Abstract
In view of the possibility that spent mushroom substrates (SMSs) may be used as agricultural materials to prevent crop diseases, we examined the effect of treatment with a hot water extract from the SMS of Lentinula edodes on plant resistance to pathogenic infection. The extract of Le. edodes SMS was sprayed onto the leaves of rice plants, followed by inoculation of the leaves with the conidia of rice blast fungus. The development of lesions was suppressed by treatment with the SMS extract. The extract markedly inhibited the germination of Pyricularia oryzae conidia. We purified compounds 1, 2, and 3, which showed inhibitory effects on conidial germination, from the Le. edodes SMS extract of by successive solvent extraction, column chromatography, and preparative HPLC. Spectroscopic analyses revealed that 1, 2, and 3 were phenolic acids with two carboxyl groups in common.
Keywords: Lentinula edodes, spent mushroom substrate, antifungal activity, rice blast, lignin
Introduction
Commercially available mushrooms are mainly produced using the mycelial block cultivation method. In 2015, 89% of fresh shiitake mushrooms (Lentinula edodes) were produced via mycelial block cultivation in Japan according to the statistical survey by the Ministry of Agriculture, Forestry, and Fisheries.1) Generally, the mushroom substrate used for mycelial block cultivation is comprised of 80–90 wt% of sawdust and tip of trees and 10–20 wt% of wheat or rice bran. In mycelial block cultivation, the spent mushroom substrate (SMS) is disposed as waste after harvesting the fruiting bodies. In Japan, the amount of fresh shiitake mushrooms produced by mycelial block cultivation was approximately 61,000 tons in 2016.2) The amount of mushroom substrate used for 350 g of shiitake is approximately 1.2 kg.2) Thus, the amount of SMS disposed per year is estimated to be approximately 210,000 tons. Other mushrooms such as Pholiota microspora (nameko), Hypsizygus marmoreus (bunashimeji), and Pleurotus eryngii (king oyster mushroom) are also produced using the mycelial block cultivation method. However, the disposal of the large amounts of SMS generated is a serious problem for the mushroom industry. Therefore, the recycling of SMS would be greatly beneficial.
The recycling of SMS has been attempted in different ways. It has been demonstrated that mixing SMS with soil improved soil quality and increased the growth and yield of sweet corn.3) SMS was used for the production of silage for cows,4) and feeding basal diet of which 12% were replaced by SMS did not affect the body weight of cows. The other approach is the utilization of SMS in agricultural materials for crop protection against diseases. Inagaki and Yamaguchi (2009) found that application of the SMS of shiitake mushrooms (Le. edodes) as compost was effective in reducing the severity of anthracnose in cucumber plants.5) Arase et al. (2013) reported that spraying a water extract of the SMS of Lyophyllum decastes (hatakeshimeji) suppressed the development of lesions caused by infection with rice blast fungus [Pyricularia oryzae (syn.: Magnaporthe grisea)].6) Parada et al. (2012) investigated the effect of SMS treatment on fungal and bacterial diseases of cucumber plants.7) They sprayed water extracts of the SMSs of Ly. decastes and Pl. eryngii onto leaves or mixed autoclaved SMS with soil. These treatments significantly reduced powdery mildew caused by Podosphaera xanthii and angular leaf spot caused by Pseudomonas syringae pv. lachrymans but not Corynespora leaf spot caused by Corynespora cassiicola or scab caused by Cladosporium cucumerinum.7) The water extracts of SMS did not exhibit antimicrobial activity against the above fungal and bacterial pathogens. Therefore, the authors suggested that the protective effects of SMS were attributable to the induction of systemic acquired resistance (SAR) by SMS treatment.7)
Almost all industrially cultivated mushrooms are wood-decay fungi, which are classified into brown- and white-rot fungi. Brown-rot fungi preferentially degrade cellulose and hemicellulose, while white-rot fungi, such as Le. edodes, efficiently degrade lignin in addition to cellulose. Because lignin is a biopolymer that is not easily degraded by chemical reactions, the mechanism of lignin degradation by white-rot fungi has attracted increased attention. Lignin peroxidase, manganese peroxidase, and laccase have been identified to be involved in lignin degradation by white-rot fungi.8–10) These enzymes degrade lignin to low-molecular-weight compounds, such as aromatic alcohols, aldehydes, and acids.11–13) The inhibitory activity of small molecules derived from lignin on yeast growth was investigated because they could potentially inhibit the fermentation of lignocellulosic biomass for biofuel production.14) However, other biological activities of lignin-derived small molecules have not been extensively investigated.
