As expected, PDGF-DD had no effects on Bw-NKp30 cells. Next, we assessed the ability of NID1 to interfere with the PDGF-DD-induced IFN-γ production in NK cells. Figure 6B shows that rPDGF-DD could efficiently stimulate IFN-γ secretion by polyclonal IL-2-activated NK cells and that this effect was inhibited by K562-NID1-SN. Taken together, these data highlight the potential role of NID1 as a decoy NKp44 ligand.
Figure 6.
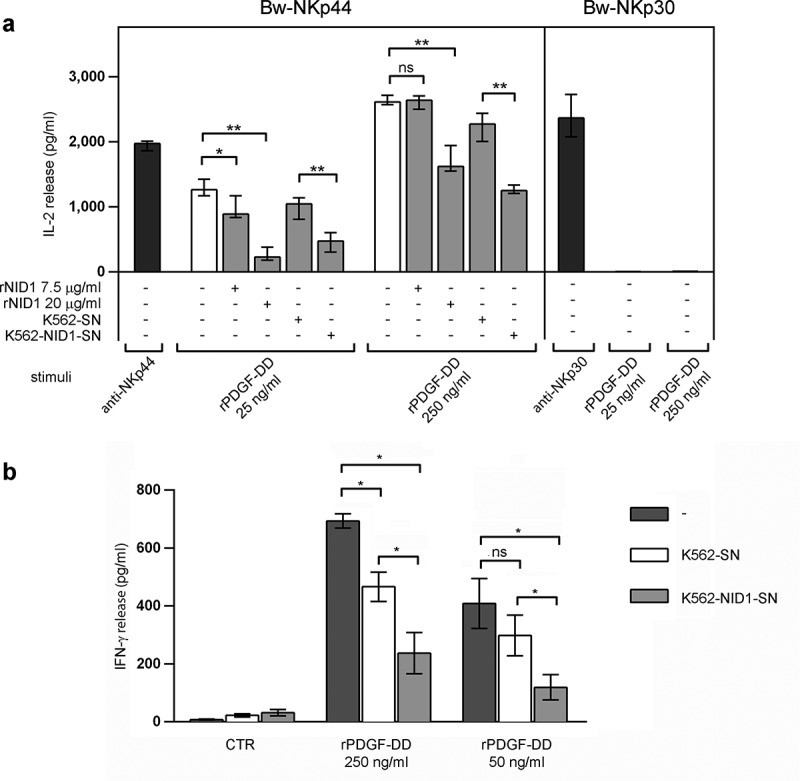
Effects of soluble NID1 on PDGF-DD-induced cytokine production in Bw-NKp44 cells and human polyclonal NK cells. (a) Bw-NKp44 and Bw-NKp30 cells were left untreated (white bars) or were pre-treated (grey bars) as indicated for 1 h at 37°C and subsequently incubated either on anti-NKp44 or anti-NKp30 mAb-coated plates, respectively, or stimulated with rPDGF-DD at 250 and 25 ng/ml for 20 h. IL-2 release in the SN was evaluated by ELISA. The background (from GAM-stimulated or unstimulated cells) was subtracted for each value. Data are medians of duplicates ± interquartile range and are the pooled results of three independent experiments. *p = 0.0152, **p = 0.0022, ns>0.9999, by two-tailed Mann-Whitney test. (b) Polyclonal NK cell lines were incubated with SN derived from wild type or NID1-transfected K562 cells and cultured for 20 h at 37°C in the presence of rPDGF-DD at 250 and 50 ng/ml. IFN-γ release in the SN was evaluated by ELISA. Data are means of six independent experiments ± SEM performed with NK cells from three donors. *p = 0.0156, *p = 0.0325 (K562-SN vs. K562-SN-NID1 PDGF-DD 50 ng/ml), ns = 0.2188 by one-tailed Wilcoxon test.
NID1 surface expression and functional effects on NKp44
Having demonstrated that soluble NID1 is able to interfere with NKp44-mediated NK cell activation, we next asked whether NID1 could be expressed also at the cell surface and be recognized as surface-associated molecule with possible functional outcomes. To this end, we assessed the cellular expression and distribution of NID1 in HEK293T, a cell line capable of binding NKp44Fc at its surface (see above). Thus, HEK293T cells were stained with a NID1-specific mAb and analyzed by imaging flow cytometry. This analysis revealed a weak but detectable surface expression of NID1. Figure 7A shows representative single cells stained with anti-NID1 mAb, while the histogram profiles in Figure 7B show the mean fluorescence intensity of all cells analyzed. The presence of NID1 protein at the cell surface suggested that it could be accessible for a direct interaction with NKp44. Thus, we analyzed by flow cytometry the surface expression of NID1 and, in parallel, the reactivity of NKp44Fc on HEK293T, K562, and K562-NID1 cells. As shown in Figure 7C, anti-NID1 mAb stained HEK293T and K562-NID1 cells, but not K562 cells. Consistently, NKp44Fc bound to HEK293T and K562-NID1 but not to K562 cells. Comparable results were obtained with Bw-NID1 cell transfectants (Figure 7C). Taken together, these data indicate that NID1 can be expressed at the cell surface and recognized by NKp44(Fc) receptor.
Figure 7.
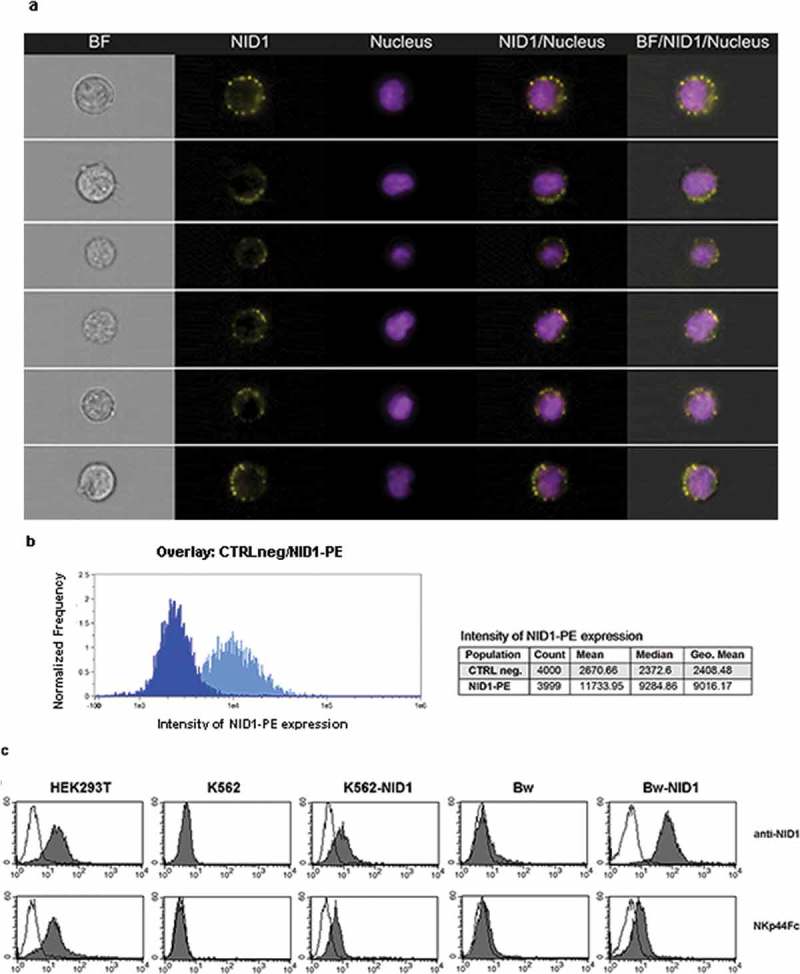
Analysis of NID1 surface expression in HEK293T cell line and in NID1 cell transfectants. (A, B) HEK293T cells were stained with a NID1-specific mAb followed by PE-conjugated anti-mouse IgG1 mAb. Prior to analysis by Amnis ImageStream, cells were incubated with Hoechst 33342 nuclear dye. NID1 expression on individual single cells is shown in (A). Data show representative images from one experiment out of three performed. Histograms in (B) represent total intensity of NID1 immunostaining for HEK293T cells as compared to isotype control. (C) HEK293T, K652, K562-NID1, Bw, and Bw-NID1 cells were stained with a NID1-specific mAb or with NKp44Fc followed by the appropriate isotype-matched PE-conjugated secondary mAb. Samples were analyzed by flow cytometry. Grey profiles represent cells stained with anti-NID1 or with NKp44Fc, while white profiles correspond to isotype control. One representative experiment of four is shown.
On the basis of these results, we further investigated the possible effect of NID1 expressed at the cell surface on NKp44-induced NK cell activation. To this end, Bw-NKp44 cells were cultured in the presence of K562 or K562-NID1 cells or in rNID1-coated plates. After 20 h, IL-2 was measured in culture SN. As shown in Figure 8A, neither K562 nor K562-NID1 cells could induce increases of IL-2 production by Bw-NKp44 cells. Similarly, no increments in IL-2 production occurred upon culture of Bw-NKp44 in rNID1-coated plates (Figure 8B). Comparable results were obtained when rNID1 coating was performed through anti-NID1 (or anti-His) mAb.
Figure 8.
