Abstract
cDNA encoding an unclassified glutathione S-transferase (GST) of the diamondback moth, Plutella xylostella, was cloned by reverse transcriptase-polymerase chain reaction. The resulting clone was sequenced and the amino acid sequence deduced, revealing 67%–73% identities with unclassified GSTs from other organisms. A recombinant protein was functionally overexpressed in Escherichia coli cells in a soluble form and purified to homogeneity. The enzyme was capable to catalyze the transformation of 1-chloro-2,4-dinitrobenzene and ethacrynic acid with glutathione. A competition assay revealed that GST activity was inhibited by insecticides, suggesting that the enzyme could contribute to insecticide metabolism in the diamondback moth.
Keywords: Plutella xylostella, glutathione, glutathione transferase, Lepidoptera
Introduction
Glutathione conjugation is known to be a major pathway for the detoxification of xenobiotics as well as for the homeostasis of endogenous compounds. Glutathione S-transferases [GSTs, EC 2.5.1.18] are enzymes that are widespread in both prokaryotic and eukaryotic cells that catalyze the glutathione conjugation reaction with reduced glutathione (GSH).1,2) We have identified several GSTs (delta, epsilon, omega, sigma, theta, zeta, and unclassified) in the silkworm, Bombyx mori, a lepidopteran model insect,3–9) while six GST classes (delta, epsilon, omega, sigma, theta, and zeta) have been identified in dipteran insects, such as Anopheles gambiae10) and Drosophila melanogaster.11,12) In addition to the silkworm, we also characterized a sigma-class GST in the fall webworm Hyphantria cunea, one of the most serious lepidopteran pests of broad-leaved trees,6) and a delta-class GST in Nilaparvata lugens, which is a rice crop pest.13) Because Lepidoptera are major agricultural pests, it is extremely important to know lepidopteran GSTs.
In this study, we focused on an unclassified GST of the diamondback moth, Plutella xylostella (pxGSTu1), that is one of main pests of Brassicaceae vegetables in the world. Because the P. xylostella population with resistance to insecticides has increased (http://www.pesticideresistance.com/index.php), it is helpful to study its detoxification capacity, particularly through the novel GST identified in the current study. To investigate its properties, cDNA encoding this enzyme was sequenced and overexpressed as a recombinant protein in Escherichia coli cells.
Materials and Methods
1. Insects
P. xylostella was kindly supplied, reared, and maintained by Dr. K. Miyamoto of the National Institute of Agrobiological Sciences, NARO. Total RNA was isolated from last instar larvae of insects using Isogen (Nippon Gene, Tokyo, Japan) and an SV Total RNA Isolation System (Promega, Madison, WI, USA) in accordance with the manufacturer’s instructions.
2. Cloning and sequencing of the cDNA encoding pxGSTu1
Total RNA was subjected to reverse transcriptase-polymerase chain reaction (RT-PCR). First-strand cDNA was produced using SuperScript II Reverse Transcriptase (Thermo Fisher Scientific, Carlsbad, CA, USA) and an oligo-dT primer. The resulting cDNA was used as a template to amplify a DNA fragment by PCR with the following two oligonucleotide primers: 5′-ATT CCA ACATATGGTG CTG AAG CTG TAC GC-3′ (sense) and 5′-AAGGATCCTCA ATG CCT AAT TGG GTG GAA G-3′ (antisense). These were designed based on partial sequences obtained from the KONAGAbase EST database.14) The underlined and double-underlined regions are NdeI and BamHI restriction enzyme sites, respectively. These were incorporated for the purpose of subcloning the PCR product into an expression plasmid vector. PCR was conducted for one cycle at 94°C for 2 min, then for 35 cycles at 94°C for 1 min, 61°C for 1 min, and 72°C for 2 min, followed by one cycle at 72°C for 10 min. The pxGSTu1 cDNA (pxgstu1) obtained was ligated into the pGEM-T Easy Vector (Promega, Madison, WI, USA). GENETYX-MAC software (ver. 14.0.12, GENETYX Corp., Tokyo, Japan) was used for sequence analysis and homology alignment. The phylogenetic tree was prepared using neighbor-joining plot software (http://www-igbmc.u-strasbg.fr/Bioinfo/ClustulX/Top.html).