In the present study, we examined the effects of treatment with water extracts of the SMS of Le. edodes on lesion formation on rice leaves caused by infection with rice blast fungus. We found that the water extract of the SMS of Le. edodes inhibited the conidial germination of Py. oryzae. Based on this finding, we purified the antifungal compounds in the extract and identified their chemical structures via spectroscopic analyses.
Materials and Methods
1. General procedures
1H- and 13C-NMR spectra and 2D-NMR spectra (COSY, HMQC, and HMBC) were recorded on an Avance II instrument (Bruker, Billerica, MA, USA). High-resolution mass spectra were recorded using an Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA), and ESI-MS and UV-Vis spectra were recorded on a Quattro Micro API mass spectrometer (Waters, Milford, MA, USA) connected to an Acquity UPLC (Waters). Preparative HPLC was performed with a 10A HPLC system (Shimadzu, Kyoto, Japan).
2. Plant material and pathogenic fungi
Rice (Oryza sativa cv. Nipponbare) seeds were placed on a moist paper and incubated at 28°C for 4 days under a 16 hr light/8 hr dark cycle. Five germinated seedlings were transplanted into a 1 : 1 mixture of vermiculite and artificial compost, Green soil (Shoei Sangyo, Okayama, Japan), in a pot (4.5 cm in diameter, 4 cm in depth). Rice blast fungus (Py. oryzae, race 007, Naga 69–150) was the stock culture of the Laboratory of Plant Pathology, Shimane University.
3. Spent mushroom substrate
The SMS of Le. edodes was kindly provided by JA Tottori Inaba, Hatto branch, Yazu Cho, Tottori, Japan. SMSs were autoclaved and preserved at 5°C until use.
4. Treatment of rice seedlings with water extracts of SMS
SMS was dried in a desiccator under reduced pressure for 12 hr. Distilled water (500 mL) was added to the dried SMS (50 g) and autoclaved at 120°C for 15 min, followed by filtration through four layers of gauze and then through filter paper (Advantec No. 2, Toyo Roshi, Tokyo, Japan). The obtained water extract of SMS was autoclaved again at 120°C for 15 min and preserved at 5°C until use. The extract (5 mL) was sprayed on five 16-day-old seedlings in a pot. As a positive control, 20 mL of BIT solution (1,1-benzisothiazol-3(H)-one-1,1-dioxide, 56 mg/L, Wako Pure Chemical Industry, Osaka, Japan) was added to a pot (50 mL) containing 13-day-old rice seedlings as described by Nakashita et al. (2002).15)
5. Inoculation with conidia of rice blast fungus
Pyricularia oryzae was cultured on oatmeal medium agar plates for 15 days. Aerial hyphae were washed away with a brush after adding distilled water, and the plates were incubated for a further 2–4 days under a black light lamp (FL15BLB, Hitachi, Tokyo, Japan). Synchronously formed conidia were used as inocula. The conidia were suspended in distilled water containing 0.25% Tween-20, washed twice by centrifugation at 1,000×g for 10 min, and resuspended in distilled water containing 0.25% Tween-20. Then, droplets (5 µL) of the conidial suspension (5×105 conidia/mL) were placed at five points on the third leaves of 18-day-old seedlings at 1 cm intervals. The inoculated seedlings were kept in a moist chamber for 12 hr. The leaves were observed 5 days after inoculation.
6. Assay for inhibition of conidia germination
Water extracts (10 µL) of SMS were added to a conidial suspension (10 µL, 5×105 conidia/mL) placed on a glass slide in a petri dish lined with moist filter paper. The conidia were incubated at 25°C in the dark for 6–10 hr. The germinated conidia were counted under a microscope (BX43, Olympus, Tokyo, Japan).
The fractions obtained during the purification process were dissolved in DMSO and subjected to the assay. Typically, DMSO solution (100 mg/mL, 1 µL) of a fraction obtained by purification process was added to the conidial suspension (99 µL). A droplet of the conidial suspension was placed on a glass slide and incubated.
7. Purification of antifungal compounds from the water extract of the SMS of Le. edodes
A water extract (10 L) of the SMS of Le. edodes was fractionated by serial solvent extraction. First, the water extract was partitioned with AcOEt (7 L×3). The AcOEt extract was dried over Na2SO4 overnight and concentrated. The concentrate was dissolved in 100 mL of MeOH, and 1.9 L of water was added to the solution. The solution was acidified to pH 3 by adding 6 M HCl and extracted with AcOEt (2 L×3). The AcOEt layer was concentrated to 2 L and extracted with 1 M NaOH (1 L×3). The solution was acidified to pH 3 by adding 6 M HCl and extraction with ether (2 L) three times. The ether layer was washed with 100 mL of water three times and dried over Na2SO4 overnight.