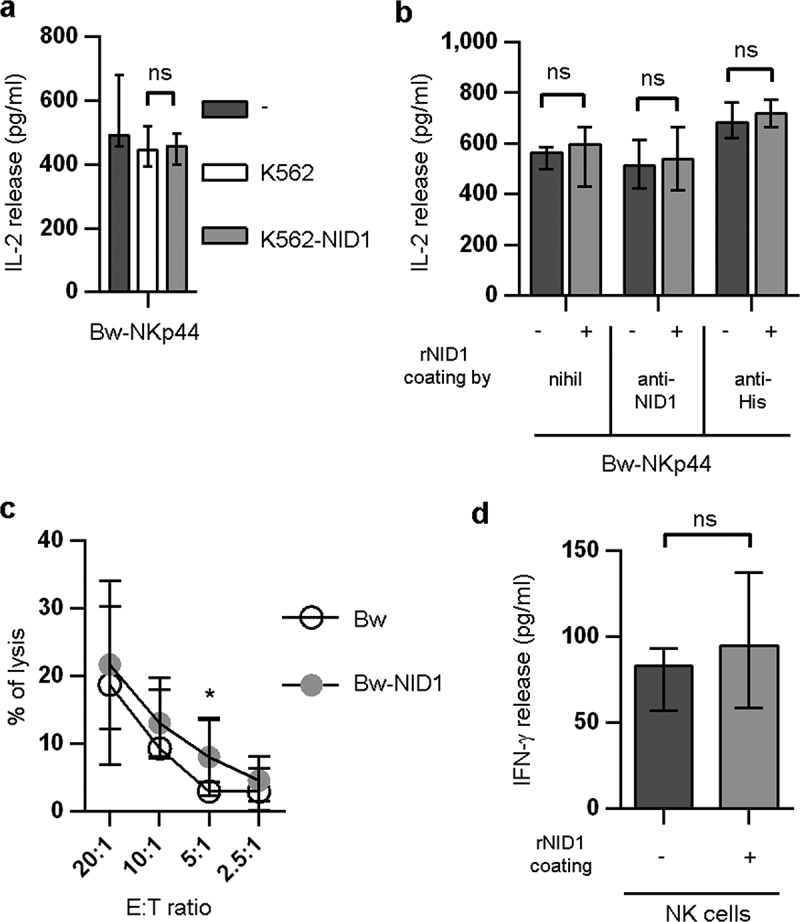
Functional effects of cell surface-associated NID1 or plastic-bound rNID1 in Bw-NKp44 and in NK cells. (A) Bw-NKp44 cells were cultured alone or in the presence of wild type or NID1-transfected K562 cells for 20 h at 37°C. IL-2 release in the SN was evaluated by ELISA. Data are medians of duplicates ± interquartile range and are the pooled results of three independent experiments. ns = 0.5714 by two-tailed Mann-Whitney test. (B) Bw-NKp44 cells were incubated on plates coated with rNID1, anti-NID1 + rNID1, or anti-His + rNID1 for 20 h at 37°C. IL-2 release in the SN was evaluated by ELISA. Data are medians of duplicates ± interquartile range and are the pooled results of two independent experiments. ns = 0.8286 (−) or ns = 0.6571 (anti-NID1, anti-His) by two-tailed Mann-Whitney test. (C) Polyclonal NK cell lines were evaluated for their cytolytic activity in a 4-h 51Cr release assay against Bw and Bw-NID1 cells at the indicated E:T ratios. Data are medians of duplicates ± interquartile range and are the pooled results of fifteen experiments performed with NK cells derived from five donors. *p = 0.0277 by one-tailed Wilcoxon test. (D) Polyclonal NK cell lines were incubated on plates coated with rNID1 for 20 h at 37°C. IFN-γ release in the SN was evaluated by ELISA. Data are medians of eight independent experiments ± interquartile range performed with NK cells derived from three donors. ns = 0.2305 by one-tailed Wilcoxon test.
We then analyzed whether cell surface NID1 had any effect on NK cells expressing NKp44 upon activation. To this end, IL-2-activated polyclonal NK cells were obtained from several healthy donors and used in functional assays. Since, in NK cells, NKp44 triggers cytotoxicity, we analyzed the susceptibility to NK cell-mediated killing of NID1+ or NID1− cells in a cytolytic assay. Because NK cells express different activating receptors that recognize ligands on human K562 cells, this assay was performed using murine Bw cell transfectants (see Fig. S4). As shown in Figure 8C, Bw-NID1 and Bw cells didn’t display significant differences in their susceptibility to NK-mediated cytolysis.
We also assessed the possible effect of plastic-bound rNID1 on IFN-γ release from NK cells cultured in rNID1-coated plates. As shown in Figure 8D, exposure to NID1 failed to induce significant increments of IFN-γ production by NK cells.
In order to confirm NKp44 interaction with NID1 exposed at the cell surface, we performed transient NID1 silencing in HEK293T cells. SiRNA-mediated silencing resulted in a reduced presence of NID1 both as surface associated and as soluble released molecule (Figure 9). Remarkably, NID1 silencing also induced a reduced binding of NKp44Fc to the cell surface (Figure 9A) and a reduced NKp44Fc reactivity with the SN derived from NID1-silenced HEK293T cells (Figure 9B). These results confirm that NKp44Fc is able to recognize NID1 exposed at the cell surface as well as NID1 released in the extracellular space.
Figure 9.
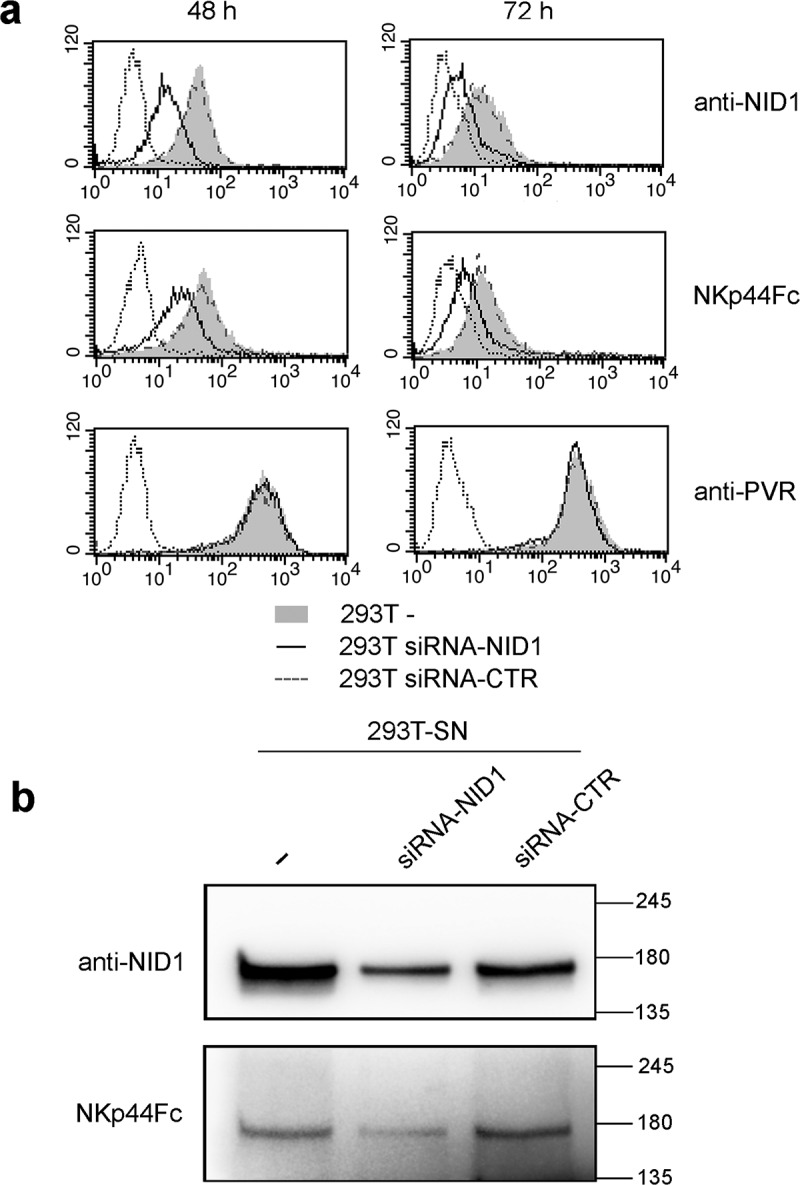
Effect of NID1 silencing on NKp44-NID1 interaction in HEK293T cells and in SN derived from NID1-silenced HEK293T cells. (a) NID1 surface expression and NKp44Fc reactivity was analyzed in untransfected HEK293T cells and in HEK293T cells transfected with siRNA-NID1 or siRNA-CTR 48 and 72 h after transfection. Samples were analyzed by flow cytometry. Dotted profiles correspond to isotype control. One representative experiment of three is shown. (b) Western blot analysis of SN derived from HEK293T cells transfected with siRNA-NID1 or siRNA-CTR. Concentrated SN (30 μg for each sample) derived from untransfected HEK293T cells and HEK293T transfected with siRNA-NID1 or siRNA-CTR cultured in protein-free medium were analyzed in SDS-PAGE on a 7.5% polyacrylamide gel; membrane was probed with mouse anti-NID1 mAb or with NKp44Fc molecule followed by the appropriate HRP-conjugated secondary mAb. MW markers (kDa) are indicated on the right. One representative experiment of three is shown.
Effect of NID1 on NK cell proteome
The above data suggest that NID1 exposed at the target cell surface or bound to a solid support could not significantly modify the cytokine release or cytotoxicity in NKp44+ cells. Thus, to gain insight on the possible effects of surface-exposed NID1 on NK cells, we used a proteomic approach. To this end, NKp44+ polyclonal NK cell populations expanded from four healthy donors were cultured for 20 h on plates coated with rNID1 (NID1) (or w/o coating as control, CTR). In parallel, NK cells were also cultured on plates coated with goat anti-mouse IgG (GAM) + anti-NKp44 mAb (NKp44) (or GAM alone as control, GAM). NK cells were then collected and lysed; total cell lysates were analyzed by high-resolution mass spectrometry.
Data processing through the MaxQuant software allowed the identification of a total of 6903 proteins (of which 5682 were quantified using a Label-Free Quantitation approach). Most of the proteins (5317) could be identified in all of the experimental conditions analyzed (i.e. CTR, NID1, GAM, and NKp44), while few proteins were exclusive of specific conditions (Fig. S6A). An initial analysis of the whole data set (i.e. without any restriction for statistical significance and fold change threshold) indicated that both stimuli could modify the NK cell proteomic profile and could induce concordant expression changes (i.e. up- or down-regulation) in a substantial number of proteins (Fig. S6B).