3. Overexpression and purification of the recombinant protein
The pxgstu1 cDNA was cloned into the pGEM-T Easy Vector, as described above. After digestion of the PCR product with NdeI and BamHI, the obtained fragment was subcloned into the NdeI–BamHI site of expression vector pET-15b (Novagen; EMD Biosciences, Inc., Darmstadt, Germany). Competent Escherichia coli Rosetta (DE3) pLysS cells (Novagen; EMD Millipore, USA) were transformed with a prepared expression plasmid that harbored pxgstu1 and were grown at 37°C on Luria–Bertani medium that contained 100 µg/mL ampicillin. After the cell density reached 0.7 OD600, isopropyl 1-thio-β-D-galactoside was added for a final concentration of 1 mM to induce the production of the recombinant protein. After further incubation overnight at 30°C, cells were harvested by centrifugation; homogenized in 20 mM Tris-HCl buffer (pH 8.0) that contained 0.5 M NaCl, 4 mg/mL of lysozyme, and 2×103 U Cryonase Cold-active Nuclease (Takara, Tokyo, Japan); and disrupted by sonication. Unless otherwise noted, all of the operations described below were conducted at 4°C. The supernatant was clarified by centrifugation at 10,000×g for 15 min and subjected to Ni2+-affinity chromatography equilibrated with 20 mM Tris-HCl buffer (pH 8.0) that contained 0.2 M NaCl. After washing with the same buffer, samples were eluted with a linear gradient of 0–0.5 M imidazole. The enzyme-containing fractions, assayed as described below, were pooled, concentrated using a centrifugal filter (Millipore Corp., Billerica, MA, USA), and applied to a Superdex 200 column (GE Healthcare Bio-Sciences, Buckinghamshire, UK) equilibrated with the same buffer, but with the addition of 0.2 M NaCl. Each fraction was assayed and analyzed by SDS-PAGE using a 15% polyacrylamide slab gel containing 0.1% SDS, in accordance with the methods of Laemmli.15) Protein bands were visualized by staining with Coomassie Brilliant Blue R-250.
4. Measuring enzyme activity
GST activity was measured spectrophotometrically using 1-chloro-2,4-dinitrobenzene (CDNB) and 5 mM GSH as standard substrates.16) Enzymatic activity was expressed as mol CDNB conjugated with GSH per min per mg of protein. Kinetic parameters (Km and kcat) were assessed with a nonlinear least-squares data fit under assay conditions with different substrate concentrations in the presence of 5 mM GSH.