The ether layer was concentrated and subjected to silica gel (300 g, Daiso gel IR-60-63/210, Daiso, Osaka, Japan) column chromatography. The compounds were eluted with 0, 20, 40, 60, 80, and 100% acetone in hexane (2 L each). The 60% acetone fraction showed the highest activity and was subjected to silica gel column chromatography (90 g, Daiso gel) again. The compounds were eluted with 60, 70, 80, 90, and 100% AcOEt in hexane (900 mL each). The 60% AcOEt fraction, which showed the highest activity, was analyzed via HPLC, and three major peaks (corresponding compounds 1, 2, and 3) were detected. The conditions for analytical HPLC were as follows: column, Cosmosil 5C18-AR-II (4.6×150 mm, Nacalai Tesque, Kyoto, Japan); temperature, 40°C; solvents, 0.1% TFA in water (A) and 0.1% TFA in acetonitrile (B); gradient, 5–100% B/(A+B) for 30 min; flow rate, 0.8 mL/min; and detection, 220 nm. The compounds were purified with preparative HPLC. The conditions for the first preparative HPLC were as follows: column, Cosmosil 5C18-AR-II (20×250 mm); temperature, 40°C; solvent, 30% acetonitrile containing 0.1% TFA; flow rate, 7 mL/min; and detection, 280 nm. The retention time of 3 was 12.4 min. Because the retention times of peaks of 1 (54.2 min) and 2 (56.5 min) were close, they were collected as a single fraction. The fraction containing 1 and 2 was subjected to second preparative HPLC. The conditions for the second preparative HPLC were as follows: column, Cosmosil 5C18-AR-II (10×250 mm); temperature, 40°C; solvent, 20% acetonitrile containing 0.1% TFA; flow rate, 3 mL/min; and detection, 280 nm. The retention times of 1 and 2 were 20.7 and 22.5 min, respectively, under these conditions.
1 (12.6 mg). Positive ESI-MS: m/z 317.1 [M−OH]+, 335.0 [M+H]+; negative ESI-MS: m/z 333.1 [M−H]−; positive HR ESI-MS: m/z 335.0761 [M+H]+ (calc. for C16H15O8: 335.0767); negative HR ESI-MS: m/z 333.0618 [M−H]− (calc. for C16H13O8: 333.0610); UV-Vis λmax: 262 nm. NMR data are shown in Table 1.
Table 1. NMR data of 1.
| Position | δH (J in Hz)(600 MHz, methanol-d4) | δC (150 MHz, methanol-d4) | δC (150 MHz, acetone-d6/D2O, 9 : 1 v/v) | δCa) (150 MHz, acetone-d6/D2O, 9 : 1 v/v) |
|---|---|---|---|---|
| 1 | 122.2 | 121.9 | 121.8 | |
| 2 | 7.19 d (1.9) | 115.7 | 115.5 | 115.4 |
| 3 | 144.4 | 143.6 | 143.6 | |
| 4 | 144.1 | 143.2 | 143.3 | |
| 5 | 150.0 | 149.5 | 149.3 | |
| 6 | 7.46 d (1.9) | 108.0 | 109.7 | 109.9 |
| 7 | 169.4 | 167.8 | 168.2±0.2 | |
| 5-OCH3 | 3.95 s | 56.6 | 56.6 | —b) |
| 1′ | 151.6 | 151.1 | 151.0 | |
| 2′ | 151.2 | 150.3 | 150.6 | |
| 3′ | 7.71 d (2.0) | 114.9 | 114.3 | 114.6 |
| 4′ | 127.3 | 126.5 | 126.6 | |
| 5′ | 7.60 dd (8.4, 2.0) | 124.5 | 123.8 | 123.9 |
| 6′ | 6.81 d (8.4) | 118.2 | 117.1 | 117.7 |
| 7′ | 169.3 | 167.6 | 168.2±0.2 | |
| 2′-OCH3 | 3.91 s | 56.3 | 56.3 | —b) |
a) Lüdemann and Nimz (1974) Makromol. Chem. 175, 2398–2470. b) Chemical shifts were not reported.