In order to analyze the statistically significant regulated proteins and to define the possible relationships between rNID1 and anti-NKp44 stimulation, two volcano plots were generated from the NID1vsCTR and NKp44vsGAM data sets, and proteins were selected on the basis of a two-sample t-test (FDR = 0.05 S0 = 0.1) (Figure 10A, B). By this selection, 112 proteins resulted modulated by rNID1 (64 up- and 44 down-regulated, as compared to CTR) (Table S2), and 129 proteins were modulated by anti-NKp44 mAb (41 up- and 88 down-regulated, as compared to GAM) (Tab S3). 15 modulated proteins were common to the two conditions (indicated in red in Figure 10A, B). The GO analysis indicated that a major fraction of NID1-modulated proteins were involved in the regulation of cell metabolism, but substantial numbers of proteins were also involved in processes related to cell proliferation, signal transduction, endo/exocytosis, and immune response, indicating that NID1 stimulation could induce effective functional responses on NK cells (Figure 10C). Interestingly, NID1- and NKp44-modulated proteins appeared to be similarly involved in the same biological processes, and were similarly distributed among the main cellular components, with the highest percentage of proteins located in membrane compartments (Figure 10C, D). In order to further compare the two stimuli, we drilled down through the GO biological processes/molecular functions and built a functional heatmap depicting the quantitative modulation of the NK cell proteotype in response to NID1 or anti-NKp44 stimuli (Fig. S7). This analysis indicates that part of the effects on the modulation of biological processes is shared by the two stimuli.
Figure 10.
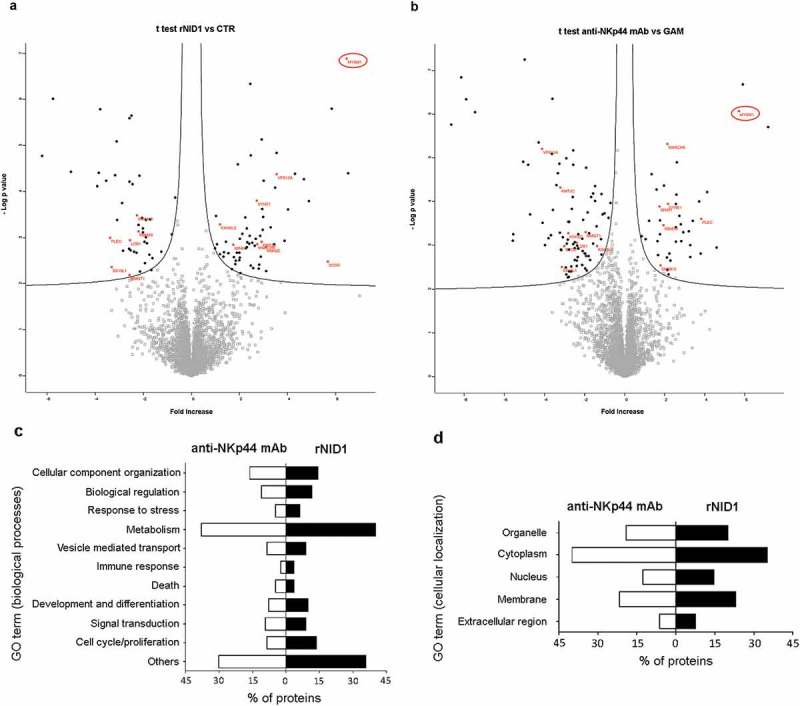
Proteomic analysis of NK cells stimulated with rNID1 or anti-NKp44 mAb. (A, B) Volcano plot representation of differentially expressed proteins. Plots represent stimulated (rNID1) vs. not stimulated (CTR) NK cells (A) or stimulated (anti-NKp44 mAb) vs. not stimulated (GAM) NK cells (B). Black dots represent proteins that display both large magnitude fold-changes (x-axis, proteins up-regulated after treatment are shown on the right) as well as high statistical significance (−log10 of P value, y-axis). The black lines show where FDR = 0.05 and s0 = 0.1. Red asterisks represent significantly modulated proteins (with the indicated gene names) which are present in both datasets. Gray squares represent non statistical significant proteins. (C, D) Gene Ontology (GO) categorization of proteins significantly modulated by rNID1 and anti-NKp44 mAb stimuli according to biological processes (C) or cellular component (D). On the x axis, the percentage of proteins associated with the indicated GO terms is shown.
On their complex our data suggest that NID1 can bind NKp44 and induce still unnoticed functional responses on NK cells. The GO analysis provides substantial information on these new putative functions, which, however, remain incompletely defined. In this context, the evaluation of the proteins modulated by both stimuli may give some hints. Within this group one of the most up-regulated proteins is represented by MYSM1 (see Figure 10). Interestingly, MYSM1 was recently demonstrated to be essential for NK cell maturation and differentiation,55,56 suggesting that the NKp44-NID1 interaction may be relevant to these processes and play a key role for the homeostasis of the mature NK cell population.
Discussion
In this study we have identified and characterized a novel extracellular ligand of the NCR NKp44. Analysis of the culture supernatant of the NKp44Fc-reactive cell line HEK293T and the use of 2-DE, combined to high-resolution mass spectrometry, revealed NID1 as an NKp44-reactive molecule. Interaction between NID1 and NKp44 was validated both by immunoprecipitation and by ELISA experiments, indicating that NKp44 recognizes NID1 in its native conformation. Soluble NID1 could down-regulate NKp44-mediated NK cell activation; in addition, plastic-bound NID1 induced significant changes in the NK cell proteomic profile, suggesting a possible effect on various NK cell functions.
NID1 glycoprotein is an essential component of the basement membrane (BM) that plays a role both in BM assembly and stabilization, and in the adhesion between cells and extracellular matrix (ECM).57,58 Therefore, NKp44-NID1 interactions are likely to occur in tissues, namely in the mucosae, which contain both NK cells and ILC3, expressing NKp44 receptor.59-61 The role of NKp44 in regulating the function of these cells has not been clearly defined. Some evidences, however, indicate that the engagement of NKp44 in ILC3 may induce TNF-α production and synergize with IL-1, IL-7, and IL-23 to induce secretion of IL-22 (a cytokine typically produced by ILC3s).62 In a recent study, the transcriptome analysis of NKp44-stimulated ILCs revealed a “genome wide regulating effect”.62 This finding is in line with our present proteomic study showing that NK cell stimulation both via mAb-mediated NKp44 cross-linking and by plastic-bound NID1 could induce significant changes in a relevant number of proteins. Consistent with the multiple ligand specificities of NKp44 and its ability to mediate different functional responses, NKp44 cross-linking and NID1-induced stimulation resulted in protein changes that were only partially overlapping. However, the GO analysis suggested some common functional effects induced by the two stimuli. Based on this analysis, NID1 recognition appeared to influence different biological processes related not only to immunologic functions but also to cell metabolism, proliferation, and development. In this context, it is of note that among the limited number of proteins up-regulated by both stimuli, the highest score was associated with MYSM1, a molecule that has been proposed to play a role in NK cell development/maturation.55,56 Thus, the finding that MYSM1 was strongly induced by both stimulation with rNID1 and mAb-mediated NKp44 cross-linking suggests a new NKp44-induced cell function and offers clues to study the role of NKp44-NID1 interaction in the process of NK cell (and/or ILC) maturation and differentiation.
Functional experiments revealed that soluble NID1 may play a role as a decoy ligand. Indeed, the addition of rNID1 to cell cultures resulted in inhibition of NKp44-mediated responses in Bw-NKp44 cells. An inhibitory effect was displayed also by soluble NID1 released from cell transfectants. Importantly, released NID1 could significantly decrease NKp44-induced IFN-γ secretion (and, marginally, cytotoxicity) in normal, polyclonal human NK cells. Remarkably, the NID1-mediated inhibition was specific for NKp44, although high concentrations of rNID1 had a slight inhibitory effect also in Bw-NKp30 cells. On the other hand, the supernatant from NID1-releasing cell transfectants did not inhibit NKp30 function in Bw-NKp30 cells. In addition, it could not inhibit NKp30 and NKp46 responses in normal, polyclonal NK cells (see Figure 5). Finally, rNID1 didn’t react with NKp30Fc in ELISA (see Figure 2C). Notably, NID1 was able to inhibit cytokine production triggered not only upon mAb-mediated NKp44 crosslinking but also by stimulation of NK cells with the recently characterized NKp44 ligand, namely PDGF-DD.52 It is conceivable that the release of NID1 in extracellular fluids may represent a regulatory mechanism that could act on NKp44+ NK cells at specific sites or in the blood stream. Notably, recent studies have highlighted the potential relevance of soluble NID1 as a biomarker in different tumors. Li and coworkers reported the presence of elevated NID1 levels in the serum of ovarian cancer patients, primarily those with an advanced stage of the disease.63 Along this line, a high content of a specific fragment of NID1, derived from its degradation by cathepsin-S (CatS) was detected in the serum of NSCLC patients.64 CatS-degraded NID1 was suggested to reflect the loss of BM integrity, an event typically associated to tumor invasion. On the other hand, loss of NID1 expression was frequently detected in human gastrointestinal tumor samples.65 In this case it was proposed that the loss of NID1 could favor tumor invasion and metastasis by destabilizing the structure of BM and loosening the cell-BM interactions. It should be considered, however, that several reports described a pro-tumoral and pro-metastatic role for NID1. Thus, NID1 has been shown to promote tumor cell migration, invasion, and chemoresistance in ovarian carcinomas via the induction of Epithelial-Mesenchymal Transition (EMT).66 Another study revealed a possible NID1 involvement in the acquisition of migratory and invasive properties by endometrial cancer cells.67 It has also been shown that NID1 was present at high levels in the secretome of lung metastases in both breast cancer and melanoma.68 This study showed that NID1 promoted cancer cell migration, invasion, and metastases in the lung. Thus, NID1 appears to be released in the extracellular environment of various tumor types. In this context, the inhibitory NID1-NKp44 interaction described in our present study may be regarded as a novel putative suppressive mechanism exploited by tumors to prevent the NK cell-mediated attack. Along this line, tumor escape mechanisms involving the release of soluble ligands have been described for different activating NK cell receptors. Thus, high levels of soluble NKG2D or DNAM-1 ligands were detected in serum of cancer patients and have been associated to advanced stages of the disease or to poor prognosis.32,33,44 Increased amounts of soluble B7-H6 were found in sera of melanoma patients and in the peritoneal fluid of ovarian carcinoma patients.47,49 Soluble BAG6/BAT3 was detected in sera of CLL patients and could modulate NKp30-mediated cytotoxicity.50 Recently, Galectin-3 has been described as a soluble molecule released from tumor cells and capable of inhibiting NKp30-induced NK cell functions.69 Soluble ligands are often released through the action of proteases, as in the case of B7-H6 shedding caused by ADAM-10 and −17 metalloproteases.47 NID1 is generally released as a whole molecule. Therefore, it is conceivable that the inhibitory effect on NK cells may be mediated by the intact molecule. On the other hand, NID1 may be modified in the ECM by the action of extracellular proteases, such as Cat-S and ADAMTS1 (A Disintegrin and Metalloproteinase with ThromboSpondin motifs).70 In addition, NID1 can interact with different BM/ECM proteins including laminin, collagen type IV, and perlecan.57,58 Thus, it is possible that the tissue microenvironment and the molecular context may further influence the functional outcome of the NID1-NKp44 interaction. The ability of NKp44 to recognize NID1 glycoprotein both in denatured and native conditions indicates that NKp44 may bind to a linear epitope. This observation suggests that NKp44 may recognize also cleaved NID1 fragments. This issue should be further investigated in view of the possible targeting of the NKp44-NID1 interaction in the context of cancer immunotherapy.