Results
1. Cloning and sequencing of cDNA that encodes pxGSTu1
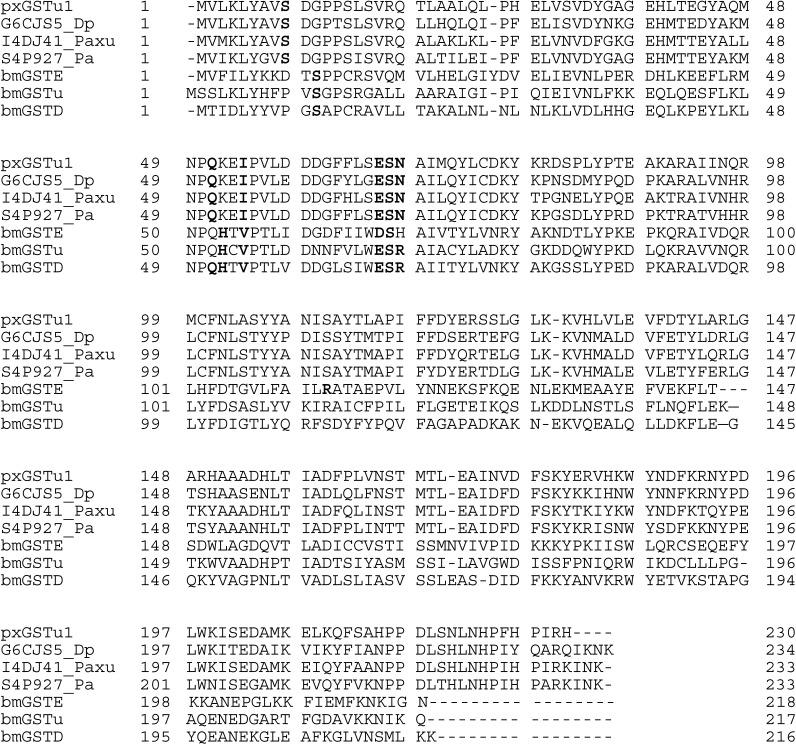
cDNA that encodes pxGSTu1 was obtained by RT-PCR using total RNA from P. xylostella. The nucleotide sequence was determined and deposited in GenBank with accession no. AB456582. It contains an open reading frame of 603 bp and encodes 230 amino acid residues (Fig. 1); its theoretical molecular mass and pI were evaluated to be 26,331 and 6.05, respectively. The deduced amino acid sequence of this putative GST showed identities 83%, 80%, 74%, and 32% homogeneous to homologs from Papilio xuthus (Asian swallowtail butterfly), Pararge aegeria (speckled wood butterfly), Danaus plexippus (monarch butterfly) and B. mori (silkmoth), respectively. In contrast, the sequence of pxGSTu1 showed homology with other classes, such as an epsilon-class GST of B. mori (bmGSTE; 30.3%), an unclassified GST (32%), and a delta-class GST of B. mori (bmGSTD; 34.4%). The GSH-binding site in the unclassified GST sequence is catalytically essential,8) and it is well conserved in pxGSTu1 (Fig. 1). The GSH-binding sites of other unclassified GSTs or classified general GSTs are highly conserved and catalytically essential and, in bmGSTD, are formed by residues Ser11, Gln51, His52, Val54, Glu66, Ser67, and Arg68.17) Equivalent residues Ser9, Gln51, Ile54, Glu66, Ser67, and Asn68 were observed in pxGSTu1 (Fig. 1).
Fig. 1. Alignment of GST amino acid sequences. Sequences of GSTs from different organisms were obtained from Swiss-Prot databases: pxGSTu1 (determined in the present study); G6CJS5_Dp (Danaus plexippus); I4DJ41_Paxu (Papilio xuthus); and S4P927_Pa (Pararge aegeria). Conserved GSH-binding site residues are shown in blue.
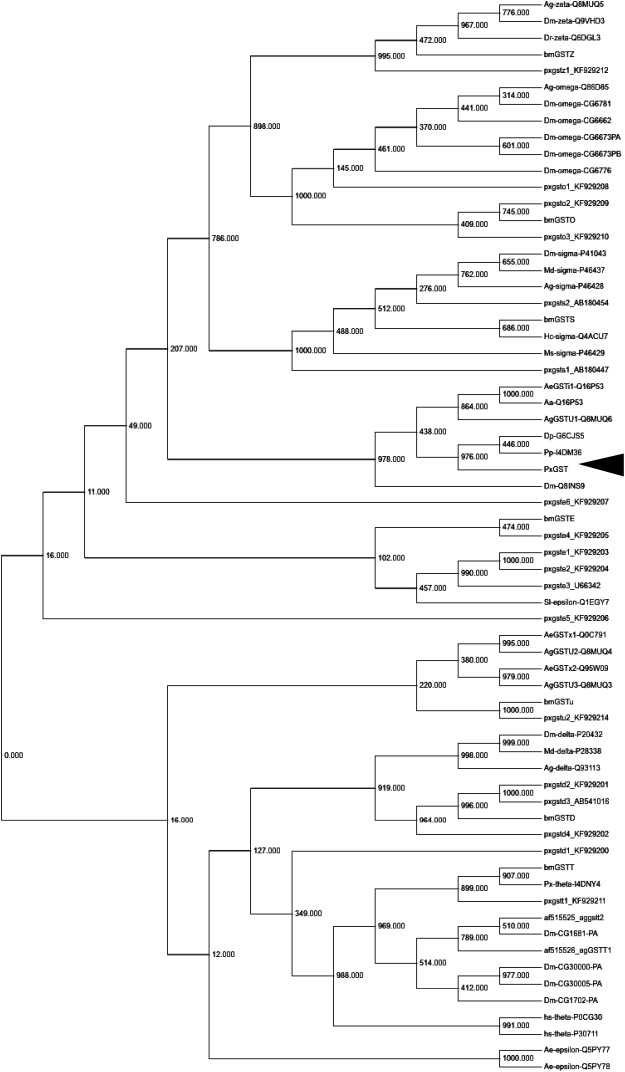
Based on the phylogenetic tree generated from the aligned amino acid sequences of the GSTs, the present GST was the closest to the unclassified GSTs of D. plexippus and P. polytes (Fig. 2). The phylogenetic trees and homogeneous amino acid identities indicated that the clone pxGSTu1 represented an unclassified GST enzyme.
