Abstract
Brassinosteroids (BRs) are plant steroid hormones involved in plant growth and environmental adaptation. It is well known that oxidation/hydroxylation steps in the BR biosynthetic pathway are catalyzed by cytochrome P450 enzymes. It has been proposed that brassinolide is biosynthesized from campesterol via campestanol (CN) in the original BR biosynthetic pathway. However, a recent enzymatic analysis of cytochrome P450 enzymes and re-evaluation of the endogenous amount of BRs in BR-deficient mutants included an investigation of the novel BR biosynthetic pathway (CN-independent pathway) not via CN. This review highlights comprehensive recent advances in the biochemical research of BR biosynthetic enzymes and the CN-independent pathway. This review also focuses the biosynthesis inhibitors and the antagonists/agonists that are utilized not only as plant growth regulators but also as tools for the chemical and biological investigation of the physiological functions of BRs.
Keywords: brassinosteroid, cytochrome P450, biosynthesis, plant growth regulator
Introduction
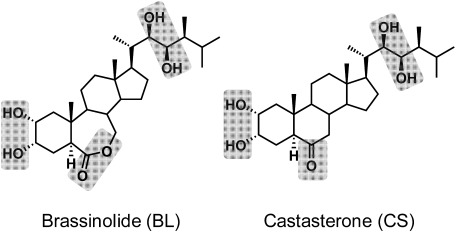
Brassinosteroids (BRs) are a class of plant steroidal hormones that have been recognized as a sixth phytohormone that plays an essential role in plant growth. BRs have been isolated from 64 plant species (53 angiosperms, 6 gymnosperms, Equisetum arvense (a pteridophyte), Marchantia polymorpha (a bryophyte), and 3 algae (Chlorella vulgaris, Cystoseira myrica and Hydrodictyon reticulatum) and have been detected in leaves, stems, roots, flowers, pollen, anthers, and seeds as recently as 2011.1) At least 69 BRs, including 65 free BRs and 5 conjugated BRs, have been identified and characterized.1) The various chemical structures of BRs can be derived from a 5α-steroid skeleton and are arranged by the functional groups on the A ring, B ring, and side chain.1,2) The most physiologically active compound, brassinolide (BL), is a C28 steroid that exists widely in the plant kingdom, together with other related BR compounds.1,2) After the isolation and structural determination of BL from the pollen of rapeseed (Brassica napus) in 1979,3) the second BR, castasterone (CS), was isolated from the insect galls of the chestnut (Castanea crenata) in 19824) (Fig. 1). Subsequently, numerous BRs have been identified in various plants.1,2,5) BR biosynthesis research is growing in both the chemical approach, including isolation and identification of BRs in plants and pulse-chase analysis using labeled synthetic BRs, and in the molecular genetic approaches of exploring BR biosynthetic pathways in BR-deficient mutants.1,2,5)
Fig. 1. Chemical structures of brassinolide (BL) and castasterone (CS). Gray shades are essential groups of BR physiological activities.1,2).
In the 1990s, BR biosynthesis-deficient mutants were discovered in various plants, and BRs were recognized as plant hormones that are indispensable for the physiology of plants.6–8) Research involving the chemical analysis of the endogenous BRs and the phenotype rescue of BR biosynthesis-deficient mutants by exogenous application of BRs suggested that several cytochrome P450 (P450) genes (CYP85A, CYP90A, CYP90B, CYP90C, CYP90D, and CYP724) and a reductase (DET2) may be involved in oxidation and reduction steps in the BR biosynthetic pathway.2,6) However, the above indirect methods have limitations when it comes to ascertaining the enzymatic function of each P450 and reductase, because the endogenous level of BRs in plants is extremely low, and the BR biosynthetic pathway forms metabolic grids.9) Therefore, an in vitro enzymatic assay with heterologously expressed proteins would be a powerful and direct method of elucidating the enzymatic functions of the BR biosynthetic pathway. Such biochemical analyses have recently revealed the functions of BR biosynthetic enzymes that catalyze 5α-reduction, C-3 oxidation (oxidation of the 3β-hydroxy group coupled with migration of the C-5 double bond), C-22 hydroxylation, C-23 hydroxylation, C-6 oxidation, and Baeyer–Villiger oxidation.
1. P450 enzymes
P450s catalyze the stereospecific oxidation of unactivated hydrocarbons. P450s exist ubiquitously in more than 5,000 species (3,651 plants, 2,960 fungi, 2,137 insects, 1,461 vertebrates, 1,042 bacteria, 27 archaea, and 2 viruses).10) In plants, there are 245 genes in Arabidopsis thaliana, 334 in Oryza sativa, 316 in Vitis vinifera, 332 in Glycine max, 71 in Physcomitrella patens, 40 in Chlamydomonas, and 19 in Volvox.10,11) The number of plant P450s is particularly large compared with number in Drosophila (87 genes) and humans (56 genes). P450 is a hemoprotein containing heme iron at the active center and a cysteine-derived thiolate anion coordinated with heme iron. The maximum absorption band (Soret band) of reduced P450 is observed at around 420 nm. When carbon monoxide is coordinated with the heme iron Fe (II) in reduced P450, ultraviolet-visible spectroscopic analysis reveals that the Soret band shifts to 450 and 380 nm (“pigment 450 nm” is the origin of the P450 name). Plant P450s localize to the endoplasmic reticulum membrane and catalyze the oxidation of substrates by activating molecular oxygen in conjunction with the NADPH-P450 reductase. P450 is a widely used target of not only fungicides but also plant growth regulators (PGRs).