Importantly, we show that NID1, besides being released in the extracellular space, can be also detected at the cell surface of NID1-releasing cells (see Figure 7). In this context, it has been reported that NID1 may associate through the interaction of its RGD sequence with α3β1 or αvβ3 integrins.53,54 Notably, we could detect surface NID1 on a panel of different NKp44Fc-binding human tumor cell lines (Figure 11). Although it has been reported that NID1 up-regulation occurs in several tumor types, information is still lacking on whether NID1 is associated to the tumor cell surface. We assessed the possible functional outcome of the interaction between cell surface-associated NID1 on NKp44+ responding cells. In the Bw-NKp44 model, no induction of IL-2 production could be observed. In NKp44+ NK cells, neither induction of IFN-γ production upon exposure to rNID1-coated plates nor increase of cytotoxic activity against NID1-expressing target cells could be detected. On the other hand, as discussed above, using a proteomic approach, we found that surface-bound rNID1 could induce changes in proteins involved in different biological processes. It is conceivable that such proteomic changes may be induced also by cell surface-associated NID1.
Figure 11.
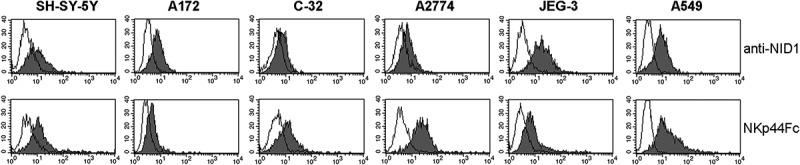
Analysis of NID1 surface expression and NKp44Fc reactivity on a panel of human cell lines. The indicated cell lines (SH-SY-5Y neuroblastoma; A172 glioblastoma; C-32 melanoma; A2774 ovarian adenocarcinoma; JEG-3 placental choriocarcinoma; A549 lung carcinoma) were stained with a NID1-specific mAb or with NKp44Fc followed by the appropriate isotype-matched PE-conjugated secondary mAb. Samples were analyzed by flow cytometry. Grey profiles represent cells stained with anti-NID1 or with NKp44Fc, while white profiles correspond to isotype control. One representative experiment of three is shown.
In conclusion, the characterization of NID1 as a ligand of NKp44 provides new interesting hints to extend our knowledge on the NK cell biology. Future investigations of the functional effects of NID1 in different molecular contexts (i.e. associated with ECM proteins or modified by ECM enzymes) may help to understand the mechanisms regulating NK cell (and ILC3) function in tissues. In addition, studies based on proteomic data may lead to the definition of new functional roles of NKp44 receptor (and, consequently, of NK cells) in different biological processes. Taken together, our present study may offer clues and open new opportunities for a more effective exploitation of NK cells for therapeutic purposes, especially (but not exclusively) in the immunotherapy of solid tumors.
Methods
Antibodies
The following monoclonal antibodies (mAbs), produced in our laboratory, were used in this study: BAB281 (IgG1, anti-NKp46), AZ20 (IgG1, anti-NKp30) and Z231 (IgG1, anti-NKp44). The following commercial antibodies were also used: mouse anti-Nidogen-1 mAb (IgG1, clone 302117, R&D, MAB2570) and goat anti-NID1 polyclonal Ab (R&D, AF2570); mouse anti-NKp46 (IgG1, clone 29A1.4.9, Miltenyi Biotec, 130-095-118); hamster anti-mouse CD29-PE (IgGλ, clone HMβ1-1, Miltenyi Biotec, 130-102-994); hamster anti-mouse CD61-PE (IgGκ, clone 2C.9.G2, Miltenyi Biotec, 130-102-851); human anti-mouse CD49a-PE (IgG1, clone REA493, Miltenyi Biotec, 130-107-632); goat anti-human IgG Horseradish Peroxidase (HRP)-conjugated mAb, (Southern Biotech, 2040-05), goat anti-mouse Ig HRP-conjugated mAb (Southern Biotech, 1031-05), mouse anti-goat IgG HRP-conjugated mAb (Southern Biotech, 6164-05), and goat anti-mouse IgG1 phycoerythrin (PE)-conjugated mAb (Southern Biotech, 1070-09); goat anti-human IgG PE-conjugated mAb (Jackson ImmunoResearch, 109-116-170); rabbit polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Ab (ThermoFisher, PA1-16780); mouse anti-His mAb (IgG1, clone BMG-His-1, Roche Diagnostics, 1 922 416).
Cell lines
The following cell lines, purchased from American Type Culture Collection (ATCC), were used in this study: HEK293T (human embryonic kidney), K562 (human erythroleukemia), Bw1547 (Bw, murine thymoma), SH-SY-5Y (human neuroblastoma), A172 (human glioblastoma), C-32 (human melanoma), A2774 (human ovarian adenocarcinoma), JEG-3 (human placental choriocarcinoma), A549 (human lung carcinoma). Cell lines were maintained either in DMEM (HEK293T) or RPMI1640 medium supplemented with 10% (v/v) fetal calf serum (FCS), L-glutamine, and antibiotics (penicillin and streptomycin). For some experiments, HEK293T and K562 cell lines were also cultured in protein-free CD medium (ThermoFisher, 11279023). Stably transfected K562 cells were selected and cultured in the presence of G418 sulphate (Calbiochem, 345810) at the final concentration of 1.2 mg/ml. All cell lines were periodically tested and were mycoplasma free.
Preparation of soluble chimeric receptors
NKp44Fc, NKp30Fc, and NKp46Fc soluble receptors were prepared as previously described.71 Briefly, the sequences coding for the extracellular portion of the different receptors were subcloned in the pRB1-2B4Fcmut vector (kindly provided by Dr. M Falco, Ist. G. Gaslini, Genova) in frame with the sequence coding for the human IgG1 portion, that was mutagenized in order to obtain a mutated Fc that does not bind to Fc receptors. These constructs were transfected into HEK293 cell line utilizing JetPEI (Polyplus, 101-10) following manufacturer’s instructions; after 48–72 h, cells were selected with 0,5 mg/ml G418 sulphate, in order to obtain stably transfected cells, and subcloned by limiting dilution. SN were collected from cell clones cultured either in DMEM/10% Ultra-low IgG FCS (ThermoFisher) or in Ex-CELL ACF CHO medium (Sigma, C5467) and the soluble Fc molecules were purified by affinity chromatography utilizing Protein A Sepharose 4 Fast Flow (G-Biosciences, 786-283). Purified recombinant proteins were checked by SDS-PAGE followed by silver staining and by ELISA utilizing mAbs specific for the different receptors.
Preparation of cell culture supernatants
Cell culture supernatants (SN) were obtained from wild type or transfected K562 or Bw cells. After 48–72 h culture in medium supplemented with 1% (v/v) FCS or in protein-free CD medium, SN were collected and utilized for subsequent experiments either as they were or following concentration (up to 30-fold) with Amicon Ultracel-10K (Millipore, UFC901024). Protein content in SN derived from culture in protein-free CD medium was determined using Bradford Protein Assay (Bio-Rad, 5000006).
Metabolic labeling
HEK293T cells were cultured in protein-free CD medium in the presence of ManAz (N-azidoacetylmannosamine-tetraacylated) (ThermoFisher, 88904) at the final concentration of 40 μM. After 72 h, SN was collected, concentrated (up to 24-fold) with Amicon Ultracel-10K, and incubated with Biotin-PEG3-Phosphine (200 μM final concentration, ThermoFisher, 88901) O/N at room temperature, in order to label secreted glycoproteins containing azido-sugars. Labeled SN was dialyzed in PBS using D-Tube Dialyzer Maxi, MWCO 6–8 kDa (Novagen, 71509-3).
Enzyme-linked immunosorbent assay (ELISA)
Direct ELISA: 100 μl of concentrated SN derived from HEK293T cells cultured in protein-free CD medium were coated on ELISA plates O/N at 4°C. Next, wells were saturated with PBS/3% (w/v) bovine serum albumin (BSA) for 3 h at r.t., washed in PBS, and incubated with Fc molecules at different concentrations ranging from 2.5 to 80 μg/ml or with 1 μg/ml anti-NID1 mAb followed by the appropriate HRP-conjugated secondary reagent. Similar experiments were carried out on ELISA plates coated with 5 μg/ml recombinant human Nidogen-1 (rNID1) (R&D, 2570-ND-050) in PBS.
