Abstract
We herein present a method for the analysis of 10 dithiocarbamate fungicides (DTCs) in beer, fruit juice, and malt samples, where the DTCs were converted into water-soluble sodium salts in the presence of NaHCO3 prior to methylation using dimethyl sulfate. Extraction of the methylated compounds from matrices was performed using a “quick, easy, cheap, effective, rugged, and safe” (QuEChERS) method. Following a dispersive solid-phase extraction as a clean-up step, the methylated compounds were detected by liquid chromatography-tandem mass spectrometry. Performance evaluation was carried out on beer, fruit juice, and malt samples using representative compounds. Accuracies of the spiked compounds from the various matrices ranged from 92.2 to 112.6%, and the limits of quantification of propineb, mancozeb, and thiuram were <0.52, <0.55, and <6.97 µg/kg, respectively. The developed method was then applied in the determination of dithiocarbamate fungicide contents in commercial beer and fruit juice samples.
Keywords: dithiocarbamate fungicide, methylation, QuEChERS, liquid chromatography-tandem mass spectrometry
Introduction
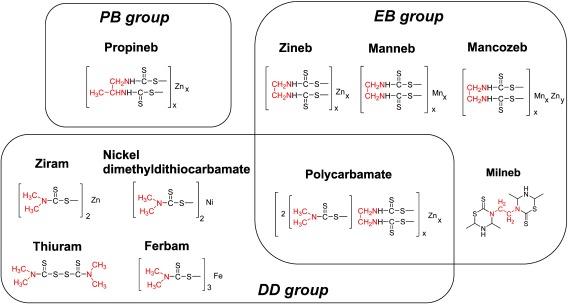
Dithiocarbamate fungicides (DTCs) have been extensively used worldwide on various crops due to their broad-spectrum control of a wide variety of fungal diseases. In the majority of countries, maximum residue levels (MRLs) of DTCs are applied to many food crops, such as vegetables, fruits, and grains, and so the precise monitoring of residue levels is of particular importance.1–3) According to the different carbon skeletons present in various DTCs (Fig. 1), the DTCs can be classified into three main groups: propylene-bis-dithiocarbamates (PBs, e.g., propineb), ethylene-bis-dithiocarbamates (EBs, e.g., mancozeb, manneb, zineb, and milneb), and dimethyl dithiocarbamates (DDs, e.g., thiuram, ziram, ferbam, and nickel dimethyldithiocarbamate). In addition, polycarbamate is classified as both an EB and a DD.
Fig. 1. Structures of the 10 dithiocarbamate fungicides of interest.
Despite their widespread use, the direct detection of DTCs is difficult, as many of these compounds are relatively unstable in aqueous solutions and exhibit low solubilities in both water and common organic solvents. Therefore, the classical method for the determination of DTCs is based on their decomposition to carbon disulfide (CS2), which is released during hot acid hydrolysis and is then detected by either spectrophotometry4–7) or gas chromatography (GC).8–10) However, this method can lead to false positives, because it is impossible to distinguish the product CS2 from that naturally present in food crops.11) Neither is it possible to discriminate between PB, EB, and DD using this method. Thus, as an alternative method, DTCs are often methylated with methyl iodide following their transformation into water-soluble sodium salts using an aqueous alkaline ethylenediaminetetraacetic acid (EDTA) solution, which allows for differentiation between PB, EB, and DD.12–14) However, the use of liquid chromatography-ultraviolet absorption (LC-UV) to detect the products obtained following this methylation process resulted in insufficient sensitivity. Therefore, a method based on the use of highly sensitive GC–mass spectrometry (GC-MS) was developed.15,16) However, the issues relating to tedious sample preparation remained for these methods, which led Hayama et al.17) to develop a simplified methylation technique by applying the “quick, easy, cheap, effective, rugged, and safe” (QuEChERS) approach. The QuEChERS method is a simple and easy method for the multiresidue analysis of pesticides in fruits and vegetables; it employs acetonitrile extraction/partitioning and dispersive solid-phase extraction.18–20) This methylation method enables simplification of the sample-preparation process but is tailored to a limited number of analytes, such as mancozeb, manneb, and zineb.