2. Campestanol (CN)-dependent pathway
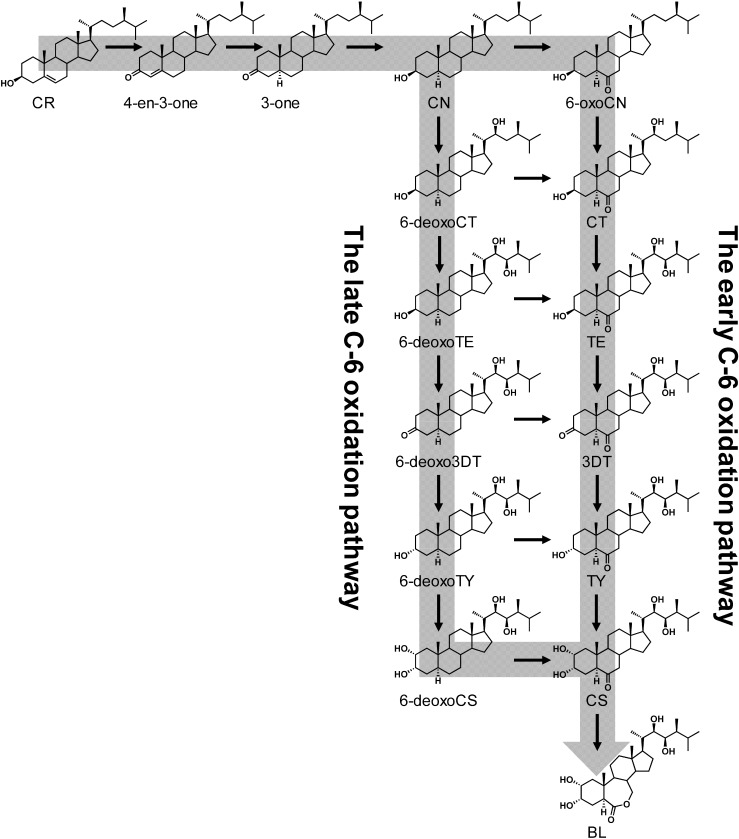
C28 BRs are synthesized from campesterol (CR), which is one of the phytosterols that possess a methyl group at the C-24 position in its side chain (Fig. 2). CR is converted to CN via campest-4-en-3-one (4-en-3-one) and 5α-campestan-3-one (3-one) by several BR biosynthetic enzymes.6) CN is then converted to CS through one of two pathways, the early C-6 oxidation pathway (where the C-6 position is oxidized early) or the late C-6 oxidation pathway (where the C-6 position is oxidized in the final step).2) CS is finally converted to BL via Baeyer–Villiger oxidation at the B ring.
Fig. 2. Conventional proposed BR biosynthetic pathway (CN-dependent pathway).
On the basis of the chemical analysis of endogenous BRs, it was found that 6-deoxoBRs (late C-6 oxidation pathway) are predominant over 6-oxoBRs (early C-6 oxidation pathway) in most plants. In the original BR biosynthetic pathway, it was proposed that BL is biosynthesized from CR via CN through the late C-6 oxidation pathway. In 1997, it was proposed that the Arabidopsis DET2 enzyme, which is homologous to mammalian steroid 5α-reductases, acts at the step in which CR is converted to CN.12) Arabidopsis det2 mutants have a small, dark-green dwarf phenotype in light-regulated development. DET2 is similar to mammalian steroid 5α-reductases (38 to 42% sequence identity with the deduced amino acid levels). The mammalian steroid 5α-reductase (Types 1 and 2) has been isolated and catalyzes the conversion of testosterone to dihydrotestosterone, which is a key step in steroid metabolism. DET2 actually catalyzes the reduction of 4-en-3-one to 3-one.13) The phenotype of Arabidopsis det2 mutants has been rescued with the overexpressed the mammalian steroid 5α-reductase gene.13) These findings opened the door for BR to be recognized as the sixth plant hormone and led to the further development of BR biosynthesis signaling researches.