Fig. 2. Phylogenetic analysis of GST amino acid sequences. The phylogenetic tree was constructed with neighbor-joining plot software using GST sequences cited from Swiss-Prot database (http://web.expasy.org/docs/swiss-prot_guideline.html). Each entry contains the species name, GST class, and accession number. Ag, A. gambiae; Md, Musca domestica, Dm, D. melanogaster; Ms, Manduca sexta; Hc, H. cunea; hs, Homo sapiens; bm, B. mori; Ae, Aedes aegypti; Sl, Spodoptera litura; Paxu, P. xuthus; px, P. xylostella; and numbers attached to nodes indicate bootstrap values. Arrow indicates pxGSTu1.
2. Overexpression and purification of pxGSTu1
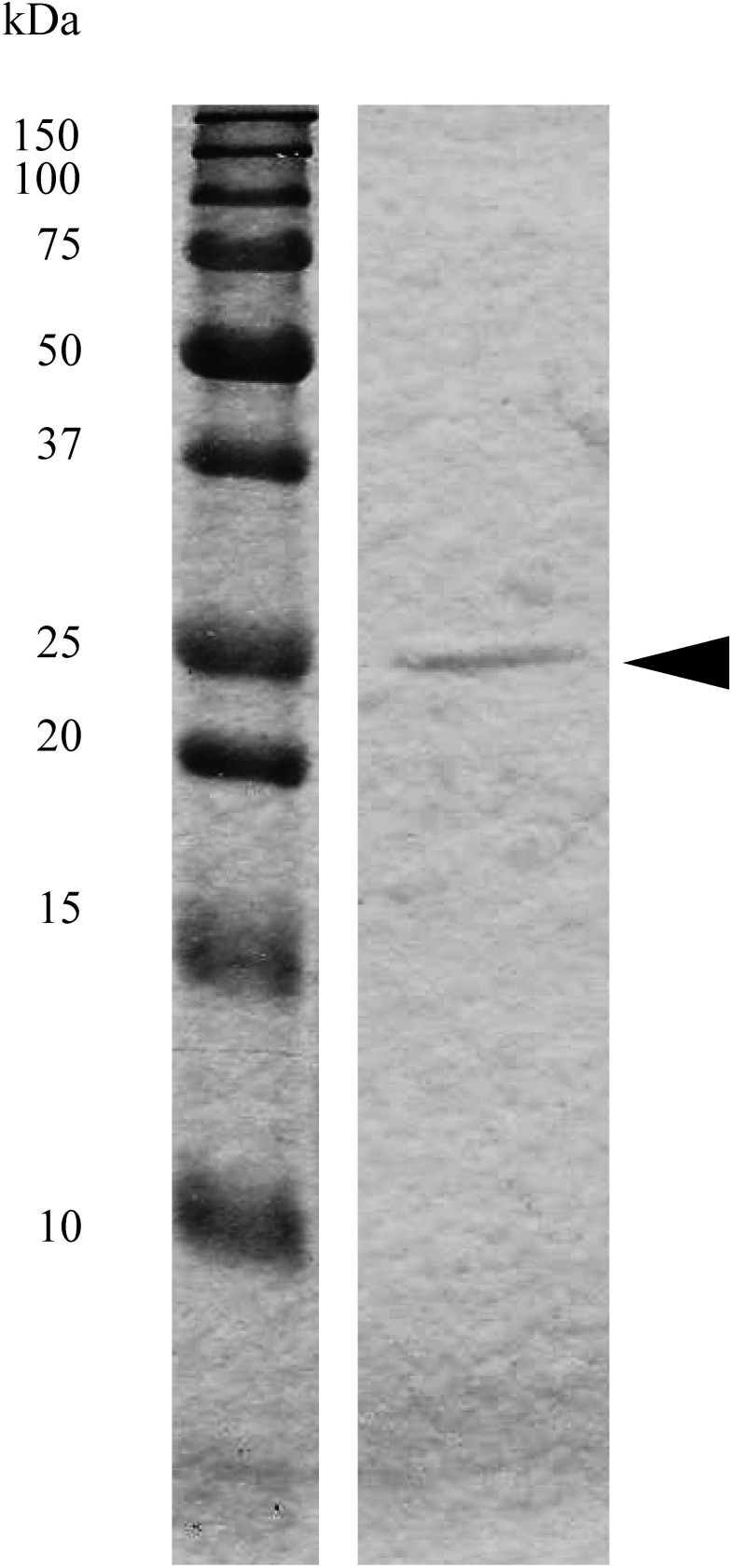
pxGSTu1 was overexpressed as a recombinant protein using an E. coli expression vector. The enzyme was in a soluble form and purified to homogeneity by affinity chromatography and gel filtration. The purified protein migrated with an apparent molecular weight of 25,000 (Fig. 3). There were insignificant differences in the molecular sizes of pxGSTu1 between the value calculated from the deduced amino acid sequence and that measured by SDS-PAGE: 26,331 and 25,000 from the sequence and SDS-PAGE, respectively. These sizes were similar to those of isolated lepidopteran GSTs.3–9) Finally, we obtained 1.2 mg of highly purified pxGSTu1 from 250 mL of Luria-Bertani growth medium. The specific activity of the final preparation toward CDNB was 0.041 µmol/min/mg (Table 1). Thus, pxGSTu1 was successfully overexpressed in a soluble form in E. coli cells.