2 (15.4 mg). Positive ESI-MS: m/z 347.1 [M−OH]+, 365.1 [M+H]+; negative ESI-MS: m/z 363.1 [M−H]−; positive HR ESI-MS: m/z 365.0878 [M+H]+ (calc. for C17H17O9: 365.0873); negative HR ESI-MS: m/z 363.0724 [M−H]− (calc. for C17H15O9: 363.0716); UV-Vis λmax: 265 nm; NMR data are shown in Table 2.
Table 2. NMR data of 2 (DMSO-d6) and 2′ (CDCl3).
| Position | 2 | 2′ | ||
|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1 | 119.0 | 124.8 | ||
| 2 | 6.56 d (1.6) | 107.6 | 6.82 d (1.9) | 108.9 |
| 3 | 145.6 | 151.2 | ||
| 4 | 140.0 | 142.1 | ||
| 5 | 148.1 | 153.2 | ||
| 6 | 7.19 d (1.6) | 107.1 | 7.30 d (1.9) | 107.4 |
| 7 | 166.7 | 166.5 | ||
| 7-COOH | 12.74 brsa) | |||
| 7-COOMe | 3.80 s | 52.1 | ||
| 5-OCH3 | 3.84 s | 48.6 | 3.93 s | 56.28 |
| 4-OH | 9.62 brs | |||
| 4-OCH3 | 4.04 s | 61.0 | ||
| 1′ | 134.8 | 135.9 | ||
| 2′,6′ | 152.7 | 153.0 | ||
| 3′,5′ | 7.36 s | 106.6 | 7.37 s | 106.8 |
| 4′ | 128.2 | 127.3 | ||
| 7′ | 166.9 | 166.8 | ||
| 7′-COOH | 12.74 brsa) | |||
| 7′-COOMe | 3.95 s | 52.4 | ||
| 2′,6′-OCH3 | 3.77 s | 56.2 | 3.82 s | 56.30 |
a) Overlapping signals.
3 (4.2 mg). Positive ESI-MS: m/z 195.0 [M−OH]+, 213.0 [M+H]+; negative ESI-MS: m/z 211.0 [M−H]−; positive HR ESI MS: m/z 213.0399 [M+H]+ (calc. for C9H9O6: 213.0399); negative HR ESI-MS: m/z 211.0254 [M−H]− (calc. for C9H7O6: 211.0242); UV-Vis λmax: 227, 267 nm; 1H-NMR (600 MHz, methanol-d4) δH: 8.20 (d, J=1.8 Hz, 1H, H-2), 7.71 (d, J=1.8 Hz, 1H, H-6), 3.92 (s, 3H, OCH3); 13C-NMR (150 MHz, methanol-d4) δC: 173.4 (6-COOH), 169.0 (1-COOH), 157.4 (C-4), 149.7 (C-5), 125.7 (C-2), 122.1 (C-3), 117.6 (C-6), 114.0 (C-1), 56.7 (OCH3).
8. Methylation of 2
A solution of (trimethylsilyl)diazomethane (2.0 M in ether, 2 mL) was added to MeOH solution (4 mL) of 2 (4.0 mg, 11.0 µmol). The reaction mixture was stirred in a sealed flask under nitrogen gas for 4 hr at ambient temperature. Excess (trimethylsilyl)diazomethane was decomposed by the addition of HOAc (ca. 100 µL). After standing for 30 min, the solvent was evaporated in vacuo and subjected to column chromatography [1.0 g, Daiso gel, AcOEt-hexane (2 : 8 v/v)] to give 2′.
2′ (yield, 3.7 mg, 91%). Positive ESI MS: m/z 407.1 [M+H]+. NMR data are shown in Table 2.
Results
1. Suppression of lesion formation by treatment of rice leaves with water extract of Le. edodes SMS
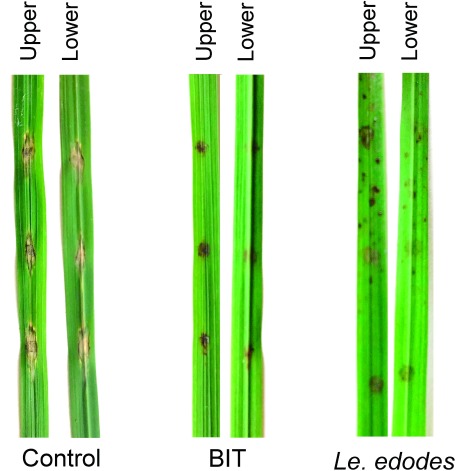
A hot water extract of the SMS of Le. edodes was sprayed onto rice leaves. After incubation for 2 days, the leaves were inoculated with Py. oryzae conidia. As a positive control, the leaves of the seedlings treated with BIT for 5 days were inoculated with conidia. Large necrotic lesions formed on the control leaves at the inoculation site as shown in Fig. 1. In contrast, the leaves sprayed with extracts of Le. edodes SMS showed brown lesions, and the size of the lesions remained unchanged thereafter. The leaves treated with BIT also showed brown lesions. The color of the lesions formed on SMS-treated leaves was pale when compared with those formed on BIT-treated leaves.