3. C-22 hydroxylation
The phenotype of the Arabidopsis dwf4 mutant exhibits a severe dwarf phenotype, and the leaves are dark green with small curls. DWF4 encodes a cytochrome P450 protein, CYP90B1.14)
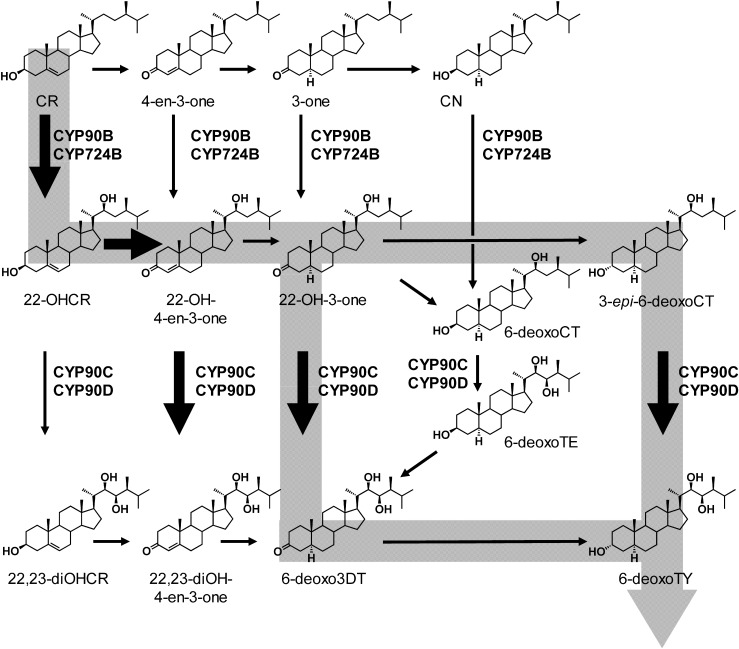
It was assumed that CYP90B1 would catalyze C-22 hydroxylation based on measurements of the endogenous BR amounts in dwf4 mutants and on phenotype rescue experiments involving the administration of BR biosynthesis intermediates. Concurrently, a novel compound, 22-OHCR with a hydroxyl group at the C-22 position, was identified in Arabidopsis, and metabolic experiments using the deuterium labeled 22-OHCR showed that it was converted to 22-OH-4-en-3-one→22-hydroxycampest-3-one (22-OH-3-one)→6-deoxocathasterone (6-deoxoCT)15) (Fig. 3). This indicated that the early C-22 oxidation pathway also functioned in Arabidopsis in parallel with the route from CR to CN. In 2006, Fujita et al. carried out enzymatic chemical analysis of the function of recombinant CYP90B1 expressed in Escherichia coli and demonstrated that CYP90B1 is a C-22 hydroxylase.16) In addition, the catalytic efficiency (kcat/Km) showed that the substrate specificity of CYP90B1 was about 300 times higher for CR than for CN, indicating that CR is a good substrate of CYP90B1.16) Furthermore, endogenous CR levels are more than 50 times higher those of than CN. Taken together, these observations suggest that the early C-22 hydroxylation is the main route for BR biosynthesis in Arabidopsis.
Fig. 3. BR C-22 and C-23 hydroxylation. CYP90B and 724B function in the BR in early C-22 hydroxylation. CYP90C/D catalyzes the C-23 hydroxylation of 22-hydroxylated BRs (22-OHCR, 22-OH-4-en-3-one, 22-OH-3-one, 3-epi-6-deoxoCT, and 6-deoxoCT). Gray band shows the main route in C-22 hydroxylation and C-23 hydroxylation.
Molecular evolutionary analysis of plant P450s has shown that CYP90B and CYP724 are located in the same cluster, which suggests that the two enzymes possess the same function.16) A rice cyp724B1 single mutant exhibits a weak phenotype, including a shorter seed length, and a cyp90b2 single mutant produces only a very weak dwarf phenotype. Additionally, a cyp90b2/cyp724B1 double mutant exhibits a remarkable dwarf phenotype. This suggests that the two P450s are functionally redundant in rice. The enzymatic characterization of CYP724B1 and CYP90B2 revealed that the two P450s are C-22 hydroxylases that convert CR to 22OH-CR and CN to 6-deoxoCT and that their substrate specificity toward CR is higher than that toward CN. Thus, from the viewpoint of the molecular evolution of P450s, it is interesting that CYP724B and CYP90B, belonging to two different P450 families, both function as C-22 hydroxylases (Fig. 3).
4. C-23 hydroxylation
Initially, CYP90A1 was inappropriately attributed to a C-23 hydroxylase that converts cathasterone (CT) to teasterone (TE) because of a quantitative analysis of endogenous steroid levels in an Arabidopsis constitutive photomorphogenesis and dwarfism (cpd) mutant that exhibited severe dwarfism due to a deficiency of CYP90A1.17) This erroneous conclusion was drawn because of the available indirect evidence that the dwarf cpd phenotype could be rescued by TE (22,23-dihydroxyBR) but not by CT (22-hydroxyBR without a C-23 hydroxy group). In addition, CYP90C1 encoded by the Arabidopsis ROTUNDIFOLIA3 (ROT3) gene, which is involved in the regulation of leaf length, was initially assigned to be a C-2 hydroxylase on the basis of phenotype rescue experiments of rot3 mutants.18,19) Finally, CYP90D1, which is most closely related to ROT3, was misunderstood to be an oxidase of the 3β-hydroxy group that produces 22-OH-4-en-3-one from 22-OHCR. However, these inappropriate attributions of enzymatic function were later corrected by biochemical analysis of CYP90C1 and CYP90D1.20) The growth-deficient phenotype of a cyp90c1cyp90d1 double mutant was rescued by exogenous feeding of only 23-hydroxylated BRs. This indicated that the cyp90c1cyp90d1 double mutant was deficient in C-23 hydroxylation and that the genes were functionally redundant. In vitro assays indicated that CYP90C1/CYP90D1 catalyzed the C-23 hydroxylation of various 22-hydroxyBRs.20) Although indirect methods such as quantitative analysis of the endogenous BR in BR-deficient mutants and phenotypic rescue experiments are valid approaches to identify of the enzymatic functions of BR biosynthetic enzymes, direct methods such as in vitro enzymatic assay tend to yield more accurate results. CYP90C1 and CYP90D1 function redundantly as BR C-23 hydroxylases (Fig. 3).