Fig. 3. Electropherograms of pxGSTu1 after purification. Purified protein was subjected to 15% SDS-PAGE followed by staining with Coomassie Brilliant blue. A, Lane 1, protein molecular size markers; lane 2, pxGSTu1 purified by the methods described in the text. Arrow indicates the purified pxGSTu1.
Table 1. Substrate specificity of pxGSTu1.
| Substrate | Concentration (mM) | Activity (µmol/min/mg) | Wavelength (nm) | Δε (/mM/cm) |
|---|---|---|---|---|
| CDNB | 1.0 | 0.041 | 340 | 9.6 |
| EPNP | 1.0 | NA | 260 | 0.5 |
| 4NBC | 1.0 | NA | 310 | 1.9 |
| 4NPB | 1.0 | NA | 310 | 1.2 |
| 4HNE | 0.1 | NA | 224 | 13.8 |
| ECA | 0.2 | 0.13 | 270 | 5.0 |
| 4NPA | 1.0 | NA | 400 | 8.3 |
| H2O2 | 0.2 | NA | 340 | −6.2 |
Activity was measured at pH 8 in the presence of 5 mM GSH. Data are expressed as means of three independent experiments. NA represents no activity. Wavelength and Δε represent maximum wavelength of the absorption and molecular coefficient, respectively.
3. Characterization of pxGSTu1
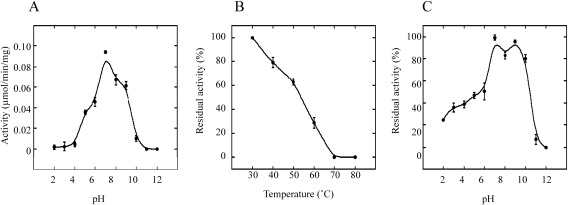
The enzymatic properties of pxGSTu1 were determined by measuring the mol CDNB biotransformed. The optimum pH of pxGSTu1 was found to be ∼7 (Fig. 4A), slightly lower than those of bmGSTD, bmGSTE, theta-class GST of B. mori (bmGSTT), and sigma-class GST of B. mori (bmGSTS), as well as an H. cunea sigma-class GST.4,6,9,17,18) pxGSTu1 was stable at temperatures below 40°C (Fig. 4B), similar to bmGSTS, whereas bmGSTT, bmGSTE, bmGSTD, omega-class GST of B. mori (bmGSTO), and zeta-class GST of B. mori (bmGSTZ) were stable at temperatures below 50°C.4,5,7,9,18) An analysis of pH stability (Fig. 4C) indicated that pxGSTu1 retained more than 80% of its original activity at pHs of 7 to 10, a narrower range than those of bmGSTT, bmGSTE, bmGSTD, bmGSTO, bmGSTS, and bmGSTZ.