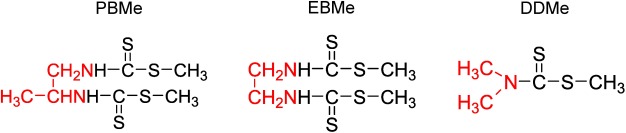
Thus, we herein aimed to develop a simple and sensitive method for the analysis of 10 DTCs in beer, fruit juice, and malt samples. The QuEChERS sample-preparation approach will be employed to achieve the simple extraction/partitioning and methylation steps in succession. This method should result in transformation of the 10 DTCs into the 3 methylated compounds shown in Fig. 2, namely dimethyl propylenebisdithiocarbamate (PBMe), dimethyl ethylenebis(dithiocarbamic acid) (EBMe), and methyl diethyldithiocarbamate (DDMe), which will then be detected by LC–tandem mass spectrometry (LC-MS/MS). We will then apply our proposed method in the analysis of DTCs present in beer, fruit juice, and malt samples.
Fig. 2. Structures of the three methylated compounds, namely PBMe, EBMe, and DDMe, which were obtained following the methylation of PB, EB, and DD, respectively.
Materials and Methods
1. Materials
The various standard reagents employed herein, i.e., propineb (84.2%), manneb (75%), zineb (96.9%), polycarbamate (102.4%), thiuram (98%), and ziram (99.5%) were purchased from Wako Pure Chemical Industries (Osaka, Japan), while mancozeb (73%) and ferbam (75%) were purchased from Dr. Ehrenstorfer GmbH (Augsburg, Germany). In addition, milneb (76%), nickel dimethyldithiocarbamate (99.9%), PBMe (99.4%), EBMe (98.7%), and DDMe (99.9%) were purchased from Hayashi Pure Chemical Industries (Osaka, Japan).
Acetonitrile (MeCN, LC/MS grade), sodium hydroxide (NaOH, special grade), dithiothreitol (DTT), and dimethyl sulfate (special grade) were supplied by Wako Pure Chemical Industries. Formic acid (LC/MS grade), sodium hydrogen carbonate (NaHCO3, special grade), L-cysteine hydrochloride monohydrate (cysteine, special grade), disodium dihydrogen ethylenediaminetetraacetate dihydrate (EDTA-2Na, special grade), and N,N-dimethylformamide (DMF, special grade) were purchased from Kanto Chemical Co. (Tokyo, Japan). All QuEChERS materials were obtained from Sigma-Aldrich (St. Louis, MO, USA). For the extraction step, we used a Supel™ QuE non-buffered tube, which contained magnesium sulfate (MgSO4, 4 g) and sodium chloride (1 g), while for the clean-up step, we employed a Supel™ QuE PSA (EN) tube containing MgSO4 (900 mg) and primary secondary amine (PSA, 150 mg). A 0.2 µm polytetrafluoroethylene (PTFE) filter (Advantech Toyo Kaisha Ltd., Tokyo, Japan) was used to filter the samples prior to analysis. Ultrapure water was obtained using a Milli-Q® Integral Q-POD EDS-Pak water purification system (Merck Millipore Japan, Tokyo, Japan).
All commercial samples were purchased from Japanese supermarkets and were stored at 4°C until employed for sample preparation.
2. Preparation of the reagents
The cysteine-EDTA solution was prepared by dissolving cysteine (50 g) and EDTA-2Na (50 g) in ultrapure water (1 L) and adjusting the pH to 9.6 using a 12 M aqueous NaOH solution. The 1 M DTT solution was prepared by dissolving DTT (0.154 g) in water (1 mL), while the 1 M NaHCO3 solution was prepared by dissolving NaHCO3 (0.84 g) in water (10 mL).
3. Preparation of the standard solutions
The solvents employed to prepare standard solutions of the various compounds were as follows: propineb for DMF; mancozeb, manneb, zineb, and polycarbamate for the cysteine-EDTA solution; thiuram, ziram, ferbam, milneb, and nickel dimethyldithiocarbamate for MeCN; PBMe, EBMe, and DDMe for 0.1 vol% formic acid in MeCN. All measured mass values were corrected according to the purity of each DTC. Standard stock solutions of PBMe, EBMe, and DDMe were prepared using the above formic acid/MeCN solution to obtain final concentrations of 200 mg/L, and these stock solutions were diluted further using 0.1 vol% formic acid in MeCN to give the desired concentrations. They were stored at −20°C in a refrigerator. The standard solutions of the 10 DTCs were prepared immediately prior to use due to issues regarding their degradation in one day.