Fig. 1. Effect of treatment with water extracts of spent mushroom substrates of Lentinula edodes on the development of lesions caused by infection with rice blast fungus. The rice leaves treated with BIT were used as the positive control.
2. Antifungal activity of water extract of SMS
To detect the antifungal activity of the SMS water extract, we performed an assay to determine its inhibitory activity on conidia germination. The germination rate of conidia in distilled water was 91.2%, while that in the solution containing SMS extract was 4.1%, indicating the presence of antifungal compounds in the SMS extract.
3. Isolation of antifungal compounds from water extract of Le. edodes SMS
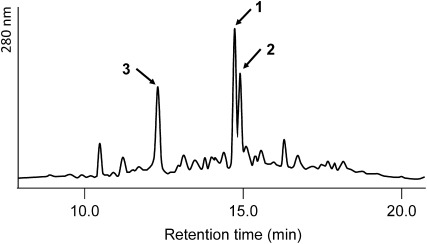
The water extract of Le. edodes SMS was partitioned with AcOEt. The AcOEt extract showed inhibitory activity against conidia germination. Successive solvent extractions revealed that the active compounds were in the acidic ether layer. The acidic ether layer was then fractionated by silica gel column chromatography with two different solvent systems. The most active fraction obtained by column chromatography was subjected to HPLC, which showed the presence of three major peaks (Fig. 2). Finally, the active compounds 1–3 were purified by preparative HPLC.
Fig. 2. HPLC analysis of the 60% AcOEt fraction obtained by silica gel column chromatography of the AcOEt layer from the water extract of spent mushroom substrates of Lentinula edodes.
4. Determination of chemical structures of 1, 2, and 3
The molecular weight of 1 was determined to be 334 based on the presence of an [M+H]+ ion at m/z 335 and an [M−H]− ion at m/z 333 on positive and negative ESI-MS, respectively. HR ESI-MS indicated that the molecular formula of 1 was C16H14O8, with a hydrogen deficiency index of 10.
The 1H-NMR signals at δH 7.46 and 7.19 ppm with a coupling constant of 1.9 Hz indicated the presence of a 1,3,4,5-tetrasubstituted benzene ring, while the coupling pattern of signals at δH 7.71, 7.60, and 6.81 ppm indicated the presence of a 1,3,4-trisubstituted benzene ring. The presence of these two benzene rings was supported by the detection of 12 13C-NMR signals in the aromatic region. The 1H-NMR signals at δH 3.95 and 3.91 ppm and the 13C-NMR signals at δC 55.6 and 55.3 ppm revealed the presence of two methoxy groups. The HMBC spectrum showed that these two methoxy groups were directly linked to the benzene rings. The 13C-NMR spectrum indicated the presence of two carbonyl carbons (δC 169.4 and 169.3 ppm) bearing an oxygen atom.
