Abstract
Purpose
Exosomes are small 30–100 nm vesicles secreted by various cell types. They are released by most cell types, indicating their important role in physiological and pathological processes, including signaling pathways, cell-to-cell communication, tumor progression, and molecule transferring. As natural nanovesicles, exosomes can be a good candidate for drug delivery due to low immunogenicity and ability to enter tissues and even cross the blood–brain barrier. In an effort to improve the efficiency of exosomes for targeted drug delivery with minimal effect on normal cells, we expressed ligands against HER2+ cells.
Methods
To purify exosomes, transduced mesenchymal stromal cells were cultured to reach 80% confluency. Next, the cells were cultured in serum-free media for 48 hours and the supernatant was harvested to purify exosomes. These exosomes were then labeled with PKH67 and added to BT-474, SKBR3 (HER2+), and MDA-MB231 (HER2−), cell lines and their binding to HER2+ was evaluated by flow cytometry. Exosomes were loaded with doxorubicin and quantified using intrinsic fluorescence of doxorubicin at 594 nm.
Results
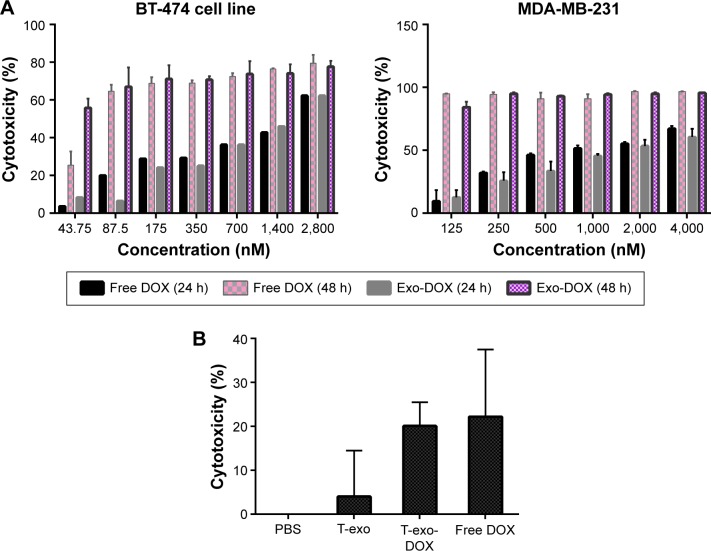
Targeted exosomes were preferably uptaken by HER2+ cells. Therefore, untargeted exosomes showed lower binding to HER2+ cells compared to their targeted counterparts. MTT assay was performed to analyze cytotoxic effect of exo-DOX (exosome encapsulated with doxorubicin). Efficiency of exo-DOX and free DOX (doxorubicin) delivery with different concentrations, to the BT-474 cell line, was compared, and no significant difference was observed.
Conclusion
Our results imply that targeted exosomes are preferentially uptaken by HER2+ cells relative to HER2− cells and have the potential to be used as an efficient drug delivery system.
Keywords: breast cancer, doxorubicin, HER2+, mesenchymal stem cell
Introduction
Extracellular vesicles (EVs) are small membrane vesicles secreted by most cells, containing proteins, lipids, and nucleic acids. Based on their biogenesis pathways and size range, EVs are generally categorized into 3 types, including exosomes, apoptotic bodies, and microvesicles. With a size rage of 30–100 nm1 and endocytic origin, exosomes are secreted by various cell types, including red blood cells, platelets, lymphocytes, dendritic cells, epithelial cells, adipocytes, fibroblasts, neural cells, stem cells, and tumor cells.2 They also commonly appear in biological fluids such as urine, breast milk, plasma,3 amniotic fluid, malignant ascites, saliva,4 synovial fluid, bronchoalveolar lavage fluid, and cerebrospinal fluid.2 This highlights their important role in physiological and pathological processes of signaling pathways, cell-to-cell communication,5 immunomodulatory functions, antigen presentation in some cells, tumor progression, coagulation, inflammation, and transferring macromolecules such as proteins and RNA.2,6
Depending on the cell of origin, exosomes contain specific protein and lipid constituents. Although most exosomes have similar components due to their endosomal origin, heat shock proteins, fusion and membrane transporters (annexins, RabGTPases, and flotilin), MVBs proteins such as TSG101, Alix, integrins, and tetraspanins (CD9, CD63, CD81, and CD82) are common exosomal proteins. Furthermore, special glycosylated motifs enriched with raft-lipids such as sphingolipids, cholesterol, and ceramide are typically observed as their lipid component.6
Exosomes affect target cells through the following mechanisms: activation of certain signaling pathways by ligand–receptor interaction (without entrance), releasing their content through extracellular proteases-mediated cleavage and subsequent binding of contents to cell surface receptors, fusion to the cell membrane and releasing their content into the cytoplasm, and entering cells by endocytic mechanisms such as receptor-mediated endocytosis, phagocytosis, and macropinocytosis.2,6