4. Instrumentation details and analytical conditions
For the LC-MS/MS procedure, we employed a Nexera UHPLC system (Shimadzu, Kyoto, Japan) and an API 4000™ triple quadrupole mass spectrometer (Sciex, Framingham, MA, USA). Analyst 1.6.2 software was used to control the instruments and to process all recorded data. Chromatographic separation was performed using an Acquity UPLC BEH C18 column (100×2.1 mm, 1.7 µm) (Waters, Milford, MA, USA) equipped with a VanGuard BEH C18 guard column (5×2.1 mm, 1.7 µm) (Waters). The mobile phases employed for separation were a 0.1 vol% aqueous solution of formic acid (A) and MeCN (B). A column temperature of 40°C was used, along with a sample injection volume of 5 µL. Elution was carried out at a flow rate of 0.4 mL/min, and a gradient was established as follows: 40%B (0–0.5 min), 40–48%B (0.5–4 min), 48–80%B (4–4.5 min), 80%B (4.5–6.5 min), 80–40%B (6.5–6.7 min), and 40%B (6.7–10 min). An electrospray ionization (ESI) source was operated for MS measurements, and the operating parameters employed were as follows: 500°C ion source temperature, 4200 V ion spray voltage, 30 psi ion source gases 1 and 2, and 10 psi curtain gas. Quantification and confirmation data were acquired in the selected reaction monitoring (SRM) mode, and all compounds were measured in positive ion mode. Finally, Table 1 lists the precursor (Q1)-to-fragment (Q3) transitions, the optimum declustering potential (DP), the optimum collision energy (CE), and the optimum collision cell exit potential (CXP) for each compound.
Table 1. SRM conditions achieved for each compounda).
| Compounds | Precursor ion (m/z) | Production (m/z) | DP (V) | CE (V) | CXP (V) |
|---|---|---|---|---|---|
| EBMe | 241.1 | 193.0b) | 41 | 11 | 16 |
| 241.1 | 134.0c) | 41 | 25 | 8 | |
| DDMe | 136.1 | 88.0b) | 26 | 15 | 6 |
| 136.1 | 73.1c) | 26 | 45 | 6 | |
| PBMe | 255.0 | 206.9b) | 51 | 11 | 18 |
| 255.0 | 148.1c) | 51 | 27 | 12 |
a) DP=declustering potential, CE=collision energy, CXP=collision cell exit potential. b) Transition 1 was used for quantification. c) Transition 2 was used for confirmation.
5. Sample pre-treatment procedures
A portion (10 mL) of the liquid sample (i.e., beer or fruit juice) was placed in a 50 mL polypropylene centrifuge tube, and portions of the DTT solution (100 µL) and the NaHCO3 solution (0.5 mL, or 1 mL for the fruit juice samples) were added. Subsequently, MeCN (10 mL) and dimethyl sulfate (47.4 µL) were added to the sample, and the centrifuge tube was shaken for 15 min by a shaker. The contents of the Supel™ QuE non-buffered tube were then added to the sample solution, and the mixture was shaken for 10 min prior to centrifugation for 5 min at 3500 rpm to obtain the extract. A sample of the extract (i.e., the upper MeCN layer, 1 mL) was then transferred to a 15 mL polypropylene centrifuge tube, and a sample of the contents of the Supel™ QuE PSA SPE clean-up tube (175 mg) was added. The centrifuge tube was then vortexed for 30 sec and subjected to centrifugation for 3 min at 1500 rpm. The resulting supernatant was passed through a PTFE filter and diluted 5-fold using a 0.1 vol% solution of formic acid in MeCN. The obtained solution was placed in the autosampler of the LC-MS/MS system for analysis.