5. C-3 oxidation
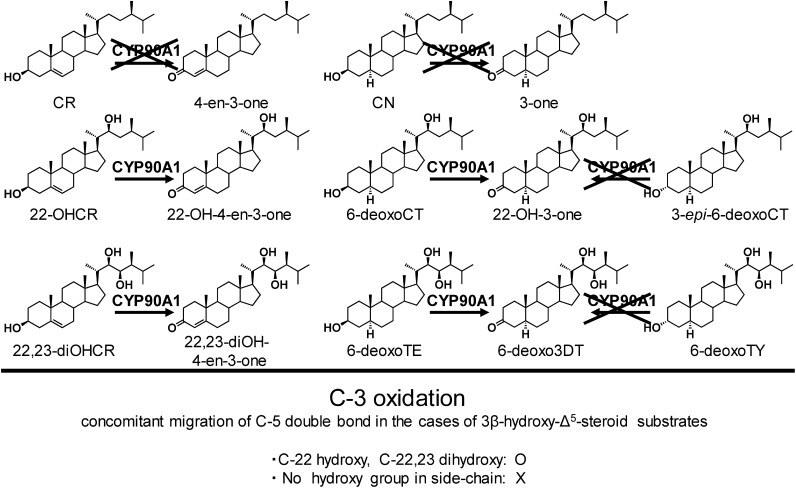
This step consists of oxidation of the 3β-hydroxy group and migration of the C-5 double bond. Therefore, the overall reaction converts 3β-hydroxy-Δ5 steroids to 3-oxo-Δ4 steroids. In case of human steroid hormone biosynthesis, the 3β-hydroxysteroid dehydrogenase/Δ5–4 isomerase, a non-P450 enzyme, catalyzes the reaction, and 3β-hydroxy-Δ5 steroids such as pregnenolone, 17α-hydroxypregnenolone and dehydroepiandrosterone are converted to corresponding 3-oxo-Δ4 steroids.21) In contrast, the conversion from (22S)-22-hydroxycampesterol (22-OHCR) and (22R,23R)-22,23-dihydroxycampesterol (22,23-diOHCR) to (22S)-22-hydroxycampest-4-en-3-one (22-OH-4-en-3-one) and (22R,23R)-22,23-dihydroxy-4-en-3-one (22,23-diOH-4-en-3-one), respectively, in BR biosynthesis is catalyzed by P450. In this reaction, a C-3 carbon atom would be directly oxidized by CYP90A1, i.e., CYP90A1 would put an additional oxygen atom on the C-3 carbon to yield gem-diol intermediate,12) which is subsequently dehydrated to furnish a carbonyl group. This mechanism suggests that CYP90A1 could essentially convert 3β-hydroxy-Δ5 steroids to 3-oxo-Δ5 steroids without double-bond migration.
As mentioned above, CYP90A1 was erroneously assigned as a C-23 hydroxylase.17) In 2006, genetic and biochemical approaches revealed that CYP90C1 and CYP90D1 catalyze the C-23 hydroxylation step of the BR biosynthetic pathway, and an Arabidopsis cyp90c1cyp90d1 double mutant exhibited remarkable dwarfism.20) If CYP90A1 is a member of C-23 hydroxylases, as originally concluded, CYP90A1 and CYP90C1/CYP90D1 would be redundant in function. However, overexpression of CYP90C1 does not revert the cpd mutant to dwarfism.22) This result clarified that CYP90A1’s function is different from that of CYP90C1/CYP90D1. To re-evaluate the CYP90A1 function, BR quantitative analyses were repeated. The endogenous BR levels of the cpd mutant had already been analyzed in 1996, but the first experiments analyzed BR biosynthesis intermediates of the late C-6 oxidation pathway.17) The second quantitative analysis focused on BR intermediates not only on the early C-6 hydroxylation pathway but also on the early C-22 hydroxylation pathway. This second study found that 6-deoxo-3-dehydroteasterone (6-deoxo3DT) and 6-deoxocastasterone (6-deoxoCS) in the early C-22 hydroxylation pathway were markedly reduced in the cpd mutant as compared with the wild type. 6-Deoxoteasterone (6-deoxoTE) and 6-deoxotyphasterol (6-deoxoTY) were found to be below the limits of detection. In contrast, the endogenous level of 22-OHCR increased 85- to 121-fold in the cpd mutant.22) These findings increased the possibilities that CYP90A1 is (1) a C-23 hydroxylase of 22-OHCR to 22,23-diOHCR or (2) an oxidase of the 3β-hydroxy group and isomerase of the C-5 double bond of 22-OHCR to 22-OH-4-en-3-one. To more directly clarify the function of CYP90A1, an in vitro enzymatic assay using a baculovirus–insect cell expression system was performed. The biochemical analysis of CYP90A122) showed that this enzyme catalyzes the C-3 oxidation of 3β-hydroxy-Δ5-steroids such as 22-OHCR and 22,23-diOHCR to form 22-OH-4-en-3-one and 22,23-diOH-4-en-3-one (Fig. 4). CYP90A1 also catalyzes the oxidation of the 3β-hydroxy group of C-5 saturated steroids such as 6-deoxoCT and 6-deoxoTE.22) Therefore, CYP90A1 is considered to be mainly responsible for C-3 oxidation in plants. However, CYP90A1 did not react with CR, which possesses no hydroxy group in the side chain. In addition, CYP90A1 did not recognize 3α-hydroxy steroids such as 3-epi-6-deoxoCT and 6-deoxoTY as substrates. These findings clearly demonstrate that CYP90A1 is specific to 3β-hydroxy steroids, and the presence of a hydroxy group at C-22 seems to be essential for the substrate recognition of CYP90A1.
Fig. 4. C-3 oxidation (oxidation of the 3β-hydroxy group coupled with migration of the C-5 double bond) of BR biosynthetic intermediates by CYP90A.