Relative to other cell types, mesenchymal stromal cells (MSCs) possess distinct advantages as an exosome source. They release higher amounts of exosome compared to other cells. MSC-derived EVs are relatively well tolerated in different animal models and show more stability and sustainability in human plasma and at −20°C. Moreover, MSC-derived exosomes release paracrine mediators and exhibit regenerative properties. As natural nanovesicles, exosomes are good candidates for drug delivery due to their low immunogenicity and ability to enter tissues and even cross the blood–brain barrier.7 However, exosomes are rapidly cleared by reticulum endothelial system and generally should be specifically targeted against desired cells; thus, the clinical usage of these EVs is still a subject of debate.
We expressed Designed Ankyrin Repeated Proteins (DARPins) as a specific ligand against HER2+ cells8 in order to improve the efficiency of exosomes and employed them as targeted drug delivery vehicles with minimal effect on normal cells. Binding efficiency of targeted exosomes to HER2+ and HER2− cells was evaluated and compared to the binding of control untargeted exosomes. Exosomes were loaded with doxorubicin (DOX) and their cytotoxicity was assessed using MTT assay. Our results can be useful data for further in vivo and clinical studies aiming to develop novel therapeutic approaches against breast cancer.
Materials and methods
Cell culture
Human embryonic kidney HEK293T, SKBR3, MDA-MB231, and BT-474 cell lines were purchased from Iranian biological resource center and grown in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich, St Louis, MO, USA) with 10% fetal bovine serum (Gibco, Waltham, MA, USA), and 1% penicillin/streptomycin solution (Gibco). MSCs were purchased from Stem Cell Technology Research Center (Tehran, Iran) and cultured in α-MEM (Sigma-Aldrich) with 10% fetal bovine serum (Gibco), 1% penicillin/streptomycin solution (Gibco), and 2 mM l-glutamine (Gibco). All cells were incubated at 37°C, 5% CO2 in a humidified atmosphere.
MSC characterization
When cultured in standard conditions, MSCs are plastic-adherent cells. Expression of CD105 and CD90 cell markers and the absence of CD45, CD31, and CD11b were analyzed by flow cytometry using respective antibodies conjugated with PE and FITC. Finally, the differentiation potency of MSCs to osteoblasts and adipocytes was tested in vitro. In brief, MSCs were seeded on a 4-well plate and the media culture was replaced with osteogenic and adipogenic media every other day. After 20 days, the cells were fixed in 4% paraformaldehyde for 20 minutes at 4°C. The cells were washed 3 times with phosphate-buffered saline (PBS). Oil Red and Alizarin red were used for staining adipocytes and osteocytes, respectively. Osteo media containing 10 mM β-glycerophosphate, 50 µg/mL ascorbate-2-phosphate, and 1% antibiotic and adipocyte media containing 0.5 mM isobutyl-methylxanthine, 10−7 M dexamethasone, 200 µM indomethacin, and 1% antibiotic were used as culture media for the respective cells.
Lentivirus production
HEK293T cell lines were seeded (3–4×106 cells/plate) on 10 cm diameter plates for 24 hours to reach 70%–80% confluency. One hour before transfection, the media was changed. HEK293T cells were transfected with 21 µg of psPAX2 (Addgene, Cambridge, MA, USA), 10.5 µg of pMD2 (Addgene), and 21 µg of pLEX-LAMP2b-DARPin8 using calcium phosphate transfection. After 17 hours, the culture media were replaced with 10–12 mL of complete media.