Indirect (sandwich) ELISA: a similar procedure was applied. Briefly, ELISA plates were coated with Fc molecules (5 μg/ml in PBS), saturated, and incubated with 100 µl HEK293T-SN-biot followed by HRP-conjugated Streptavidin (Southern Biotech, 7100-05). Alternatively, ELISA plates were coated with mouse anti-NID1 mAb or Fc molecules (5 μg/ml in PBS), saturated, and incubated with 100 µl SN derived from wild type or NID1-transfected K562 or Bw cells cultured in medium with 1% (v/v) FCS or in protein-free CD medium. Next, a polyclonal goat anti-NID1 Ab was added, followed by a HRP-conjugated anti-goat Ig mAb.
In all experiments a final incubation with the HRP substrate ABTS (2,2ʹ-azino-di-(3-ethylbenzthiazoline sulfonic acid) (Roche Diagnostics, 102 946) was performed. The colorimetric signal was measured with a microplate reader (TECAN Sunrise) at an optical density (OD) of 405 nm.
Western blot analysis
Samples (6–28 μg SN obtained in protein-free medium, 30 μl SN obtained in medium/1% FCS, 160 ng rND1) were run on 7,5% or 10% polyacrylamide gels under non-reducing conditions and transferred to Immobilon-P PVDF membranes (Millipore, IPVH00010). For some experiments, gels were prepared with 7.5% TGX Stain-Free Acrylamide Solutions (Bio-Rad, 161-0181). Membranes were blocked with 5% BSA in Tris-buffered-saline containing 0.05% Tween-20 (TBS-T) and probed with anti-NID1 mAb, a rabbit polyclonal anti-GAPDH Ab, or Fc molecules (7 μg/ml) followed by the appropriate HRP-conjugated secondary reagent. The SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher, 34,080) was used for detection. Images were acquired with ChemiDoc Touch Imaging System (Bio-Rad) and analyzed with Image Lab software (Bio-Rad).
Two-dimensional electrophoresis (2-DE) and western blot
Concentrated HEK293T-SN and HEK293T-SN-biot (300 μg for Western blot and 600 μg for preparative gels) were solubilized in the reduction/alkylation solution containing 8 M urea, 4% CHAPS, 5 mM tributylphosphine (TBP), 20 mM iodoacetamide (IAA), 40 mM Tris, and 0.1 mM EDTA for 1 h. To prevent over-alkylation during the isoelectro focusing (IEF) step, excess of IAA was neutralized by adding an equimolar amount of DTT. Finally, samples were dissolved in the focusing/re-hydration solution, i.e. 7 M urea, 2 M thiourea, 4% CHAPS, and 15 mM dithioerythritol (DTE) and a 0.6% (v/v) carrier ampholyte cocktail, containing 40% of the pH 3.5–10 and 60% of the pH 4–8 intervals (BDH Biochemical, 44430 2F) and loaded onto home-made non-linear pH 3–10 strips.72 After IEF runs, the strips were equilibrated in 6 M Urea, 50 mM Tris-HCl pH 8.8, 2% (w/v) SDS, 30% (v/v) glycerol, and traces of bromophenol blue; the proteins were separated using a SDS-PAGE (T% 8–16) and transferred onto nitrocellulose membranes (Protran BA85, Whatman, 10402588) with a semidry system. The membranes were saturated with 3% w/v polyvinyl-pyrrolidone (PVP) in TBS and incubated overnight separately with NKp44Fc, NKp30Fc, or NKp46Fc in 3% w/v BSA in TBS-Tween 0.15% v/v (TBS-T). Membranes were then rinsed in TBS-T and incubated with anti-human IgG HRP-conjugated mAb. HEK293T-SN-biot was subjected to the same procedure and immunoblotted with Neutravidin-HRP (ThermoFisher, 31001). For preparative experiments, SDS-gels were stained with “blue silver” colloidal Coomassie.73 Images were digitalized using ChemiDoc Touch (Bio-Rad) and analyzed with PDQuest software (Bio-Rad).
2-DE spot identification
Spots excised from 2D-PAGE were fully discolored and digested with trypsin. All mass spectrometric measurements were performed using a LTQ linear ion trap mass spectrometer (Thermo Electron) coupled to a HPLC Surveyor (Thermo Electron) equipped with a Jupiter C18 column 250 mm × 1 mm (Phenomenex). Protein identification was performed using SEQUEST software and searched against a Human protein database. Peptide MS/MS assignments were filtered following very high stringent criteria: Xcorr ≥1.9 for the singly charged ions, Xcorr ≥2.2 for doubly charged ions, Xcorr ≥3.7 for triply charged ions, peptide probability ≤0.01, delta Cn ≥0.1 and Rsp ≤4.26.
Immunoprecipitation
Immunoprecipitation experiments were carried out using 12 µg NKp44Fc, NKp30Fc, or DNAM-1Fc molecules linked to 50 μl Dynabeads Protein G (ThermoFisher, 10003D) and incubated O/N at 4°C with 400 µl concentrated HEK293T SN (corresponding to 500 µg proteins). Samples were eluted using non-reducing sample buffer (10% glycerol, 2% SDS, 62.5 mM Tris-HCl pH 6.8, 0.01% bromophenol blue) for 5ʹ at 60°C and subsequently analyzed by SDS-PAGE using TGX Stain-Free Acrylamide Solutions; membrane was probed with mouse anti-NID1 mAb followed by HRP-conjugated anti-mouse Ig mAb.
RT-PCR analysis
Total RNA was extracted using RNAeasy Mini Kit (Qiagen, 74,104) from the following cells: K562, K562-NID1, Bw, Bw-NID1. Oligo(dT)-primed cDNA was prepared by standard technique using a Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics, 04379012001) following manufacturer’s instructions. Amplifications were performed for 30 cycles utilizing Platinum TAQ DNA Polymerase (ThermoFisher, 10966034) with an annealing T of 58°C (β-actin) or 62°C (NID1). Primers used were: β-actin for 5ʹ ACTCCATCATGAAGTGTGACG and β-actin rev 5ʹ CATACTCCTGCTTGCTGATCC; NID1 for 5ʹ CTCCATTGGGCCTGTGAGG and NID1 rev 5ʹ AGACACGGGGGC GTCATC. PCR products (249 bp fragment for β-actin and 795 bp for NID1) were separated by electrophoresis on a 1.5% (w/v) agarose gel and visualized by ethidium bromide staining.
Stable cell transfectants
K562 cell line was transfected with pcDNA3.1-NID1 construct (Geneart, ThermoFisher) using JetPEI following manufacturer’s instructions. After 72 h cells were cultured in medium containing 1.2 mg/ml G418 sulphate. At the end of the selection period, surviving cells were sub-cloned by limiting dilution.
Bw-NID1 cells were prepared by retrovirus gene transfer. NID1 ORF cDNA was sub-cloned in pMXs-IG (IRES-GFP) retrovirus vector (kindly provided by Dr. Kitamura, Tokyo, Japan). The pMXs-IG-NID1 construct was transiently transfected into Plat-E packaging cell line in order to generate viral particles that were used to infect Bw cells. NID1-positive cells were sorted according to GFP expression; subsequently, NID1-positive cells were sub-cloned by limiting dilution. Bw-NKp44/DAP12 cells (Bw-NKp44) were obtained by a similar approach, utilizing pMXS-IG vector, in which NKp44 ORF and DAP12 ORF cDNAs were sub-cloned in the two available cloning sites (GFP-encoding sequence was replaced by DAP12 ORF cDNA). NKp44-expressing cells were sorted and sub-cloned by limiting dilution. Bw-NKp30-CD3ζ cells (Bw-NKp30), expressing a chimeric molecule composed of the extracellular region of NKp30 fused to murine CD3ζ, were kindly provided by Eric Vivier (Marseille, France), and were sorted and sub-cloned by limiting dilution.
For the recovery of SN from K562 and BW cells and transfectants, cell culture was carried out either in RPMI/1% FCS or in protein-free CD medium and SN was collected after 48–72 h. In the latter case, SN were concentrated up to 10-fold using Amicon Ultracel-10K.
Functional assays on Bw cells
1 × 105 Bw cells were pre-treated 1 h at 37°C with 20 or 7.5 μg/ml rNID1 or with 100 μl SN obtained from wild type or transfected K562 and Bw cells cultured in RPMI/1% FCS, and subsequently plated on 96-well cell culture plates (1 × 105 cells/well) coated with 5 μg/ml goat anti-mouse IgG (GAM, MP Cappel, 55481) alone or with GAM plus anti-NKp44 or -NKp30 mAbs. Alternatively, pretreated cells were stimulated with rPDGF-DD (R&D, 1159-SB) at 250 and 25 ng/ml. For co-culture experiments, Bw cells were pre-incubated 1 h at 37°C with wild type or NID1-transfected K562 and Bw cells at an effector/target (E/T) ratio of 1:1 and then transferred on mAb-coated plates. In other experiments, Bw-NKp44 cells were incubated on plates in which rNID1 was coated either directly or through anti-NID1 or anti-His mAbs. For all these assays, after 20 h at 37°C, SN were collected and analyzed for their IL-2 content by ELISA using the Mouse IL-2 ELISA Ready-SET-Go (eBioscience, BMS88-7024-77) according to manufacturer’s instructions. Each sample was run in duplicate.
Generation of polyclonal NK cell lines
NK cells were derived from healthy donors. Approval was obtained from the ethical committee of IRCCS Ospedale Policlinico San Martino (39/2012) of Genova (Italy), and informed consent was provided according to the Declaration of Helsinki. NK cells were purified from peripheral blood using the RosetteSep™ NK Cell Enrichment Cocktail (StemCell Technologies, 15025). Those populations displaying more than 95% of CD56+CD3−CD14− NK cells were selected. Polyclonal NK cell lines were obtained by culturing purified NK cells at appropriate dilutions on irradiated feeder cells in the presence of 100 U/mL rhIL-2 (Proleukin, Novartis) and 1,5 ng/mL phytohemagglutinin (PHA, Gibco Ltd, 10576-015) in round-bottomed 96-well microtiter plates. After 3/4 weeks of culture the expanded NK cells were used for NK cell stimulation experiments.