A portion (1 g) of the crushed solid sample (i.e., malt) was homogenized in water (10 mL), and solutions of DTT (100 µL) and NaHCO3 (0.5 mL) were added prior to the addition of MeCN (10 mL) and dimethyl sulfate (47.4 µL). The extraction and clean-up steps described above were then employed for the obtained solution. Finally, the supernatant obtained following the clean-up stage was passed through a PTFE filter, and formic acid (1 µL) was added prior to analysis of the clear filtrate by LC-MS/MS.
6. Performance evaluation
The experimental method was validated using representative compounds for each group, namely propineb for PB, mancozeb for EB, and thiuram for DD. The repeatability [relative standard deviation (RSD) %] and accuracy (%) were assessed using samples spiked with the analytes at a level of 10 µg/kg for propineb and mancozeb and 100 µg/kg for thiuram. Six pre-spiked samples were prepared and analyzed separately on the same day. The linearities of the standard addition calibration curves were then estimated from the six obtained points, and the linearity range for each component is listed in Table 3. Furthermore, the limits of quantification (LOQs) were calculated from the chromatograms and were defined as a signal-to-noise (S/N) ratio of 10.
Table 3. Linearity, repeatability, accuracy, and LOQs of the DTCs for the various samples.
| Sample | Compounds | Linearity (r) | RSD (%) | Recovery (%) | LOQ (mg/kg) |
|---|---|---|---|---|---|
| Beer | Propineb | 0.9999a) | 8.0 | 102.9 | 0.52 |
| Mancozeb | 0.9996a) | 7.8 | 92.2 | 0.55 | |
| Thiuram | 0.9993a) | 4.5 | 99.4 | 6.97 | |
| Apple juice | Propineb | 0.9993b) | 4.0 | 97.5 | 0.40 |
| Mancozeb | 0.9988b) | 9.6 | 98.5 | 0.50 | |
| Thiuram | 0.9990b) | 3.2 | 102.1 | 4.89 | |
| Grape juice | Propineb | 0.9969c) | 5.1 | 107.6 | 0.14 |
| Mancozeb | 0.9987c) | 4.9 | 108.2 | 0.13 | |
| Thiuram | 0.9997c) | 6.4 | 101.4 | 6.55 | |
| Malt | Propineb | 0.9991d) | 7.0 | 112.6 | 0.23 |
| Mancozeb | 0.9997d) | 6.4 | 108 | 0.49 | |
| Thiuram | 0.9999d) | 4.3 | 99.6 | 6.27 |
a) The concentrations were 5, 10, 20, 50, 80, and 100 µg/kg for propineb and mancozeb and 50, 100, 200, 500, 800, 1000, and 1500 µg/kg for thiuram. b) The concentrations were 5, 10, 20, 50, 80, and 100 µg/kg for propineb; 5, 10, 20, 50, 80, 100, and 150 µg/kg for mancozeb; and 50, 100, 200, 500, 800, 1000, and 1500 µg/kg for thiuram. c) The concentrations were 10, 20, 50, 80, 100, and 150 µg/kg for propineb; 5, 10, 20, 50, 80, 100, and 150 µg/kg for mancozeb; and 100, 200, 500, 800, 1000, and 1500 µg/kg for thiuram. d) The concentrations were 5, 10, 20, 50, and 80 µg/kg for propineb; 5, 10, 20, 50, 80, 100, and 150 µg/kg for mancozeb; and 50, 100, 200, 500, 800, and 1000 µg/kg for thiuram.
7. Quantification
To determine the concentration of each sample, calibration curves were obtained using the standard addition method to ensure the accuracy of the quantitative values. As the MRLs of DTCs are normally expressed as a measure of the CS2 content, the values calculated using the peak areas from the calibration curves corresponding to propineb, mancozeb, and thiuram were converted to CS2 contents by multiplying the following factors: PB by 0.60 (two molecules of CS2 generated from one molecule of PBMe); EB by 0.63 (two molecules of CS2 generated from one molecule of EBMe); and DD by 0.56 (one molecule of CS2 generated from one molecule of DDMe).