6. C-6 oxidation
CS is recognized as an active BR, and its bioactivity is estimated to be as high as one-fourth of that of BL based on results from a lamina joint inclination test and on their dissociation constants for the BR receptor (BRASSINOSTEROID-INSENSITIVE I; BRI1).23,24) The tomato d mutant with a defect in the CYP85A1 gene exhibits a severe dwarf phenotype resembling Arabidopsis cpd and dwarf4 mutants, and so CYP85 was predicted to be involved in BR biosynthesis.25) In 1999, the enzymatic characterization of tomato CYP85A1 demonstrated that it is a C-6 oxidase catalyzing the C-6 oxidation of 6-deoxoCS to 6α-hydroxyCS and the sequential oxidation of 6α-hydroxyCS to CS.26) This is the first P450 that was enzymatically characterized in the BR biosynthetic pathway. In addition, it was revealed that Arabidopsis CYP85A1 and CYP85A2 and rice OsDWARF are BR C-6 oxidases producing CS from 6-deoxoCS as a substrate.27,28) In contrast, CYP85A expressed in yeast did not catalyze the C-6 oxidation step of CN to 6-oxocampestanol (6-oxoCN). In addition, the endogenous levels of 6-oxoCN in a cyp85A1cyp85A2 double mutant were almost the same as in the wild type. These results indicate that the C-6 oxidation step of CN is catalyzed by an unidentified enzyme that is not CYP85A.
7. 7-oxa-6-oxolactone formation
The conversion from CS to BL introduces an oxygen atom into the C–C bond of the B ring to form 7-oxa-6-oxolactone. Such lactone formation is similar to a Baeyer–Villiger oxidation reaction, and flavin-containing oxygenase catalyzes this reaction in fungi, but in Arabidopsis, P450 catalyzes 7-oxa-6-oxolactone formation. A large amount of BL was detected in the tomato fruit, and the CYP85A3 gene, which is different from the CYP85A1 gene, was strongly expressed. CYP85A3 expressed in yeast catalyzes the C-6 oxidation of 6-deoxoCS to produce CS and continuously catalyzes further oxidation to produce BL.29) Similarly, Arabidopsis CYP85A2 was shown to catalyze this oxidation from CS to BL.30) In contrast, CYP85A1 in tomato plants and Arabidopsis did not catalyze the lactonization of CS. Thus, the difference in enzyme function among the CYP85A subfamily is interesting with respect to the evolution of P450.
8. CN-independent pathway
The originally proposed BR biosynthetic pathway was initiated by the C-3 oxidation of CR to 4-en-3-one, which was then converted by 5α-reductase to 3-one, which was then converted to CN (Fig. 2); however, CYP90A1 does not convert CR to 4-en-3-one.22) The endogenous level of CN in cpd mutant is equivalent to that in wild type in spite of the mutant’s severe BR deficiency. These findings indicate that CYP90A1 is not involved in CN synthesis and the biosynthetic route from CR to CN is not essential for BR biosynthesis. The early C-22 hydroxylation holds a prominent position in BR biosynthesis. CR is initially converted to 22-OHCR, and because 22-OHCR is not a favored substrate of the C-23 hydroxylase (CYP90C1 and CYP90D1), the endogenous levels of 22,23-diOHCR are below the detection limit in cpd mutants. Then CYP90A1 converts 22-OHCR to 22-OH-4-en-3-one. CYP90C1 and CYP90D1 prefer 22-OH-4-en-3-one, 22-OH-3-one, and 3-epi-6-deoxoCT as substrates. The catalytic efficiency (kcat/Km) of CYP90C1 and CYP90D1 toward these three 22-hydroxyBRs is 60 to 120 times higher than that for 6-deoxoCT. The endogenous amounts of intermediates in the early C-22 hydroxylation step demonstrated that 22-OH-3-one, 3-epi-6-deoxoCT, and 6-deoxoCT are comparable, whereas 22-OH-4-en-3-one was not detected at all. On the basis of the above, CYP90C1 and CYP90D1 directly cause the C-23 hydroxylation of 22-OH-3-one and 3-epi-6-deoxoCT to 6-deoxo3DT and 6-deoxoTY, respectively.21) These findings suggest that the main biosynthetic pathway of BR in plants is a pathway leading to a BR shortcut via the C-22 early hydroxylation route, but the contribution of the CN-dependent pathway via CN has not yet been clarified. However, despite the marked dwarfism of the cpd mutant, the endogenous levels of CR and CN are almost the same in it and the wild type. Therefore, the biosynthetic pathway from CR to CN normally functions even in the severe dwarf cpd mutant. Based on these findings, CN is not the main BR biosynthetic pathway, and the CN-independent pathway (CR→22-OHCR→22-OH-4-en-3-one→22-OH-3-one→3-epi-6-deoxoCT/6-deoxo3DT→6-deoxoTY→6-deoxoCS→CS→BL), which does not pass through CN, is considered to be the main BR biosynthetic route. It is not currently known how grid-like routes are used by plants and whether they are properly used. Future identification of these unidentified enzyme genes will further clarify the full picture of BR biosynthesis (Fig. 5).
Fig. 5. The campestanol (CN) independent route (the novel BR biosynthetic pathway). Gray band shows the proposed main BR biosynthetic pathway.