Functional assays on NK cells
Evaluation of NK cell-mediated cytotoxicity against Bw and Bw-NID-1 transfected cells or against the P815 FcγR+ murine cell line (redirected killing assay) was done in a 4-h 51Cr-release test. In the redirected killing assay mAbs were added at the final concentration of 1.25 μg/ml (anti-NKp30 and anti-NKp46 mAbs) or 0.5 μg/ml (anti-NKp44 mAb) (appropriate concentration was determined after titration to induce specific functional response in NK cells). When indicated, K562-SN or K562-NID1-SN was added to NK cells (50% v/v) 1h before the onset of the test.
For the IFN-γ secretion assay, NK cells (10 × 104/well) were cultured overnight in 96-well microtiter plates pre-coated with GAM either in the absence or in the presence of anti-NKp44, -NKp30, -NKp46 mAbs. Alternatively, NK cells were stimulated with rPDGF-DD at 250 and 50 ng/ml. When indicated, K562-SN or K562-NID1-SN was added at the onset of culture. The culture SN were then collected and analyzed for the presence of IFN-γ using the IFN gamma Human ELISA Kit (ThermoFisher, EHIFNG).
Imaging flow cytometry
HEK293T cells were incubated with anti-NID1 mAb followed by PE-conjugated anti-IgG1 mAb. Prior to analysis, cells were stained with the nuclear dye Hoechst 33342 (ThermoFisher, 62249) (1:1000 dilution). Cells were acquired with a 12 channel MultiMag system ImageStreamX Mark II imaging flow cytometer (IFC) (Merck) using INSPIRE acquisition software (Amnis Corporation). Three ImageStream channels were used: channel 1 for the brightfield, channel 3 for NID1-PE (excited by a 488-nm laser), channel 7 for Hoechst 33342 (excited by a 405-nm laser). In order to collect only on focus-single-cells, we first restricted our attention on the brightfield (BF) parameters analysis. Single cells were selected using a biparametric dot plot representing BF aspect ratio (i.e. width/height) versus BF cell area then, by mean of the BF gradient RMS (root mean square), which measures the sharpness quality of an image, we gated only on focus cells. For each staining condition, 4,000 raw images were collected using a 40x objective. To avoid spectral overlaps, single-color compensation controls (500 cells each) were gathered and a compensation matrix was generated. The raw image files were then analyzed using IDEAS 6.0.3 software (Amnis Corporation) setting compensation on the basis of the calculated matrix. To verify the correspondence between data derived from traditional cytometry and IFC, we compared NID expression with its negative control (i.e. cells labeled only with secondary PE-conjugated mAb) by mean of their MFI (mean fluorescence intensity). To grant a better visualization of these data, we merged the two files.
Flow cytometry
Cells were incubated with the primary mAbs or Fc molecules (20 μg/ml) for 30 minutes at 4°C, washed, and stained with the appropriate PE- isotype-matched secondary mAbs for 30ʹ at 4°C. Bw-NKp44 and Bw-NKp30 cells were incubated with PE-conjugated anti-integrin mAbs for 30ʹ at 4°C. All samples were analyzed using a FACSCalibur flow cytometer and CellQuest Pro software (BD Biosciences).
Silencing experiments
NID1 expression was transiently silenced in HEK293T cells by siRNA transfection using iBONi siRNA 4-duplexes Plus (Riboxx Life Sciences). In particular, the following siRNA were used: siRNA-NID1 (ID4811_4): Sense 5ʹ-UAAACCAUCUUGUCCACGCCCCC-3ʹ, Antisense 5ʹ-GGGGGCGUGGACAAGAUGGUUUA-3ʹ; siRNA-CTR (iBONi Negative Control-N3): Sense 5ʹ-ACAACAUUCAUAUAGCUGCCCCC-3ʹ, Antisense 5ʹ-GGGGGCAGCUAUAUGAAUGUUGU-3ʹ.
siRNA (300 nM) were introduced in HEK293T cells by Nucleofector technology (Lonza) using Amaxa Cell Line Nucleofector Kit V (Lonza, VACA-1003); after 48 and 72 h cells were analyzed by flow cytometry with anti-NID1 mAb and NKp44Fc followed by appropriate PE-conjugated secondary reagents. For the analysis of SN derived from NID1-silenced cells, siRNA (50 nM) were transfected in HEK293T cells with Interferin (Polyplus, 409-10) following manufacturer’s instructions. Cells were then cultured in CD medium; after 72 h SN were collected, concentrated, and analyzed by Western blot with anti-NID1 mAb and NKp44Fc followed by appropriate HRP-conjugated secondary reagents.
Proteomic analysis of polyclonal NK cell populations
Polyclonal IL-2-activated NK cells derived from four different healthy donors were cultured for 20 h at 37°C in FCS- and IL-2-deprived medium in the absence or in the presence of rNID1 (20 μg/ml) directly coated on the plate or of anti-NKp44 mAb coated via GAM. Next, for subsequent proteomic analysis, cells were processed by in-StageTip (iST) method.74 Briefly, the pellets were lysed, reduced, alkylated in a single step using a buffer containing 2% (w/v) SDC (sodium deoxycholate), 10 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride), 40 mM CAA (chloroacetamide), 100 mM Tris HCl pH 8.0, and loaded into StageTip. The lysates were diluted with 25 mM Tris pH 8.5 containing 1 µg of trypsin. Samples were acidified with 100 µl of 1% (v/v) TFA (Trifluoroacetic Acid) and washed three times with 0.2% (v/v) TFA. Elutions were performed with 60 µl of 5% (v/v) ammonium hydroxide, 80% (v/v) ACN.
Samples were loaded from the sample loop directly into a 75-μm ID × 50 cm 2 μm, 100 Å C18 column mounted in the thermostated column compartment and the peptides were separated with increasing organic solvent at a flow rate of 250 nl/min using a non-linear gradient of 5–45% solution B (80% CAN and 20% H2O, 5% DMSO, 0.1% FA) in 180 min.
Eluting peptides were analyzed using an Orbitrap Fusion Tribrid mass spectrometer (ThermoFisher Scientific). Orbitrap detection was used for both MS1 and MS2 measurements at resolving powers of 120 K and 30 K (at m/z 200), respectively. Data dependent MS/MS analysis was performed in top speed mode with a 2 sec. cycle time, during which precursors detected within the range of m/z 375 − 1500 were selected for activation in order of abundance. Quadrupole isolation with a 1.8 m/z isolation window was used, and dynamic exclusion was enabled for 30s. Automatic gain control targets were 2.5 × 105 for MS1 and 5 × 104 for MS2, with 50 and 54 ms maximum injection times, respectively. The signal intensity threshold for MS2 was 1 × 104. HCD was performed using 28% normalized collision energy. One microscan was used for both MS1 and MS2 events.
MaxQuant software,75 version 1.6.0.1, was used to process the raw data, setting a false discovery rate (FDR) of 0.01 for the identification of proteins, peptides and PSM (peptide-spectrum match), a minimum length of 6 amino acids for peptide identification was required. Andromeda engine, incorporated into MaxQuant software, was used to search MS/MS spectra against Uniprot human database (release UP000005640_9606 February 2017). In the processing the variable modifications are Acetyl (Protein N-Term) Oxidation (M), Deamidation (NQ); on the contrary the Carbamidomethyl (C) was selected as fixed modification. The intensity values were extracted and statistically evaluated using the ProteinGroup Table and Perseus software. Algorithm MaxLFQ was chosen for the protein quantification with the activated option ‘match between runs’ to reduce the number of the missing proteins.
Statistical analyses
Data were analyzed using GraphPad Prism 6, R, or Perseus software. Details of statistical analysis for each experiment are indicated in the corresponding figure legend.
Data availability statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE76 partner repository with the dataset identifier PXD008252.
Project Name: Nidogen-1 is a novel extracellular ligand for the NKp44 activating receptor
Project accession: PXD008252
Funding Statement
This work was supported by grants awarded by Associazione Italiana Ricerca sul Cancro AIRC: IG 2014 project n. 15283 (L.M.), “Special Program Molecular Clinical Oncology 5x1000” project n. 9962 (A.M., L.M.), IG 2014 project n. 15428 (M.V.) and by funds from Renal Child Foundation (M.B., L.S., G.C.). M.P. is recipient of the AIRC fellowship n. 18274, year 2016
Acknowledgments
While this paper was under revision, Alessandro Moretta passed away on February 17th 2018. His groundbreaking discoveries of KIRs and NCR (including NKp44, the focus of this contribution) are true milestones in Immunology and Medicine. The great success in the clinical outcome of patients with otherwise lethal leukemia in the haploidentical HSCT setting are largely based on Alessandro’s discoveries. We sadly miss his lighted scientific insight and, even more, his generosity, humanity, irony, and smile. We want to dedicate the present study to Alessandro’s memory.
Competing financial interests
A.M. was a co‐founder and shareholder of Innate-Pharma (Marseille, France). The other authors declare no competing financial interests.
Author contributions
M.B., A.P., G.C., M.V., C.C designed and coordinated research; S.G., performed most of the experiments; M.P. performed experiments on NK cells; M.B., A.P., C.L., and L.S. performed proteomic experiments; G.DZ. performed imaging flow cytometry and cell sorting experiments; C.P. and A.B. prepared cell transfectants and expression constructs; S.G., M.B., A.P., C.B., G.C., A.M., M.V., C.C analyzed data; A.M., M.V., and L.M. provided financial support; A.M., M.V., L.M., and C.C. wrote the paper.
All Authors reviewed the manuscript.
Supplemental Material
Supplemental data for this article can be accessed here.