The supernatant containing viral particles was harvested every 24 hours for up to 72 hours. Media were centrifuged at 500 g for 5 minutes and cell pellet was discarded. The harvested supernatant was subsequently centrifuged (2,000 g for 20 minutes) to remove cell debris, and then filtered through a 0.45 µm PVDF (low protein attachment) membrane. Finally, centrifugation at 19,000 g for 1:30 hours was applied to concentrate viral particles and Multiplicity Of Infection (MOI) (number of viral particles per target cell) was measured for HEK293T cells transduction.
Real-time PCR
Total RNA was isolated form MSC-transduced cells and cDNA was synthesized using RNeasy mini kit (Qiagen, Hilden, Germany). PCR amplification was performed using DARPin primer sets outlined in Table 1. Primer blast was used to check for nonspecific binding to human sequences.
Table 1.
Primer sequences
| Forward | Reverse | |
|---|---|---|
| DARPin | 5′-CTCGAGATGAGAGGTAGTC-3′ | 5′-CTACCCCCGTTGAGTTTC-3′ |
| Puromycin | 5′-GCAGCAACAGATGGAAGG-3′ | 5′-GAGGTCTCCAGGAAGGC-3′ |
Abbreviation: DARPin, Designed Ankyrin Repeated Protein.
Furthermore, DNA was purified from MSC-transduced cells and quantitative real-time PCR was established using puromycin primers (Table 1) to calculate the number of insertion and determine MOI for MSCs.
MSC transduction
MSCs were seeded in a 24-well plate at a cell density of 1–2×104/well. Viral particles with MOI of 100 and protamine sulfate were added to each well, and media were changed after 24 hours. The transduction procedure was repeated 3 times to increase efficiency. After 1 week, the transduced MSCs were selected by appropriate concentration (2 µg/mL) of puromycin.
Exosome isolation
To extract exosomes, cells were cultured to reach 80% confluency. The cells were then cultured in serum-free media for 48 hours and the supernatant was harvested to purify exosomes using Cell Guidance Systems (Cambridge, UK) exosome isolation kit with minor changes to the manufacturer’s protocol. In brief, supernatant was centrifuged to remove cell debris, followed by filtration through a 0.22 µm membrane to remove large vesicles. The media was concentrated using 100 kDa centrifugal filtering columns (Merck Millipore, Burlington, MA, USA) according to the manufacturer’s protocol.
Exosome characterization
Transmission electron microscopy
Purified exosomes from MSCs were resuspended in PBS and fixed with 2% paraformaldehyde for 30 minutes at room temperature. Fixed exosomes were deposited on transmission electron microscopy (TEM) grids treated with UV light to decrease static electricity.
Size distribution and zeta potential of exosomes
To determine size and zeta potential of exosomes, exosome solution diluted in PBS was applied on a Zetasizer (Malvern Corp., Malvern, UK) and the results were analyzed using the Zetasizer software v7.11 (Malvern Corp.).
Immunoblotting
Transduced cells were lysed in RIPA buffer containing PMSF protease inhibitor (Roche, Mannheim, Germany) and resolved on a 4%–12% polyacrylamide sodium dodecyl sulfate gel. Western blotting and enhanced chemiluminescence (ECL) detection system (Pars Toos biotechnology Co., Mashhad, Iran) were used with HRP-conjugated anti-His tag antibody to detect N-terminal His tag on chimeric LAMP2b-DARPin protein.
DOX loading
To load exosomes with DOX, 100 µg (total protein) of purified exosomes and 100 µg of DOX were gently mixed in 200 µL of electroporation buffer at 4°C. Electroporation was performed at 600 V in 0.4 cm cuvettes in a multiporator (Eppendorf, Hamburg, Germany) and the mixture was incubated at 37°C for 30 minutes, to recover membrane of exosomes.9 Exosomes were then passed through a 100 kDa Amicon filter (Merck Millipore) to remove unincorporated DOX. Finally, drug loading was quantified through the intrinsic fluorescence of DOX using a spectrophotometer (BioTek®, Cell Imaging Multi-Mode Reader, Guangdong, China) at 594 nm with excitation at 480 nm.