Results and Discussion
1. Optimization of the sample-preparation methods
As previously mentioned, DTCs, which form complexes with metal species, exhibit low solubilities in the majority of solvents, and so the addition of NaHCO3 was employed to convert these species to their corresponding water-soluble sodium salts. In this case, the addition of a conventional EDTA solution12–17,21–25) was not appropriate, as the LC-MS/MS peak corresponding to EDTA overlapped with the peak originating from PBMe. We therefore investigated the effect of the NaHCO3 concentration on the sample prior to methylation, using propineb, mancozeb, and thiuram as representative compounds for PB, EB, and DD, respectively, as previously mentioned. Thus, the beer samples spiked with propineb, mancozeb, thiuram, and their corresponding methylated compounds were analyzed and the methylation rate was calculated according to the following formula:
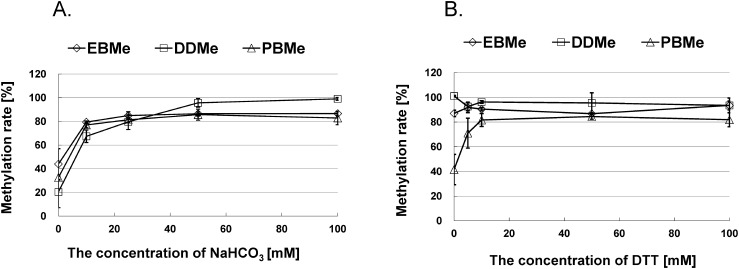
where Ax is the measured value of propineb, mancozeb, or thiuram as determined by LC-MS/MS; Am is the corresponding value for the methylated compounds; Mx is the molecular weight of propineb, mancozeb, or thiuram; and Mm is the molecular weight of the corresponding methylated compounds. It was assumed that the solubilities of these species in water increased upon the addition of NaHCO3, thus enhancing the methylation rate. Indeed, this is supported by the plots shown in Fig. 3A, where an optimized NaHCO3 concentration of 50 mM was apparent, as no further increase in methylation rate was observed for the various samples upon further increasing the NaHCO3 concentration.
Fig. 3. Effects of the concentrations of (A) NaHCO3 and (B) DTT on the methylation rate (%). The spiked concentrations of mancozeb, propineb, EBMe, and PBMe in the beer samples were 10 µg/kg, while those of thiuram and DDMe were 100 µg/kg. Data are presented as the mean±the standard error (n=3).
However, as DTCs are unstable in water, their calculated quantities can be underestimated. Thus, we attempted to prevent degradation by adding DTT as a stabilizer, as it has been reported that reducing agents are effective stabilizers.26) In this case, we could not select cysteine,12–17,22–25) as the retention times for the methylated form of cysteine and DDMe were comparable under the same MS/MS conditions due to their similar molecular weights. Thus, we also investigated the effect of the DTT concentration on the samples and found that degradation was prevented in a number of cases, resulting in enhanced methylation rates. As indicated in Fig. 3B, the optimized DTT concentration for the methylation procedure was 10 mM.
2. Methylation rates of the DTCs
The beer sample spiked with 10 DTCs and their corresponding methylated compounds was analyzed and the methylation rates were calculated. The intraday and interday repeatability values were then evaluated, and methylation rates of 91.6, 87.8–94.1, and 87.8–94.9% were obtained for the intraday repeatability of PB, EB, and DD, respectively. These results confirmed that no significant difference in methylation rates existed between the various compounds. Furthermore, the RSD values obtained for the intraday repeatability were 2.0, 2.4–9.6, and 1.6–6.5% for PB, EB, and DD, respectively. Moreover, for the interday repeatability, satisfactory RSD values of 9.4, 10.2–13.2, and 4.6–10.3% were obtained for the PB, EB, and DD samples, respectively (Table 2).
Table 2. Methylation rates and relative standard deviations for the beer samples spiked using a range of DTCsa).