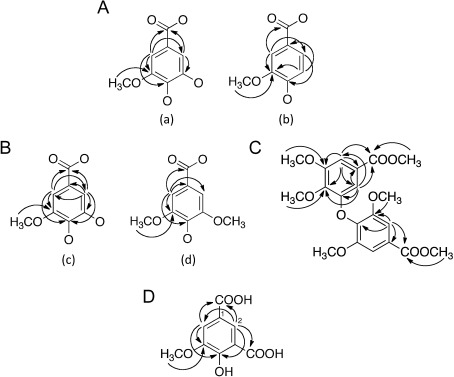
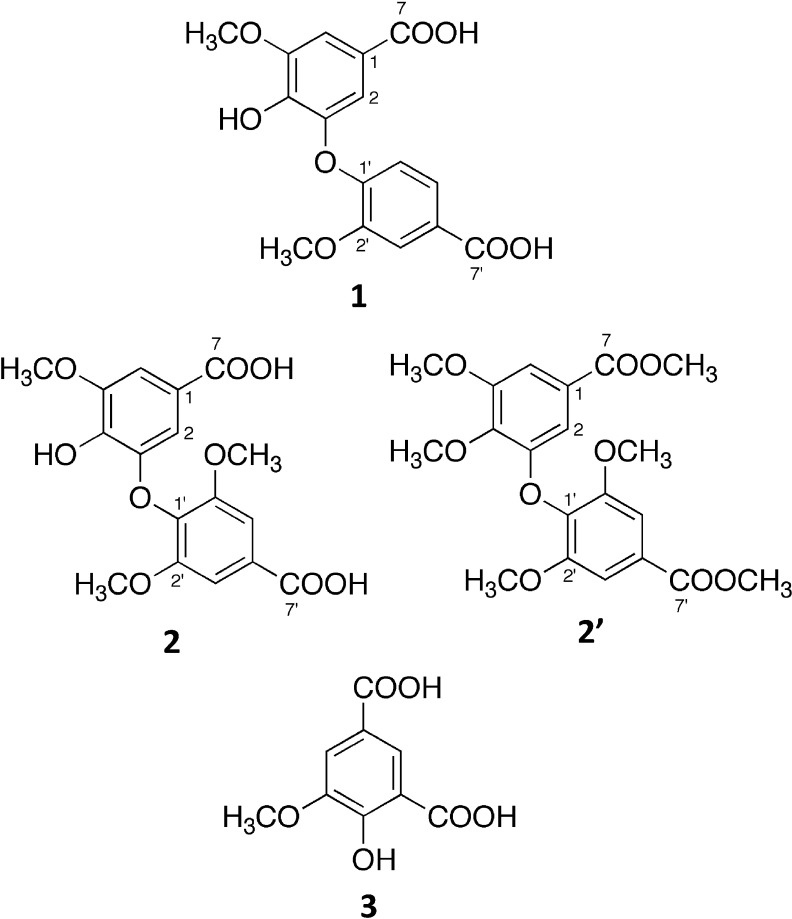
Based on correlations detected on the HMBC spectrum and chemical shifts of the 13C-NMR signals, the presence of partial structures was revealed [Fig. 3A (a) and (b)]. A database search of compounds with these partial structures found the compound 3-(4-carboxy-2-methoxyphenoxy)-4-hydroxy-5-methoxybenzoic acid.16) We concluded that this compound was 1 (Fig. 4) because the 13C NMR data of this compound and that of 1 were identical, although the chemical shifts of the two methoxy groups were not reported in the literature (Table 1).
Fig. 3. HMBC correlations detected for 1 (A), 2 (B), 2′ (C), and 3 (D).
Fig. 4. Chemical structures of 1, 2, 2′, and 3.
The molecular weight of 2 was determined to be 364 based on the presence of an [M+H]+ ion at m/z 365 and an [M−H]− ion at m/z 363 on positive and negative ESI MS, respectively. The molecular formula C17H16O9 was established based on the [M+H]+ ion, and the index of hydrogen deficiency was 10. From this molecular formula, it can be suggested that 2 possesses an additional methoxy group when compared with 1.
The 13C-NMR spectrum showed 14 signals, with three overlapping signals. Three 1H- (for four protons) and 10 13C-NMR signals in the aromatic region suggested the presence of two tetrasubstituted benzene rings. The small coupling constant (1.6 Hz) between the signals at δH 6.56 and δH 7.19 indicated that one benzene ring was substituted at the 1-, 3-, 4-, and 5-positions. Six of the 10 13C-NMR signals in the aromatic region were considered to correspond to this benzene ring. The singlet signal at δH 7.36 (for two protons) and four aromatic carbon signals suggested that the other benzene ring is 1,3,4,5-tetrasubstituted benzene, which has a symmetric structure with the same substituents at the 3- and 5-positions. 1H-NMR and 13C-NMR spectra revealed the presence of three methoxy groups, two of which overlapped each other. The HMBC spectrum revealed that all three methoxy groups were directly linked to the benzene rings. The 13C-NMR spectrum showed the signals of two carbonyl carbons bearing an oxygen atom. Based on these findings and further consideration of correlations observed in the HMBC spectrum, we constructed partial structures (c) and (d) as shown in Fig. 4B.