References
- 1.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. 2013. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol 13(2):145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 2.Montaldo E, Vacca P, Moretta L, Mingari MC.. 2014. Development of human natural killer cells and other innate lymphoid cells. Semin Immunol 26(2):107–113. doi: 10.1016/j.smim.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S.. 2011. Innate or adaptive immunity? The example of natural killer cells. Science 331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caligiuri MA. 2008. Human natural killer cells. Blood 112(3):461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moretta A, Marcenaro E, Sivori S, Chiesa MD, Vitale M, Moretta L. 2005. Early liaisons between cells of the innate immune system in inflamed peripheral tissues. Trends Immunol 26(12):668–675. doi: 10.1016/j.it.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Thorén FB, Riise RE, Ousbäck J, Della Chiesa M, Alsterholm M, Marcenaro E, Pesce S, Prato C, Cantoni C, Bylund J, et al. 2012. Human NK Cells induce neutrophil apoptosis via an NKp46- and Fas-dependent mechanism. J Immunol Baltim Md 1950 188(4):1668–1674. doi: 10.4049/jimmunol.1102002. [DOI] [PubMed] [Google Scholar]
- 7.Pesce S, Thoren FB, Cantoni C, Prato C, Moretta L, Moretta A, Marcenaro E. 2017. The innate immune cross talk between NK cells and eosinophils is regulated by the interaction of natural cytotoxicity receptors with eosinophil surface ligands. Front Immunol 8:510. doi: 10.3389/fimmu.2017.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellora F, Castriconi R, Dondero A, Reggiardo G, Moretta L, Mantovani A, Moretta A, Bottino C. 2010. The interaction of human natural killer cells with either unpolarized or polarized macrophages results in different functional outcomes. Proc Natl Acad Sci USA 107(50):21659–21664. doi: 10.1073/pnas.1007654108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martín-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, Sallusto F. 2004. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol 5(12):1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 10.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, Biassoni R, Moretta L. 2001. Activating receptors and co-receptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol 19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 11.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. 2013. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol 31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch J, Steinle A, Watzl C, Mandelboim O. 2013. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol 34(4):182–191. doi: 10.1016/j.it.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, Biassoni R, Moretta A. 1998. Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J Exp Med 188(5):953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, Marcenaro E, Accame L, Malaspina A, Biassoni R, et al. 1999. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med 190(10):1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, Parolini S, Moretta L, Moretta A, Biassoni R. 1999. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med 189(5):787–796. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terme M, Ullrich E, Delahaye NF, Chaput N, Zitvogel L. 2008. Natural killer cell-directed therapies: moving from unexpected results to successful strategies. Nat Immunol 9(5):486–494. doi: 10.1038/ni1580. [DOI] [PubMed] [Google Scholar]
- 17.Burke S, Lakshmikanth T, Colucci F, Carbone E. 2010. New views on natural killer cell-based immunotherapy for melanoma treatment. Trends Immunol 31(9):339–345. doi: 10.1016/j.it.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Moretta L, Pietra G, Montaldo E, Vacca P, Pende D, Falco M, Del Zotto G, Locatelli F, Moretta A, Mingari MC. 2014. Human NK cells: from surface receptors to the therapy of leukemias and solid tumors. Front Immunol 5. doi: 10.3389/fimmu.2014.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohrt HE, Thielens A, Marabelle A, Sagiv-Barfi I, Sola C, Chanuc F, Fuseri N, Bonnafous C, Czerwinski D, Rajapaksa A, et al. 2014. Anti-KIR antibody enhancement of anti-lymphoma activity of natural killer cells as monotherapy and in combination with anti-CD20 antibodies. Blood 123(5):678–686. doi: 10.1182/blood-2013-08-519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knorr DA, Bachanova V, Verneris MR, Miller JS. 2014. Clinical utility of natural killer cells in cancer therapy and transplantation. Semin Immunol 26(2):161–172. doi: 10.1016/j.smim.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantoni C, Grauwet K, Pietra G, Parodi M, Mingari MC, Maria AD, Favoreel H, Vitale M. 2015. Role of NK cells in immunotherapy and virotherapy of solid tumors. Immunotherapy 7(8):861–882. doi: 10.2217/imt.15.53. [DOI] [PubMed] [Google Scholar]
- 22.Kruse PH, Matta J, Ugolini S, Vivier E. 2014. Natural cytotoxicity receptors and their ligands. Immunol Cell Biol 92(3):221–229. doi: 10.1038/icb.2013.98. [DOI] [PubMed] [Google Scholar]
- 23.Arnon TI, Markel G, Mandelboim O. 2006. Tumor and viral recognition by natural killer cells receptors. Semin Cancer Biol 16(5):348–358. doi: 10.1016/j.semcancer.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Brandt CS, Baratin M, Yi EC, Kennedy J, Gao Z, Fox B, Haldeman B, Ostrander CD, Kaifu T, Chabannon C, et al. 2009. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med 206(7):1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogge von Strandmann E, Simhadri VR, von Tresckow B, Sasse S, Reiners KS, Hansen HP, Rothe A, Böll B, Simhadri VL, Borchmann P, et al. 2007. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity 27(6):965–974. doi: 10.1016/j.immuni.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Baychelier F, Sennepin A, Ermonval M, Dorgham K, Debré P, Vieillard V. 2013. Identification of a cellular ligand for the natural cytotoxicity receptor NKp44. Blood 122(17):2935–2942. doi: 10.1182/blood-2013-03-489054. [DOI] [PubMed] [Google Scholar]
- 27.Hecht M-L, Rosental B, Horlacher T, Hershkovitz O, De Paz JL, Noti C, Schauer S, Porgador A, Seeberger PH. 2009. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J Proteome Res 8(2):712–720. doi: 10.1021/pr800747c. [DOI] [PubMed] [Google Scholar]
- 28.Brusilovsky M, Radinsky O, Cohen L, Yossef R, Shemesh A, Braiman A, Mandelboim O, Campbell KS, Porgador A. 2015. Regulation of natural cytotoxicity receptors by heparan sulfate proteoglycans in -cis: A lesson from NKp44. Eur J Immunol 45(4):1180–1191. doi: 10.1002/eji.201445177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosental B, Brusilovsky M, Hadad U, Oz D, Appel MY, Afergan F, Yossef R, Rosenberg LA, Aharoni A, Cerwenka A, et al. 2011. Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. J Immunol Baltim Md 1950 187(11):5693–5702. doi: 10.4049/jimmunol.1102267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton NC, Mathew SO, Mathew PA. 2013. Novel interaction between proliferating cell nuclear antigen and HLA I on the surface of tumor cells inhibits NK cell function through NKp44. PloS One 8(3):e59552. doi: 10.1371/journal.pone.0059552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simhadri VR, Reiners KS, Hansen HP, Topolar D, Simhadri VL, Nohroudi K, Kufer TA, Engert A, Pogge von Strandmann E. 2008. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PloS One 3(10):e3377. doi: 10.1371/journal.pone.0003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Gazzar A, Groh V, Spies T. 2013. Immunobiology and conflicting roles of the human NKG2D lymphocyte receptor and its ligands in cancer. J Immunol Baltim Md 1950 191(4):1509–1515. doi: 10.4049/jimmunol.1301071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanier LL. 2015. NKG2D receptor and its ligands in host defense. Cancer Immunol Res 3(6):575–582. doi: 10.1158/2326-6066.CIR-15-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibuya A, Campbell D, Hannum C, Yssel H, Franz-Bacon K, McClanahan T, Kitamura T, Nicholl J, Sutherland GR, Lanier LL, et al. 1996. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 4(6):573–581. doi: 10.1016/S1074-7613(00)70060-4. [DOI] [PubMed] [Google Scholar]
- 35.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. 2013. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol 31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, Cantoni C, Grassi J, Marcenaro S, Reymond N, et al. 2003. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med 198(4):557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mamessier E, Sylvain A, Bertucci F, Castellano R, Finetti P, Houvenaeghel G, Charaffe-Jaufret E, Birnbaum D, Moretta A, Olive D. 2011. Human breast tumor cells induce self-tolerance mechanisms to avoid NKG2D-mediated and DNAM-mediated NK cell recognition. Cancer Res 71(21):6621–6632. doi: 10.1158/0008-5472.CAN-11-0792. [DOI] [PubMed] [Google Scholar]
- 38.Stojanovic A, Correia MP, Cerwenka A. 2013. Shaping of NK cell responses by the tumor microenvironment. Cancer Microenviron. Off J Int Cancer Microenviron Soc 6(2):135–146. doi: 10.1007/s12307-012-0125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vitale M, Cantoni C, Pietra G, Mingari MC, Moretta L. 2014. Effect of tumor cells and tumor microenvironment on NK-cell function. Eur J Immunol 44(6):1582–1592. doi: 10.1002/eji.201344272. [DOI] [PubMed] [Google Scholar]