Fig. 4. Enzymatic properties of pxGSTu1 were assayed with CDNB and GSH as substrates. GST activity was assayed under standard conditions, as described in Materials and Methods, unless otherwise indicated. The maximum value obtained was set to 100%. (A) Optimum pH levels for the activities were assayed using citrate-phosphate-borate buffer at various pH levels with a fixed ionic strength of 0.25. (B) Thermostability was determined by the preincubation of the enzyme solution at various temperatures for 30 min before the residual activity was assayed. (C) pH stability was assessed by preincubation of the enzyme solution at various pH levels at 4°C for 24 hr before the residual activity was assayed.
The substrate specificity of pxGSTu1 with various substrates was examined at pH 7.0 and 30°C, as summarized in Table 1. Under these conditions, pxGSTu1 has activity toward CDNB with Km=1.57 mM. We found that pxGSTu1 possesses detectable activity toward ECA (Km=2.1 mM). When 1,2-epoxy-3-(4-nitrophenoxy) propane (EPNP), 4-nitrobenzyl chloride (4NBC), 4-nitrophenethyl bromide (4NPB), 4-hydroxynonenal (4HNE), 4-nitrophenyl acetate (4NPA), and hydrogen peroxide were used as a substrate, the activity of pxGSTu1 was undetectable.
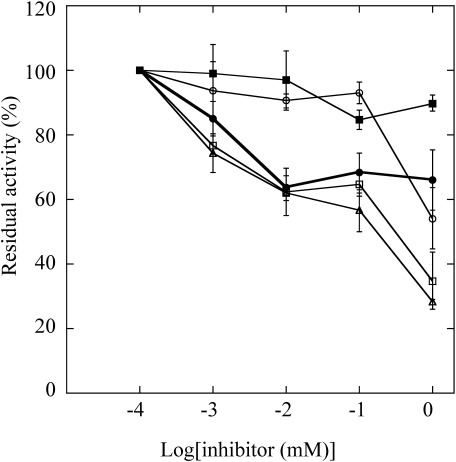
In further activity testing, the inhibitory effects of model insecticides on pxGSTu1 were examined using CDNB as a substrate. The competitive assay revealed that pxGSTu1 activity was inhibited by permethrin, bendiocarb, imidacloprid, diazinon, and chlorofenapyr as substrates (Fig. 5). Residual activity decreased in proportion to increasing amounts of each insecticide. In the presence of 1 mM diazinon or imidacloprid, pxGSTu1 showed ∼30% of its original activity. Permethrin caused inhibition at a concentration of 1 mM, while pxGSTu1 activity was decreased to ∼50% of its original activity.
Fig. 5. Effects of insecticides on pxGSTu1 activity. Enzymatic activity was measured in the presence of various concentrations of insecticides: diazinon (open triangle), chlorophenapyr (closed square), imidacloprid (open square), bendiocarb (closed circle), or permethrin (open circle). The value from the assay with 1–10−4 M of insecticide was set to 100%. Data represent averages with standard deviations from three independent experiments.
Discussion
Glutathione S-transferases have been implicated in catalyzing the conjugation of GSH to exogenous compounds for detoxification. We found 19 GST homologs of P. xylostella in the KONAGAbase EST database.14) It is possible that at least two of them are isoforms of unclassified GSTs in the diamondback moth. Previously, we identified an unclassified GST of B. mori that could play a part in insecticide resistance in B. mori.8) Since pxGSTu1 has not been characterized, its enzymatic property might provide insight into the physiological roles, including detoxification of insecticides, in the diamondback moth.
In the present study, we cloned and sequenced cDNA that encoded pxGSTu1. The deduced amino acid sequence was homologous to unclassified GSTs from D. plexippus, P. Xuthus, and P. aegeria. The phylogenetic tree showed that pxGSTu1 is closely related to unclassified insect GSTs, including Pp_I4DM36, Dp_G6CJS5, Ag_Q8MUQ6, Aa_Q16P53, Ae_Q16P53, and Dm_Q8INS9 (Fig. 2). In the amino acid sequence of pxGSTu1, a serine residue in the N-terminus region was well conserved among unclassified GSTs, as well as in bmGSTE and bmGSTD (Fig. 1). It is reported that the residue is also present in the region of the N-terminus of theta-class GSTs, whereas a tyrosine is substituted for a serine in mammalian sigma-, alpha-, mu-, and pi-class GSTs.19–21) The amino acid sequence pxGSTu1 includes a GSH-binding site (Fig. 1) that was also found in other unclassified GSTs and bmGSTE and bmGSTD.