To determine the position of the linkage between these two partial structures, we methylated 2 with (trimethylsilyl)diazomethane. The molecular weight of the methylated compound 2′ was 406, indicating the introduction of three additional methyl groups. The HMBC spectrum showed two methyl esters and a methoxy group directly linked to a benzene ring (Fig. 3C). The NMR analyses indicated that 2′ still has an asymmetrical 1,3,4,5-tetrasubstituted benzene ring. If partial structures (c) and (d) were connected by a 4-O-1′ linkage, methylation would result in two symmetrical 1,3,4,5-tetrasubstituted benzene rings. Thus, the connection between (c) and (d) was established to be the 3-O-1′ linkage. Accordingly, 2 was identified as 3-(4-carboxy-2,6-dimethoxyphenoxy)-4-hydroxy-5-methoxybenzoic acid (Fig. 4), which has not been reported previously.
The molecular weight of 3 was determined to be 212 because of the presence of an [M+H]+ ion at m/z 213 and an [M−H]− ion at m/z 211 on ESI MS, and the molecular formula C9H8O6 was established based on HR ESI-MS. The index of hydrogen deficiency was 6. Two doublet signals (δH 7.71 and 8.20 ppm, J=1.8 Hz) on the 1H-NMR spectrum and six signals (δC 114.1, 125.7, 122.1, 157.4, 149.7, and 117.6 ppm) on the 13C-NMR spectrum revealed the presence of a 1,3,4,5-tetrasubstituted benzene ring. Furthermore, 1H-NMR indicated the presence of a methoxy group. The 13C-NMR spectrum showed two signals (δC 173.4 and 169.0 ppm) of carbonyl carbons bearing an oxygen atom. Based on the molecular formula, 3 was deduced to have two carboxyl groups and one hydroxy group. The substituted position was established based on the correlations detected in the HMBC spectrum (Fig. 3D), and 3 was identified as 4-hydroxy-5-methoxyisophthalic acid (Fig. 4).
5. Inhibition of germination of Py. oryzae conidia by 1, 2, and 3
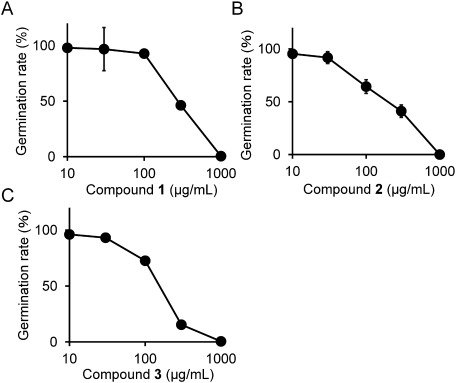
We determined the germination rate Py. oryzae conidia in the presence of different concentrations of 1, 2, and 3 using compounds purified from the SMS of Le. edodes. As shown in Fig. 5, 1, 2, and 3 inhibited conidia germination in a dose-dependent manner. The IC50 values of 1, 2, and 3 were determined to be 280, 200, and 150 µg/mL, respectively.
Fig. 5. Inhibition of Pyricularia oryzae conidia germination by 1 (A), 2 (B), and 3 (C). Data are presented as means of three replicates. Error bars indicate standard deviations.
Discussion
We successfully purified the antifungal compounds from the extract and identified them as 3-(4-carboxy-2-methoxyphenoxy)-4-hydroxy-5-methoxybenzoic acid (1), 3-(4-carboxy-2,6-dimethoxyphenoxy)-4-hydroxy-5-methoxybenzoic acid (2), and 4-hydroxy-5-methoxyisophthalic acid (3). To the best of our knowledge, compound 2 has not been previously reported. These compounds are likely the degradation products of lignin, because all of them have the 4-hydroxy-5-methoxy-benzene ring possibly derived from the coniferyl alcohol unit in lignin. Compounds 1 and 3 were detected as degradation products of lignin formed by chemical oxidation with potassium permanganate17) and by the action of the white-rot fungus Phanerochaete chrysosporium.11) Since 3 is an intermediate compound in the degradation pathway of lignin, the enzymes involved in the catabolism of 3 have attracted attention and have been analyzed in detail from the viewpoint of a catalytic mechanism.18,19)
The SMSs of Le. edodes, Ly. decastes, and Pl. eryngii have been shown to be effective in suppressing the symptoms caused by phytopathogenic fungi and bacteria.5–7) These effects have been attributed to the induction of plant resistance to pathogenic fungi by the constituents of SMS. Considering that these mushrooms and phytopathogenic fungi belong to the fungi kingdom, it is reasonable to assume that some components derived from the mushrooms are recognized by plant cells as a part of the pathogen-associated molecular pattern, thereby inducing resistance responses.