Exosome labeling and cell uptake assay
Exosomes were labeled with the green lipid membrane dye PKH67 (Sigma-Aldrich), according to the manufacturer’s protocol, with some modifications. Briefly, exosomes and PKH67 were separately diluted in 100 µL diluent C. Exosomes were mixed with the staining solution and incubated for 5 minutes at room temperature. Labeling was stopped by adding an equal volume of 1% BSA and the mixture was subsequently passed through 100 kDa Amicon filter to remove unincorporated dye. SKBR3 and MDA-MB231 cells (2×104 cells/well) were seeded in a 24-well plate and 25 µg labeled exosome was added to each well. After 3 hours, cells were washed with PBS and fixed using 4% paraformaldehyde. The nuclei were stained by DAPI fluorescent stain (Sigma-Aldrich), according to the manufacturer’s protocol and examined using an inverted fluorescence microscope (Nikon TE300; Nikon, Tokyo, Japan).
The uptake of exosome was quantified via flow cytometry. Briefly, 6×104 SKBR3, BT-474, and MDA-MB231 cells were seeded in a 24-well plate. After 24 hours, 5 µg of labeled exosome was added to each well for 24 hours. Cells were then trypsinized,6 and used for flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA). MDA-MB231 cells and untargeted exosomes were used as untargeted cell and exosome binding controls, respectively. The data were analyzed using Flow Jo (FlowJo, LLC, Ashland, OR, USA) and Flowing software 2.4.1.
Cell treatment and cytotoxicity
The antitumor effect of engineered exosomes loaded with DOX was evaluated by the standard MTT assay on BT-474 HER2+ cells and MDA-MB231 HER2− cells. Briefly, tumor cells (5,000 cells/well) were seeded in 100 µL of media in a 96-well plate overnight. Tumor cells were treated with various concentrations of exo-DOX for 24, 48, and 72 hours at 37°C and 5% CO2. After incubation, media were removed and cells were incubated with MTT reagent for 3–4 hours. Subsequently, 100 µL of DMSO was added to solubilize purple formazan crystals.10 Cytotoxic activity of free DOX and exo-DOX was then evaluated by standard MTT assay. Absorbance was measured by spectrophotometer at 545–630 nm. Survival rates were assessed compared to the negative control (wells containing only PBS). All experiments were repeated 3 times, and IC50 of loaded exosomes and free DOX were measured and compared with each other.11
Statistical analysis
Data were analyzed using Student’s t-test. Probability values lower than 0.05 were considered significant.
Results
Characterization of MSCs
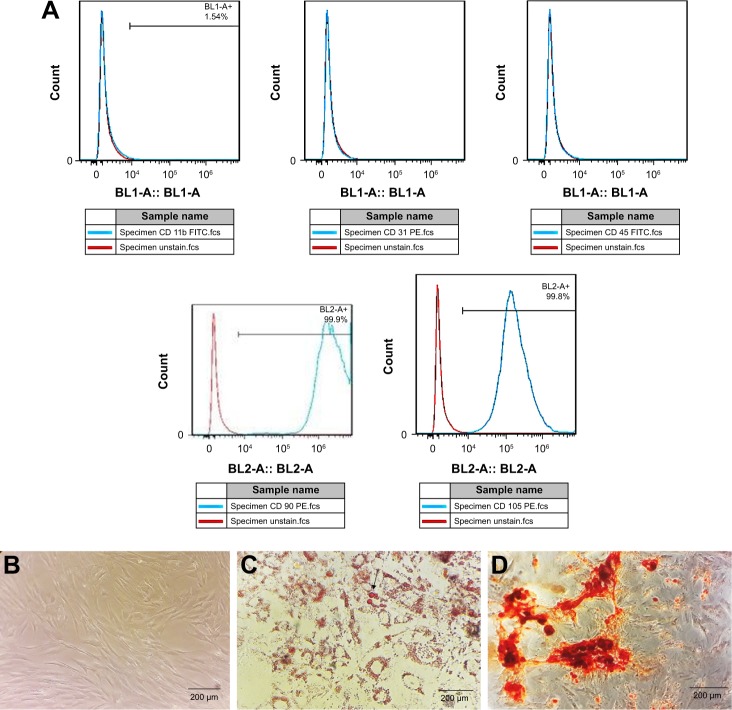
Expression of MSC markers (CD105 and CD90) and absence of CD45, CD31, and CD11b markers were confirmed by flow cytometry (Figure 1A). Differentiation of MSCs to osteoblasts and adipocytes was determined by Alizarin red and Oil Red staining, respectively (Figure 1B–D).