| Compounds | Intra-day | Inter-day | ||
|---|---|---|---|---|
| Methylation rate (%) | RSDb) (%) | Methylation rate (%) | RSDc) (%) | |
| Propineb | 91.6 | 2.0 | 80.2 | 9.4 |
| Mancozeb | 87.8 | 3.2 | 88.6 | 12.1 |
| Manneb | 85.8 | 9.6 | 89.8 | 10.2 |
| Zineb | 87.2 | 9.2 | 84.4 | 10.7 |
| Polycarbamate as EB | 87.1 | 2.4 | 82.9 | 13.2 |
| Milneb | 94.1 | 9.1 | 96.4 | 10.3 |
| Thiuram | 91.5 | 3.7 | 93.5 | 10.3 |
| Nickel dimethyldithiocarbamate | 94.9 | 6.5 | 92.1 | 7.7 |
| Polycarbamate as DD | 92.3 | 1.6 | 94.2 | 4.6 |
| Ferbam | 94.7 | 5.0 | 99.5 | 8.4 |
| Ziram | 87.8 | 5.1 | 89.2 | 8.4 |
a) Spiked levels were 10 µg/kg for EB and PB and 100 µg/kg for DD. b) The intraday RSD value represents the repeatability obtained by analyzing. c) The interday RSD value represents the repeatability obtained by analyzing samples over five consecutive days.
3. Performance evaluation results
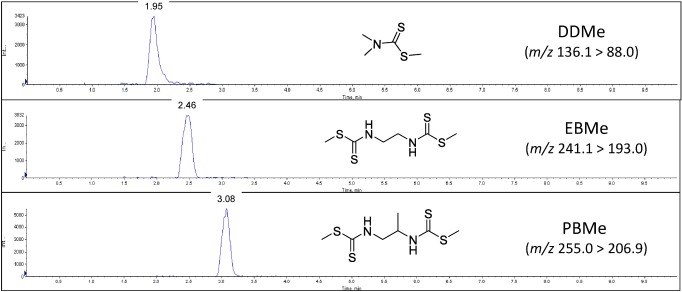
Performance evaluation was carried out using representative compounds, as previously mentioned. The typical chromatograms of a beer sample spiked with the PBMe, EBMe, and DDMe standards are shown in Fig. 4. In this case, sample preparation was relatively simple, and no background signals were observed in the chromatograms of these samples.
Fig. 4. LC-MS/MS chromatograms from the analysis of beer samples spiked with EBMe, PBMe, and DDMe. The spiked concentrations were 10 µg/kg for EBMe and PBMe and 100 µg/kg for DDMe.
Indeed, as outlined in Table 3, satisfactory performance evaluation results were obtained for the beer, apple juice, grape juice, and malt samples. More specifically, the repeatability was <9.6%, the accuracy ranged from 92.2 to 112.6%, and the linearity (r) was ≥0.99. Furthermore, the LOQs of propineb, mancozeb, and thiuram were 0.14–0.52, 0.13–0.55, and 4.89–6.97 µg/kg, respectively, which is low enough to comply with the MRL values established by a number of regulatory organizations. Moreover, all validated results satisfied the EU criteria of SANTE/11945/201527) and ensured an adequate quantitation of the analytes.
4. Sample analysis
We employed the developed method to analyze 9 commercial beer samples, 10 grape juice samples, and 5 apple juice samples. As the DTCs appeared to be present at levels lower than the LOQs for all samples, their quantification was not possible. These results therefore confirmed that no significant risk of DTC contamination exists in the Japanese market.
Conclusion
We herein reported the establishment of a method for the simultaneous analysis of 10 dithiocarbamate (DTC) fungicides in beer, fruit juice, and malt samples, wherein the application of a “quick, easy, cheap, effective, rugged, and safe” (QuEChERS) method following sample preparation allowed one-pot successive extraction and methylation of the water-soluble sodium salts, thus rendering our method simple and rapid. In addition, the liquid chromatography-tandem mass spectrometry procedure employed for detection of the methylated DTCs was highly sensitive, resulting in limits of quantification <1 µg/kg for dimethyl ethylenebis(dithiocarbamic acid) and methyl dimethyldithiocarbamate and <7 µg/kg for dimethyl propylenebisdithiocarbamate. The developed method was also applied to the determination of DTC contents in commercial beer and fruit juice samples, although the DTCs appeared to be present at levels lower than the limits of quantification. We therefore believe that this method will be suitable for quality control of our products such as beer and fruit juice.
References
- 1) European Commission: Official Journal of the European Union, Regulation No. 2016/1 (2015).
- 2) European Commission: Official Journal of the European Union, Regulation No. 2016/567 (2016).