The soluble fraction of E. coli cells successfully yielded active pxGSTu1; it was then efficiently purified to homogeneity. The theoretical molecular mass of pxGSTu1 is substantially identical to that shown by SDS-PAGE and is also consistent with those of GSTs isolated in B. mori, H. cunea, and N. lugens. We found that pxGSTu1 exhibited GSH-dependent activities in the biotransformation of CDNB and ethacrynic acid (ECA) (Table 1). CDNB is a universal substrate for GSTs. ECA is a substrate for pi-, mu-, and alpha-class GSTs but not for bmGSTS. However, bmGSTD conjugated GSH to ECA.3) 4-Hydroxynonenal (4HNE) is a cytotoxic product of lipid peroxidation under conditions of oxidative stress.22) Hydrogen peroxide is a product of active oxygen. Our results indicated that pxGSTu1 is not involved in antioxidant-related reactions. With CDNB as the substrate, the Km value was similar to those of delta-, epsilon-, and unclassified GST1 (bmGSTu), but with 3.5-, 7.9-, and 8.3-fold the value of omega-, theta-, and sigma-class GSTs, respectively.4–6,8,18,23) Although the epsilon-class GST was able to conjugate GSH to ECA, the ECA activity did not fit the Michaelis–Menten equation.4)
GSTs play a crucial role in resistance to the organophosphate (fenitrothion) and pyrethroids (deltamethrin and permethrin).24) To examine the interaction of insecticides and pxGSTu1, inhibition assays were performed. In the present study, we showed that various insecticides inhibited the activity of pxGSTu1. This result is consistent with those found in the mosquito, Anopheles dirus, and ticks, Haemaphysalis longicornis and Rhipicephalus appendiculatus.25,26) The activity levels of GSTs in these species were reduced to ∼85% and ∼60%, respectively, by 0.1 mM of deltamethrin,27) while bmGSTO exhibited ∼20% of its maximum activity under the same conditions.5)
Here, we provide the first evidence that a novel GST (unclassified GST) is present in the diamondback moth, P. xylostella. We hypothesize that pxGSTu1 functions in response to the detoxification of insecticides as xenobiotics. The existence of all GSTs in P. xylostella must be determined to understand their correlation to the insecticide detoxification system in this species. It may be useful to compare detailed properties, such as expression rates, activities, substrate specificities, and resistance spectra among all GSTs as well as the related enzymes in P. xylostella. We are currently conducting investigations along these lines in our laboratories.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (KAKENHI, 15H04611) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1).I. Listowsky, M. Abramovitz, H. Homma and Y. Niitsu: Drug Metab. Rev. 19, 305–318 (1988). [DOI] [PubMed] [Google Scholar]
- 2).R. N. Armstrong: Chem. Res. Toxicol. 10, 2–18 (1997). [DOI] [PubMed] [Google Scholar]
- 3).K. Yamamoto, P. Zhang, F. Miake, N. Kashige, Y. Aso, Y. Banno and H. Fujii: Comp. Biochem. Physiol. B 141, 340–346 (2005). [DOI] [PubMed] [Google Scholar]
- 4).K. Yamamoto, Y. Aso and N. Yamada: Insect Mol. Biol. 22, 523–531 (2013). [DOI] [PubMed] [Google Scholar]
- 5).K. Yamamoto, S. Nagaoka, Y. Banno and Y. Aso: Comp. Biochem. Physiol. C 149, 461–467 (2009). [DOI] [PubMed] [Google Scholar]