We could not exclude the possibility of resistance induction by treatment with the extract of Le. edodes SMS. Notably, compounds 1, 2, and 3 have common structural features with salicylic acid, a plant hormone responsible for the induction of SAR.20,21) In particular, 3 is a salicylic acid derivative with additional functional groups, carboxyl and methoxy groups. Therefore, these compounds might play a role as signal molecules in the induction of resistance of rice plants to pathogens. In this context, it would be interesting to examine the induction of SAR by these lignin-derived small molecules.
Acknowledgments
We would like to express our sincere gratitude to JA Tottori Inaba (Hatto Branch, Tottori City) for supplying the SMS of Le. edodes.
References
- 1) Ministry of agriculture, forestry and fisheries industries: Statistical analysis on special forestry products. http://www.maff.go.jp/j/tokei/kouhyou/tokuyo_rinsan/index.html (Accessed 7 Dec., 2017)
- 2) K. Masuno: “Mushroom Yearbook, 2015,” ed. by Mushroom Yearbook Editorial Department, Tokusan Jyoho Inc. Purantsuwarudo Inc., Tokyo, Japan, pp. 191–195, 2015.
- 3) K. Kato, U. Matsushima, Y. Muto, F. Tatsuzawa and M. Okada: Hort. Res. (Japan) 12, 381–387 (2013). [Google Scholar]
- 4) A. Koike, E. Kouno and M. Yokoo: Akita Pref. Univ. Web J. B2, 111–116 (2015). [Google Scholar]
- 5) H. Inagaki and A. Yamaguchi: Mushroom Sci. Biotechnol. 17, 113–115 (2009). [Google Scholar]
- 6) S. Arase, Y. Kondo, R. Y. Parada, H. Otani, M. Ueno and J. Kihara: Mushroom Sci. Biotechnol. 21, 79–83 (2013). [Google Scholar]
- 7) R. Y. Parada, S. Murakami, N. Shimomura and H. Otani: J. Phytopathol. 160, 390–396 (2012). [Google Scholar]
- 8) C. H. Vane, T. C. Drage and C. E. Snape: J. Agric. Food Chem. 51, 947–956 (2003). [DOI] [PubMed] [Google Scholar]
- 9) A. T. Martínez, M. Speranza, F. J. Ruiz-Dueñas, P. Ferreira, S. Camarero, F. Guillén, M. J. Martínez, A. Gutiérrez and J. C. del Río: Int. Microbiol. 8, 195–204 (2005). [PubMed] [Google Scholar]
- 10) A. B. Fisher and S. F. Fong: AIMS Bioeng. 1, 92–112 (2014). [Google Scholar]
- 11) C. Chen, H. Chang and T. K. Kirk: Holzforschung 36, 3–9 (1982). [Google Scholar]
- 12) C. Chen, H. Chang and T. K. Kirk: J. Wood Chem. Technol. 3, 35–57 (1983). [Google Scholar]
- 13) E. Masai, Y. Katayama and M. Fukuda: Biosci. Biotechnol. Biochem. 71, 1–15 (2007). [DOI] [PubMed] [Google Scholar]
- 14) E. Palmqvist and B. Hahn-Hägerdal: Bioresour. Technol. 74, 25–33 (2000). [Google Scholar]
- 15) H. Nakashita, M. Yasuda, M. Nishioka, S. Hasegawa, Y. Arai, M. Uramoto, S. Yoshida and I. Yamaguchi: Plant Cell Physiol. 43, 823–831 (2002). [DOI] [PubMed] [Google Scholar]
- 16) H. D. Lüdemann and H. Nimz: Makromol. Chem. 175, 2393–2407 (1974). [Google Scholar]
- 17) F. de Sousa, L. M. Strömberg and K. P. Kringstad: Water Sci. Technol. 20, 153–160 (1988). [Google Scholar]
- 18) X. Peng, E. Masai, H. Kitayama, K. Harada, Y. Katayama and M. Fukuda: Appl. Environ. Microbiol. 68, 4407–4415 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19) A. Vladimirova, Y. Patskovsky, A. A. Fedorov, J. B. Bonanno, E. V. Fedorov, R. Toro, B. Hillerich, R. D. Seidel, N. G. J. Richards, S. C. Almo and F. M. Raushel: J. Am. Chem. Soc. 138, 826–836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20) H. Kessmann, T. Staub, C. Hofmann, T. Maetzke, J. Herzog, E. Ward, S. Uknes and J. Ryals: Annu. Rev. Phytopathol. 32, 439–459 (1994). [DOI] [PubMed] [Google Scholar]
- 21) W. E. Durrant and X. Dong: Annu. Rev. Phytopathol. 42, 185–209 (2004). [DOI] [PubMed] [Google Scholar]