9. PGRs for regulating BR activities
BL was isolated and structurally determined, and BRs, 24-epicastasterone and 24-epibrassinolide, were synthesized soon afterward.31) As a result of intensive BR synthetic research, numerous synthetic analogs including natural and artificial chemicals have been developed and have successfully revealed various BR physiological functions, such as cell elongation, cell division, and stress tolerance. Even today, the study of the structure–activity relationships of BR analogs to BRI1 plays an important role in understanding the physiological functions of BRs in developing PGRs.31,32)
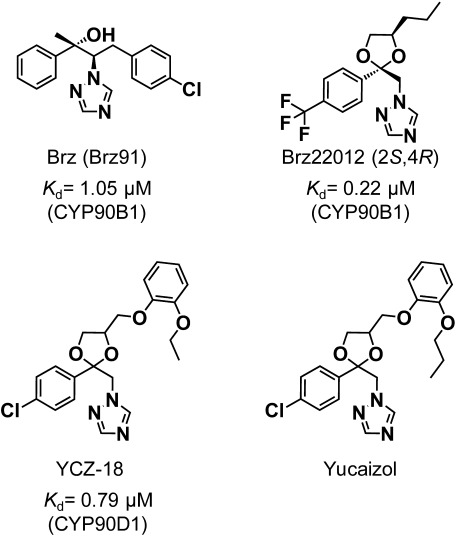
Triazole-type chemicals have been widely utilized as PGRs.33) Some triazole-type chemicals have fungicidal properties associated with the inhibition of sterol biosynthesis, whereas others are potent inhibitors of elongation growth in plants. Triazole-type PGRs such as uniconazole (UNI), paclobutrazol (PBZ), flurprimidol, and triapenthenol reduce shoot growth in plants by inhibiting a gibberellin biosynthetic P450 (CYP701). CYP701 catalyzes the three oxidation steps of ent-kauren to ent-kaurenoic acid. These triazole-type compounds are known to have affinity for the heme contained in P450s. Phenotype rescue experiments and analysis of the transcript levels of BR biosynthetic genes demonstrated that UNI and PBZ inhibit BR biosynthesis. However, although these triazole-type PGRs mostly inhibit P450s in plants, there are many reports that they also alter endogenous levels of primary and specialized metabolites such as phytosterols, phenylpropanoids, and other plant hormones. This is due to the low affinity of each P450 active site for the PGRs. To find and develop specific inhibitors of BR biosynthesis, a chemical screening assay was performed and brassinazoles (Brz91, Brz2001, Brz220, etc.) were identified as potent inhibitors (Fig. 6).34–37) To date, Brz91 (normally named brassinazole or “Brz”) is currently marketed as a reagent. The chemical structure of Brz is similar to triazole-type PGRs such as UNI and PBZ. Brz induces a severe dwarf phenotype that resembles BR biosynthesis mutants such as cpd and dwf4. Biochemical inhibition analysis indicates that Brz inhibits CYP90B1 from catalyzing the oxidation of CR to 22-OHCR. It has been recently reported that new triazole-type inhibitors based on a ketoconazole scaffold (YCZ18) inhibit a new target, CYP90D1, in Arabidopsis.38) Aside from BR synthesis inhibitors, BR antagonists have recently been developed. BL binds to BRI1 and functions as its most potent agonist. BR-structure-mimicking agonists and antagonists recently have been developed. Brassinolide-2,3-acetonide strongly inhibited the agonistic effect of BL in a rice lamina joint inclination test.39) Brassinolide-2,3-acetonide reduces BR activity because of BR’s low affinity for BRI1 and also interferes with its interactions with other BR signaling factors in rice. Two other BR-mimicking chemicals, iso-carbabrassinolide (iso-carbaBL) and 6-deoxoBL were reported. Interestingly, these two chemicals showed opposite BR activities in Arabidopsis and rice.40) Iso-carbaBL was a relatively strong agonist in Arabidopsis but worked as an antagonist that competitively inhibits BR activity. Besides BR-structure-mimicking chemicals, nonsteroidal BL-like compounds were designed in silico and synthesized.41,42) N-(3,4-dihydroxybenzoyl)-N′-(4-butanoyl-2-fluorophenyl)piperazine (NSBR1) showed BL-agonistic activity based on an Arabidopsis hypocotyl elongation assay and a rice lamina joint inclination test.41) These chemicals have high potential as agrochemicals to improve the phenotype through BR regulation.
Fig. 6. Chemical structure of BR biosynthetic triazole-type inhibitors and the dissociation constant (Kd) against P450s on the BR biosynthetic pathway.
Dwarf phenotypes in BR-deficient plants are sometimes useful agricultural traits. Semi-dwarf phenotypes of crops are an agronomically important trait that improves lodging resistance and contributes to yield increases. The semi-dwarf barley variety, uzu, exhibits dark green leaves and short coleoptiles and is the result of a mutation in a BR receptor kinase gene (HvBRI1).43) The Vicia faba dwarf mutant “Rinrei” is a BR-deficient mutant with reduced metabolism of 24-methylenecolesterol to CR. The grass height of “Rinrei” is about 70% of that of the wild type, and this cultivar exhibits markedly stronger traits with respect to snow resistance as compared with the wild-type cultivar.44) These agronomic instances indicate that modifying BR receptors or BR biosynthetic genes has high potential as a tool for controlling plant size and increasing yield and stress resistance.45) In fact, overexpression of CYP90B1 in Arabidopsis increases seed weight.46) The rice osdwarf4 mutant, which is deficient in BR C-22 hydroxylation, exhibits semi-dwarfism and erect leaves.47) The erect leaves phenotype has an advantage in photosynthetic light capture, which is associated with enhanced grain yields under dense planting conditions; the osdwarf4 mutant increases the total and fertile grain numbers in dense plantations. Thus, these BR inhibitors and antagonists are useful chemical tools not only for investigation of the protein–protein interactions in BR signaling but also for practical utilization in agriculture.