- 6).K. Yamamoto, H. Fujii, Y. Aso, Y. Banno and K. Koga: Biosci. Biotechnol. Biochem. 71, 553–560 (2007). [DOI] [PubMed] [Google Scholar]
- 7).K. Yamamoto, Y. Shigeoka, Y. Aso, Y. Banno, M. Kimura and T. Nakashima: Pestic. Biochem. Physiol. 94, 30–35 (2009). [Google Scholar]
- 8).K. Yamamoto, H. Ichinose, Y. Aso, Y. Banno, M. Kimura and T. Nakashima: Biochim. Biophys. Acta 1810, 420–426 (2011). [DOI] [PubMed] [Google Scholar]
- 9).K. Yamamoto, P. B. Zhang, Y. Banno and H. Fujii: J. Appl. Entomol. 130, 515–522 (2006). [Google Scholar]
- 10).H. Ranson and J. Hemingway: Methods Enzymol. 401, 226–241 (2005). [DOI] [PubMed] [Google Scholar]
- 11).C.-P. Tu and B. Akgul: Methods Enzymol. 401, 204–226 (2005). [DOI] [PubMed] [Google Scholar]
- 12).R. Sawicki, S. P. Singh, A. K. Mondal, H. Benes and P. Zimniak: Biochem. J. 370, 661–669 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).K. Yamamoto, A. Higashiura, M. T. Hossain, N. Yamada, T. Shiotsuki and A. Nakagawa: Arch. Biochem. Biophys. 566, 36–42 (2015). [DOI] [PubMed] [Google Scholar]
- 14).A. Jouraku, K. Yamamoto, S. Kuwazaki, M. Urio, Y. Suetsugu, J. Narukawa, K. Miyamoto, K. Kurita, H. Kanamori, Y. Katayose, T. Matsumoto and H. Noda: BMC Genomics 14, 464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).U. K. Laemmli: Nature 227, 680–685 (1970). [DOI] [PubMed] [Google Scholar]
- 16).W. H. Habig, M. J. Pabst and W. B. Jakoby: J. Biol. Chem. 249, 7130–7139 (1974). [PubMed] [Google Scholar]
- 17).K. Yamamoto, K. Usuda, Y. Kakuta, M. Kimura, A. Higashiura, A. Nakagawa, Y. Aso and M. Suzuki: Biochim. Biophys. Acta 1820, 1469–1474 (2012). [DOI] [PubMed] [Google Scholar]
- 18).M. D. Hossain, N. Yamada and K. Yamamoto: PLoS ONE 9, e97740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).P. G. Board, M. Coggan, M. C. J. Wilce and M. W. Parker: Biochem. J. 311, 247–250 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).J. Rossjohn, W. J. McKinstry, A. J. Oakley, D. Verger, J. Flanagan, G. Chelvanayagam, K.-L. Tan, P. G. Board and M. W. Parker: Structure (London) 6, 309–322 (1998). [DOI] [PubMed] [Google Scholar]
- 21).P. Reinemer, L. Prade, P. Hof, T. Neuefeind, R. Huber, R. Zettl, K. Palme, J. Schell, I. Koelln, H. D. Bartunik and B. Bieseler: J. Mol. Biol. 255, 289–309 (1996). [DOI] [PubMed] [Google Scholar]
- 22).S. P. Singh, J. A. Coronella, H. Beneš, B. J. Cochrane and P. Zimniak: Eur. J. Biochem. 268, 2912–2923 (2001). [DOI] [PubMed] [Google Scholar]
- 23).K. Yamamoto, A. Higashiura, M. Suzuki, K. Aritake, Y. Urade, N. Uodome and A. Nakagawa: Biochim. Biophys. Acta 1830, 3711–3718 (2013). [DOI] [PubMed] [Google Scholar]
- 24).J. Hemingway: Insect Biochem. Mol. Biol. 30, 1009–1015 (2000). [DOI] [PubMed] [Google Scholar]
- 25).L. Prapanthadara, H. Ranson, P. Somboon and J. Hemingway: Insect Biochem. Mol. Biol. 28, 321–329 (1998). [DOI] [PubMed] [Google Scholar]
- 26).I. da Silva Vaz Jr., T. Torino Lermen, A. Michelon, C. A. Sanchez Ferreira, D. R. Joaquim de Freitas, C. Termignoni and A. Masuda: Vet. Parasitol. 119, 237–245 (2004). [DOI] [PubMed] [Google Scholar]
- 27).T. Kozaki, T. Shono, T. Tomita and Y. Kono: Insect Biochem. Mol. Biol. 31, 991–997 (2001). [DOI] [PubMed] [Google Scholar]