Figure 1.
Characterization of MSCs.
Notes: (A) Flow cytometry analysis of surface markers expressed on MSCs CD90 and CD105 are positive markers and CD31, CD11b, and CD45 were negative. (B) Morphology of MSCs in culture. (C) Adipocyte differentiation in adipogenic media for 21 days and Oil Red staining. (D) Alizarin red staining in osteogenic media for 21 days.
Abbreviation: MSC, mesenchymal stromal cell.
Lentivirus production
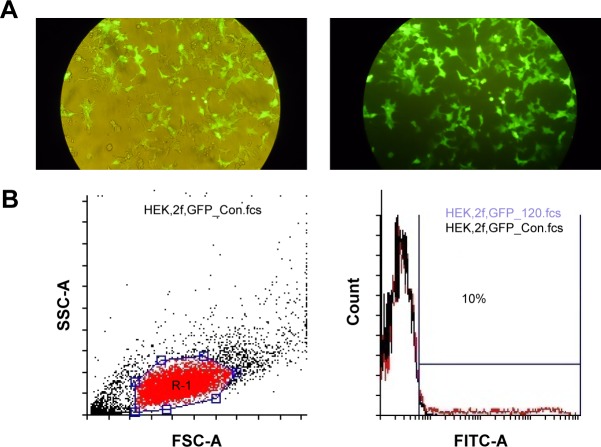
Since pLEX-LAMP2b-DARPin contains no reporter genes, TurboGFP vector was cotransfected to estimate the efficiency of transfection. About 80% of HEK293T cells expressed GFP, considered as an estimation of transfection rate (Figure 2A). Virus activity in transduced cells was validated using flow cytometry (Figure 2B) and the titer was estimated to be 1.2×109 TdU/mL.
Figure 2.
GFP expression analysis.
Notes: (A) Fluorescent microscopy of HEK cells transfected with GFP plasmid. (B) Analysis of GFP production using flow cytometry after viral transduction of HEK cells, indicating viral titration. Ten percent of HEK cells were transduced by GFP virus.
MSCs transduction
In order to kill nontransduced cells and select a stable cell line containing a genomic copy of the LAMP2b-DARPin gene, viral particles containing the gene along with puromycin were added to the cell culture and expression of LAMP2b-DARPin was analyzed by RT-PCR. Presence of a 430-bp PCR product confirmed integration of viral DNA into the host genome (Figure 3A). Furthermore, a 70 kDa protein (LAMP2b-DARPin) was identified by Western blot using anti-His tag antibody (Figure 3B).
Figure 3.
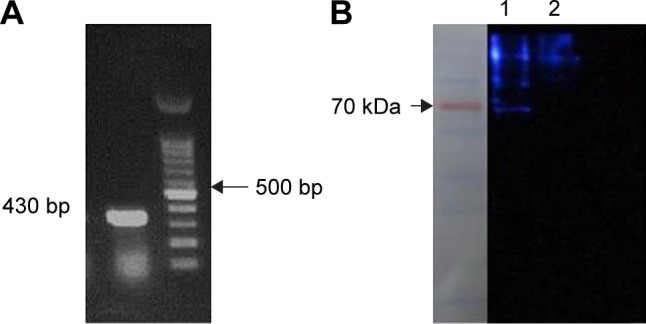
(A) Presence of LAMP2b-DARPins mRNA was determined with specific primers by RT-PCR; (B) Western blotting was performed to confirm the expression of LAMP2b-DARPins protein. Lane 1 and lane 2 are cell lysate of transduced and nontransduced cells, respectively, which was detected by anti-His tag antibody.
Abbreviation: RT-PCR, real-time PCR.
Characterization of exosomes
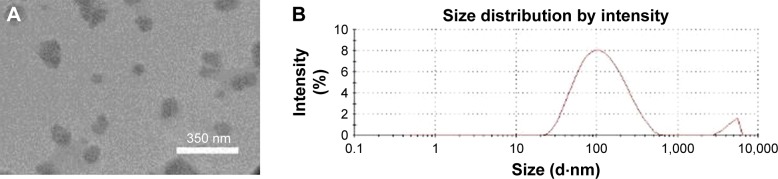
Purified exosomes were observed in TEM micrographs (Figure 4A). The size of exosomes (100 nm) was determined using a Zetasizer (Malvern Corp.) (Figure 4B).