Conclusions
BR biosynthesis research has greatly advanced by making use of chemical, enzymatic, and molecular genetic analyses. In particular, enzymatic analysis that elucidates each enzymatic reaction step in the BR biosynthetic pathway has proven to be a robust research strategy. Two reaction steps (C-2 hydroxylation and the oxidation/reduction of oxygen functionality at C-3) remain unelucidated in the BR biosynthetic pathway. Previous studies on structure–activity relationships with rice lamina joint inclination tests indicated that the vicinal hydroxyl groups at the 2α and 3α positions are important for BR physiological activities. Therefore, the identification of C-2 hydroxylase(s) and C-3 oxidoreductase(s) will be important in clarifying the entire picture of BR biosynthesis.
References
- 1).A. Bajguz: “Brassinosteroids: A Class of Plant Hormone”, ed. by S. Hayat and A. Ahmad, Springer, Dordrecht, pp. 1–27, 2011.
- 2).A. Bajguz and A. Tretyn: “Brassinosteroids: Bioactivity and Crop Productivity”, ed. by S. Hayat and A. Ahmad, Springer, Dordrecht, pp. 1–44, 2003.
- 3).M. D. Grove, G. F. Spencer, W. K. Rohwedder, N. Mandava, J. F. Worley, J. D. Warthen, G. L. Steffens, J. L. Flippen-Anderson and J. C. Cook: Nature 281, 216–217 (1979). [Google Scholar]
- 4).T. Yokota, M. Arima and N. Takahashi: Tetrahedron Lett. 23, 1275–1278 (1982). [Google Scholar]
- 5).A. Bajguz: Plant Physiol. Biochem. 45, 95–107 (2007). [DOI] [PubMed] [Google Scholar]
- 6).G. J. Bishop and T. Yokota: Plant Cell Physiol. 42, 114–120 (2001). [DOI] [PubMed] [Google Scholar]
- 7).S. Fujioka and T. Yokota: Annu. Rev. Plant Biol. 54, 137–164 (2003). [DOI] [PubMed] [Google Scholar]
- 8).T. Asami, T. Nakano and S. Fujioka: Vitam. Horm. 72, 479–504 (2006). [DOI] [PubMed] [Google Scholar]
- 9).T. Ohnishi, T. Yokota and M. Mizutani: Phytochemistry 70, 1918–1929 (2009). [DOI] [PubMed] [Google Scholar]
- 10).D. Nelson and D. Werck-Reichhart: Plant J. 66, 194–211 (2011). [DOI] [PubMed] [Google Scholar]
- 11).F. P. Guengerich and A. W. Munro: J. Biol. Chem. 288, 17065–17073 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).J. Li, P. Nagpal, V. Vitart, T. C. McMorris and J. Chory: Science 272, 398–401 (1996). [DOI] [PubMed] [Google Scholar]
- 13).J. Li, M. G. Biswas, A. Chao, D. W. Russell and J. Chory: Proc. Natl. Acad. Sci. U.S.A. 94, 3554–3559 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).S. Choe, B. P. Dilkes, S. Fujioka, S. Takatsuto, A. Sakurai and K. A. Feldmann: Plant Cell 10, 231–243 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).S. Fujioka, S. Takatsuto and S. Yoshida: Plant Physiol. 130, 930–939 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).S. Fujita, T. Ohnishi, B. Watanabe, T. Yokota, S. Takatsuto, S. Fujioka, S. Yoshida, K. Sakata and M. Mizutani: Plant J. 45, 765–774 (2006). [DOI] [PubMed] [Google Scholar]
- 17).M. Szekeres, K. Németh, Z. Koncz-Kálmán, J. Mathur, A. Kauschmann, T. Altmann, G. P. Rédei, F. Nagy, J. Schell and C. Koncz: Cell 85, 171–182 (1996). [DOI] [PubMed] [Google Scholar]
- 18).G.-T. Kim, S. Fujioka, T. Kozuka, F. E. Tax, S. Takatsuto, S. Yoshida and H. Tsukaya: Plant J. 41, 710–721 (2005). [DOI] [PubMed] [Google Scholar]
- 19).G. T. Kim, H. Tsukaya and H. Uchimiya: Genes Dev. 12, 2381–2391 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).T. Ohnishi, A. M. Szatmari, B. Watanabe, S. Fujita, S. Bancos, C. Koncz, M. Lafos, K. Shibata, T. Yokota, K. Sakata, M. Szekeres and M. Mizutani: Plant Cell 18, 3275–3288 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).M. K. Rasmussen, B. Ekstrand and G. Zamaratskaia: Int. J. Mol. Sci. 14, 17926–17942 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).T. Ohnishi, B. Godza, B. Watanabe, S. Fujioka, L. Hategan, K. Ide, K. Shibata, T. Yokota, M. Szekeres and M. Mizutani: J. Biol. Chem. 287, 31551–31560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Z. Y. Wang, H. Seto, S. Fujioka, S. Yoshida and J. Chory: Nature 410, 380–383 (2001). [DOI] [PubMed] [Google Scholar]