- 40.Rusakiewicz S, Perier A, Semeraro M, Pitt JM, Pogge Von Strandmann E, Reiners KS, Aspeslagh S, Pipéroglou C, Vély F, Ivagnes A, et al. 2017. NKp30 isoforms and NKp30 ligands are predictive biomarkers of response to imatinib mesylate in metastatic GIST patients. Oncoimmunology 6(1):e1137418. doi: 10.1080/2162402X.2015.1137418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groh V, Wu J, Yee C, Spies T. 2002. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature 419(6908):734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 42.Fernández-Messina L, Ashiru O, Boutet P, Agüera-González S, Skepper JN, Reyburn HT, Valés-Gómez M. 2010. Differential mechanisms of shedding of the glycosylphosphatidylinositol (GPI)-anchored NKG2D ligands. J Biol Chem 285(12):8543–8551. doi: 10.1074/jbc.M109.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baury B, Masson D, McDermott BM, Jarry A, Blottière HM, Blanchardie P, Laboisse CL, Lustenberger P, Racaniello VR, Denis MG. 2003. Identification of secreted CD155 isoforms. Biochem Biophys Res Commun 309(1):175–182. doi: 10.1016/S0006-291X(03)01560-2. [DOI] [PubMed] [Google Scholar]
- 44.Iguchi-Manaka A, Okumura G, Kojima H, Cho Y, Hirochika R, Bando H, Sato T, Yoshikawa H, Hara H, Shibuya A, et al. 2016. Increased Soluble CD155 in the Serum of Cancer Patients. PloS One 11(4):e0152982. doi: 10.1371/journal.pone.0152982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasero C, Gravis G, Guerin M, Granjeaud S, Thomassin-Piana J, Rocchi P, Paciencia-Gros M, Poizat F, Bentobji M, Azario-Cheillan F, et al. 2016. Inherent and tumor-driven immune tolerance in the prostate microenvironment impairs natural killer cell antitumor activity. Cancer Res 76(8):2153–2165. doi: 10.1158/0008-5472.CAN-15-1965. [DOI] [PubMed] [Google Scholar]
- 46.Galluzzi L, Zitvogel L, Kroemer G. 2016. Immunological mechanisms underneath the efficacy of cancer therapy. Cancer Immunol Res 4(11):895–902. doi: 10.1158/2326-6066.CIR-16-0197. [DOI] [PubMed] [Google Scholar]
- 47.Schlecker E, Fiegler N, Arnold A, Altevogt P, Rose-John S, Moldenhauer G, Sucker A, Paschen A, von Strandmann EP, Textor S, et al. 2014. Metalloprotease-mediated tumor cell shedding of B7-H6, the ligand of the natural killer cell-activating receptor NKp30. Cancer Res 74(13):3429–3440. doi: 10.1158/0008-5472.CAN-13-3017. [DOI] [PubMed] [Google Scholar]
- 48.Semeraro M, Rusakiewicz S, Minard-Colin V, Delahaye NF, Enot D, Vély F, Marabelle A, Papoular B, Piperoglou C, Ponzoni M, et al. 2015. Clinical impact of the NKp30/B7-H6 axis in high-risk neuroblastoma patients. Sci Transl Med 7(283):283ra55. doi: 10.1126/scitranslmed.aaa2327. [DOI] [PubMed] [Google Scholar]
- 49.Pesce S, Tabellini G, Cantoni C, Patrizi O, Coltrini D, Rampinelli F, Matta J, Vivier E, Moretta A, Parolini S, et al. 2015. B7-H6-mediated downregulation of NKp30 in NK cells contributes to ovarian carcinoma immune escape. Oncoimmunology 4(4):e1001224. doi: 10.1080/2162402X.2014.1001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reiners KS, Topolar D, Henke A, Simhadri VR, Kessler J, Sauer M, Bessler M, Hansen HP, Tawadros S, Herling M, et al. 2013. Soluble ligands for NK cell receptors promote evasion of chronic lymphocytic leukemia cells from NK cell anti-tumor activity. Blood 121(18):3658–3665. doi: 10.1182/blood-2013-01-476606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narni-Mancinelli E, Gauthier L, Baratin M, Guia S, Fenis A, Deghmane A-E, Rossi B, Fourquet P, Escalière B, Kerdiles YM, et al. 2017. Complement factor P is a ligand for the natural killer cell-activating receptor NKp46. Sci Immunol 2:10. doi: 10.1126/sciimmunol.aam9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barrow AD, Edeling MA, Trifonov V, Luo J, Goyal P, Bohl B, Bando JK, Kim AH, Walker J, Andahazy M, et al. 2018. Natural killer cells control tumor growth by sensing a growth factor. Cell 172(3):534–548.e19. doi: 10.1016/j.cell.2017.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dedhar S, Jewell K, Rojiani M, Gray V. 1992. The receptor for the basement membrane glycoprotein entactin is the integrin alpha 3/beta 1. J Biol Chem 267(26):18908–18914. [PubMed] [Google Scholar]
- 54.Yelian FD, Edgeworth NA, Dong LJ, Chung AE, Armant DR. 1993. Recombinant entactin promotes mouse primary trophoblast cell adhesion and migration through the Arg-Gly-Asp (RGD) recognition sequence. J Cell Biol 121(4):923–929. doi: 10.1083/jcb.121.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alsultan A, Shamseldin HE, Osman ME, Aljabri M, Alkuraya FS. 2013. MYSM1 is mutated in a family with transient transfusion-dependent anemia, mild thrombocytopenia, and low NK- and B-cell counts. Blood 122(23):3844–3845. doi: 10.1182/blood-2013-09-527127. [DOI] [PubMed] [Google Scholar]
- 56.Nandakumar V, Chou Y, Zang L, Huang XF, Chen S-Y. 2013. Epigenetic control of natural killer cell maturation by histone H2A deubiquitinase, MYSM1. Proc Natl Acad Sci USA 110(41):E3927–3936. doi: 10.1073/pnas.1308888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho MSP, Böse K, Mokkapati S, Nischt R, Smyth N. 2008. Nidogens-extracellular matrix linker molecules. Microsc Res Tech 71(5):387–395. doi: 10.1002/jemt.20567. [DOI] [PubMed] [Google Scholar]
- 58.Kruegel J, Miosge N. 2010. Basement membrane components are key players in specialized extracellular matrices. Cell Mol Life Sci CMLS 67(17):2879–2895. doi: 10.1007/s00018-010-0367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JKM, Doherty JM, Mills JC, Colonna M. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457(7230):722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montaldo E, Juelke K, Romagnani C. 2015. Group 3 innate lymphoid cells (ILC3s): origin, differentiation, and plasticity in humans and mice. Eur J Immunol 45(8):2171–2182. doi: 10.1002/eji.201545598. [DOI] [PubMed] [Google Scholar]
- 61.Björkström NK, Ljunggren H-G, Michaëlsson J. 2016. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol 16(5):310–320. doi: 10.1038/nri.2016.34. [DOI] [PubMed] [Google Scholar]
- 62.Glatzer T, Killig M, Meisig J, Ommert I, Luetke-Eversloh M, Babic M, Paclik D, Blüthgen N, Seidl R, Seifarth C, et al. 2013. RORγt+ innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. Immunity 38(6):1223–1235. doi: 10.1016/j.immuni.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 63.Li L, Zhang Y, Li N, Feng L, Yao H, Zhang R, Li B, Li X, Han N, Gao Y, et al. 2015. Nidogen-1: a candidate biomarker for ovarian serous cancer. Jpn J Clin Oncol 45(2):176–182. doi: 10.1093/jjco/hyu187. [DOI] [PubMed] [Google Scholar]
- 64.Willumsen N, Bager CL, Leeming DJ, Bay-Jensen A-C, Karsdal MA. 2017. Nidogen-1 degraded by cathepsin S can be quantified in serum and is associated with non-small cell lung cancer. Neoplasia N. Y. N 19(4):271–278. doi: 10.1016/j.neo.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ulazzi L, Sabbioni S, Miotto E, Veronese A, Angusti A, Gafà R, Manfredini S, Farinati F, Sasaki T, Lanza G, et al. 2007. Nidogen 1 and 2 gene promoters are aberrantly methylated in human gastrointestinal cancer. Mol Cancer 6:17. doi: 10.1186/1476-4598-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou Y, Zhu Y, Fan X, Zhang C, Wang Y, Zhang L, Zhang H, Wen T, Zhang K, Huo X, et al. 2017. NID1, a new regulator of EMT required for metastasis and chemoresistance of ovarian cancer cells. Oncotarget 8(20):33110–33121. doi: 10.18632/oncotarget.16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedrola N, Devis L, Llauradó M, Campoy I, Martinez-Garcia E, Garcia M, Muinelo-Romay L, Alonso-Alconada L, Abal M, Alameda F, et al. 2015. Nidogen 1 and nuclear protein 1: novel targets of ETV5 transcription factor involved in endometrial cancer invasion. Clin Exp Metastasis 32(5):467–478. doi: 10.1007/s10585-015-9720-7. [DOI] [PubMed] [Google Scholar]
- 68.Alečković M, Wei Y, LeRoy G, Sidoli S, Liu DD, Garcia BA, Kang Y. 2017. Identification of nidogen 1 as a lung metastasis protein through secretome analysis. Genes Dev 31(14):1439–1455. doi: 10.1101/gad.301937.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang W, Guo H, Geng J, Zheng X, Wei H, Sun R, Tian Z. 2014. Tumor-released galectin-3, a soluble inhibitory ligand of human NKp30, plays an important role in tumor escape from NK cell attack. J Biol Chem 289(48):33311–33319. doi: 10.1074/jbc.M114.603464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martino-Echarri E, Fernández-Rodríguez R, Rodríguez-Baena FJ, Barrientos-Durán A, Torres-Collado AX, Plaza-Calonge Mdel C, Amador-Cubero S, Cortés J, Reynolds LE, Hodivala-Dilke KM, et al. 2013. Contribution of ADAMTS1 as a tumor suppressor gene in human breast carcinoma. Linking its tumor inhibitory properties to its proteolytic activity on nidogen-1 and nidogen-2. Int J Cancer 133(10):2315–2324. doi: 10.1002/ijc.28271. [DOI] [PubMed] [Google Scholar]
- 71.Vacca P, Cantoni C, Prato C, Fulcheri E, Moretta A, Moretta L, Mingari MC. 2008. Regulatory role of NKp44, NKp46, DNAM-1 and NKG2D receptors in the interaction between NK cells and trophoblast cells. Evidence for divergent functional profiles of decidual versus peripheral NK cells. Int Immunol 20(11):1395–1405. doi: 10.1093/intimm/dxn105. [DOI] [PubMed] [Google Scholar]
- 72.Bruschi M, Musante L, Candiano G, Ghiggeri GM, Herbert B, Antonucci F, Righetti PG. 2003. Soft immobilized pH gradient gels in proteome analysis: a follow-up. Proteomics 3(6):821–825. doi: 10.1002/pmic.200300361. [DOI] [PubMed] [Google Scholar]
- 73.Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. 2004. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25(9):1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 74.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11(3):319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 75.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 76.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016. update of the PRIDE database and its related tools. Nucleic Acids Res 44(D1):D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE76 partner repository with the dataset identifier PXD008252.
Project Name: Nidogen-1 is a novel extracellular ligand for the NKp44 activating receptor
Project accession: PXD008252