- 3) Ministry of Health, Labour and Welfare: The Maximum Residue Limits of Substance used as Ingredient of Agricultural Chemical in Foods. Document No. 499 (2005).
- 4) T. E. Cullen: Anal. Chem. 36, 221–224 (1964). [Google Scholar]
- 5) G. E. Keppel: J. AOAC Int. 54, 528–532 (1971). [Google Scholar]
- 6) E. D. Caldas, M. H. Conceição, M. C. C. Miranda, L. C. K. R. Souza and J. F. Lima: J. Agric. Food Chem. 49, 4521–4525 (2001). [DOI] [PubMed] [Google Scholar]
- 7) A. K. Malik, V. Sharma, V. K. Sharma and A. L. J. Rao: J. Agric. Food Chem. 52, 7763–7767 (2004). [DOI] [PubMed] [Google Scholar]
- 8) T. K. McGhie and P. T. Holland: Analyst (Lond.) 112, 1075–1076 (1987). [Google Scholar]
- 9) A. Royer, M. Ménand, A. Grimault and P. Y. Communal: J. Agric. Food Chem. 49, 2152–2158 (2001). [DOI] [PubMed] [Google Scholar]
- 10) N. Ahmad, L. Guo, P. Mandarakas and S. Appleby: J. AOAC Int. 78, 1238–1243 (1995). [PubMed] [Google Scholar]
- 11) R. C. Perz, H. Lishaut and W. Schwack: J. Agric. Food Chem. 48, 792–796 (2000). [DOI] [PubMed] [Google Scholar]
- 12) K. H. Gustafsson and R. A. Thompson: J. Agric. Food Chem. 29, 729–732 (1981). [DOI] [PubMed] [Google Scholar]
- 13) K. H. Gustafsson and C. H. Fahlgren: J. Agric. Food Chem. 31, 463–466 (1983). [DOI] [PubMed] [Google Scholar]
- 14) N. Kibune, N. Higashisaka, M. Nakamura and Y. Maekawa: Food Hyg. Saf. Sci. 36, 244–251 (1995). [Google Scholar]
- 15) T. Kawamoto, M. Yano and N. Makihata: J. Chromatogr. A 1074, 155–161 (2005). [DOI] [PubMed] [Google Scholar]
- 16) M. Nakamura, S. Noda, M. Kosugi, N. Ishiduka, L. Mizukoshi, M. Taniguchi and S. Nemoto: Food Hyg. Saf. Sci. 51, 213–219 (2010). [DOI] [PubMed] [Google Scholar]
- 17) T. Hayama and M. Takada: Anal. Bioanal. Chem. 392, 969–976 (2008). [DOI] [PubMed] [Google Scholar]
- 18) M. Anastassiades, S. J. Lehotay, D. Štajnbaher and F. J. Schenck: J. AOAC Int. 86, 412–431 (2003). [PubMed] [Google Scholar]
- 19) S. J. Lehotay: J. AOAC Int. 90, 485–520 (2007). [PubMed] [Google Scholar]
- 20) European Committee for Standardization: Standard Method EN 15662 (2008).
- 21) C. Lo, M. Ho and M. Hung: J. Agric. Food Chem. 44, 2720–2723 (1996). [Google Scholar]
- 22) O. L. Fernández, R. R. Otero, C. G. Barreiro and J. S. Gándara: Food Chem. 134, 366–374 (2012). [Google Scholar]
- 23) O. H. J. Szolor: Anal. Chim. Acta 582, 191–200 (2007). [DOI] [PubMed] [Google Scholar]
- 24) G. Crnogorac and W. Schwack: Trends Analyt. Chem. 28, 40–50 (2009). [Google Scholar]
- 25) Y. Hanada, T. Tanizaki, M. Koga, H. Shiraishi and M. Soma: Anal. Sci. 18, 441–444 (2002). [DOI] [PubMed] [Google Scholar]
- 26) H. Kobayashi, M. Nishida, O. Matano and S. Goto: J. Agric. Food Chem. 40, 76–80 (1992). [Google Scholar]
- 27) European Commission: Guidance Document on Analytical Quality Control and Method Validation Procedures for Pesticides Residues Analysis in Food and Feed, Document No. SANTE/11945/2015.