- 24).D. M. Friedrichsen, C. A. Joazeiro, J. Li, T. Hunter and J. Chory: Plant Physiol. 123, 1247–1256 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).G. J. Bishop, K. Harrison and J. D. Jones: Plant Cell 8, 959–969 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).G. J. Bishop, T. Nomura, T. Yokota, K. Harrison, T. Noguchi, S. Fujioka, S. Takatsuto, J. D. Jones and Y. Kamiya: Proc. Natl. Acad. Sci. U.S.A. 96, 1761–1766 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Z. Hong, M. Ueguchi-Tanaka, S. Shimizu-Sato, Y. Inukai, S. Fujioka, Y. Shimada, S. Takatsuto, M. Agetsuma, S. Yoshida, Y. Watanabe, S. Uozu, H. Kitano, M. Ashikari and M. Matsuoka: Plant J. 32, 495–508 (2002). [DOI] [PubMed] [Google Scholar]
- 28).Y. Shimada, S. Fujioka, N. Miyauchi, M. Kushiro, S. Takatsuto, T. Nomura, T. Yokota, Y. Kamiya, G. J. Bishop and S. Yoshida: Plant Physiol. 126, 770–779 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).T. Nomura, T. Kushiro, T. Yokota, Y. Kamiya, G. J. Bishop and S. Yamaguchi: J. Biol. Chem. 280, 17873–17879 (2005). [DOI] [PubMed] [Google Scholar]
- 30).T.-W. Kim, J.-Y. Hwang, Y.-S. Kim, S.-H. Joo, S. C. Chang, J. S. Lee, S. Takatsuto and S.-K. Kim: Plant Cell 17, 2397–2412 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).J. Oklestkova, L. Rárová, M. Kvasnica and M. Strnad: Phytochem. Rev. 14, 1053–1072 (2015). [Google Scholar]
- 32).B. Watanabe, S. Yamamoto, T. Yokoi, A. Sugiura, S. Horoiwa, T. Aoki, H. Miyagawa and Y. Nakagawa: Bioorg. Med. Chem. 81, 4566–4578 (2017). [DOI] [PubMed] [Google Scholar]
- 33).W. Rademacher: Plant Physiol. 100, 625–629 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).N. Nagata, T. Asami and S. Yoshida: Plant Cell Physiol. 42, 1006–1011 (2001). [DOI] [PubMed] [Google Scholar]
- 35).T. Asami, Y. K. Min, N. Nagata, K. Yamagishi, S. Takatsuto, S. Fujioka, N. Murofushi, I. Yamaguchi and S. Yoshida: Plant Physiol. 123, 93–100 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).T. Asami, M. Mizutani, S. Fujioka, H. Goda, Y. K. Min, Y. Shimada, T. Nakano, S. Takatsuto, T. Matsuyama, N. Nagata, K. Sakata and S. Yoshida: J. Biol. Chem. 276, 25687–25691 (2001). [DOI] [PubMed] [Google Scholar]
- 37).K. Sekimata, T. Ohnishi, M. Mizutani, Y. Todoroki, S.-Y. Han, J. Uzawa, S. Fujioka, K. Yoneyama, Y. Takeuchi, S. Takatsuto, K. Sakata, S. Yoshida and T. Asami: Biosci. Biotechnol. Biochem. 72, 7–12 (2008). [DOI] [PubMed] [Google Scholar]
- 38).K. Oh, T. Matsumoto, A. Yamagami, A. Ogawa, K. Yamada, R. Suzuki, T. Sawada, S. Fujioka, Y. Yoshizawa and T. Nakano: PLoS One 10, e0120812 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).T. Muto and Y. Todoroki: Bioorg. Med. Chem. 21, 4413–4419 (2013). [DOI] [PubMed] [Google Scholar]
- 40).A. Nakamura, N. Tochio, S. Fujioka, S. Ito, T. Kigawa, Y. Shimada, M. Matsuoka, S. Yoshida, T. Kinoshita, T. Asami, H. Seto and T. Nakano: PLoS One 12, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).A. Sugiura, S. Horoiwa, T. Aoki, S. Takimoto, A. Yamagami, T. Nakano, Y. Nakagawa and H. Miyagawa: J. Pestic. Sci. 42, 105–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).B. Lei, N. Heng, X. Dang, J. Liu, X. Yao and C. Zhang: Mol. Biosyst. 13, 1364–1369 (2017). [DOI] [PubMed] [Google Scholar]
- 43).M. Chono, I. Honda, H. Zeniya, K. Yoneyama, D. Saisho, K. Takeda, S. Takatsuto, T. Hoshino and Y. Watanabe: Plant Physiol. 133, 1209–1219 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).N. Fukuta, K. Fukuzono, H. Kawaide, H. Abe and M. Nakayama: Ann. Bot. 97, 65–69 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).G. J. Bishop: J. Plant Growth Regul. 22, 325–335 (2003). [DOI] [PubMed] [Google Scholar]
- 46).S. Choe, S. Fujioka, T. Noguchi, S. Takatsuto, S. Yoshida and K. A. Feldmann: Plant J. 26, 573–582 (2001). [DOI] [PubMed] [Google Scholar]
- 47).T. Sakamoto, Y. Morinaka, T. Ohnishi, H. Sunohara, S. Fujioka, M. Ueguchi-Tanaka, M. Mizutani, K. Sakata, S. Takatsuto, S. Yoshida, H. Tanaka, H. Kitano and M. Matsuoka: Nat. Biotechnol. 24, 105–109 (2005). [DOI] [PubMed] [Google Scholar]