Figure 4.
Characterization of purified exosomes.
Notes: (A) TEM image shows the size of exosomes. (B) Analyzed peak rendered by Zetasizer indicating the size of exosomes. The main peak is around 100 nm, and the second one is related to accumulation of small number of exosomes.
Abbreviation: TEM, transmission electron microscopy.
DOX loading
Electroporation was used to load DOX into exosomes. The amount of encapsulated DOX was determined by measuring the auto-fluorescent property of DOX at 595 nm (excitation at 480 nm) against a serial dilution of known standards (Figure 5). Our results showed that ~13% of DOX were loaded into the exosomes.
Figure 5.
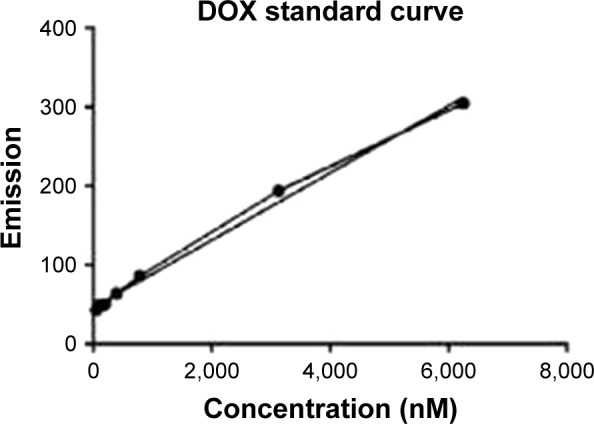
DOX standard curve. Excitation and emission were measured by fluo-rimetry at 480 and 595 nm, respectively.
Abbreviation: DOX, doxorubicin.
Exosome uptake
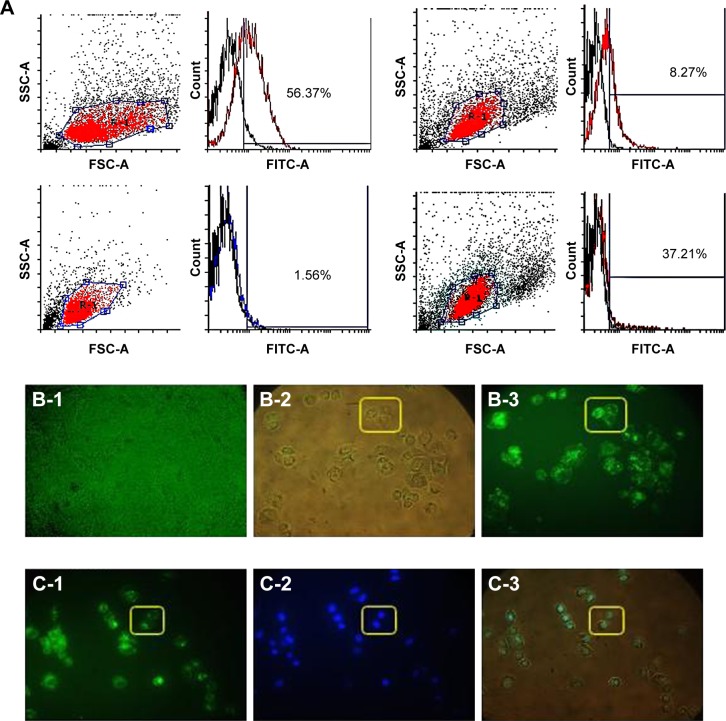
To investigate whether LAMP2b-DARPin-targeted exosomes are capable of binding to HER2+ cancer cells, MDA-MB-231 (HER2−) and SKBR3, and BT-474 (HER2+) cells were treated with targeted and untargeted exosomes previously labeled with PKH67. Flow cytometry results demonstrated that targeted exosomes bound to SKBR3 (56.37%) and BT-474 (37.21%) more efficiently compared to MDA-MB-231 cells (1.56%). As a negative control, untargeted exosomes showed relatively low binding (8.27%) to BT-474 cells. (Figure 6A). Fluorescent image of targeted exosomes labeled by PKH-67 and SKBR3 cells are depicted in Figure 6B and C.
Figure 6.
In vitro binding of targeted exosomes to human breast cancer cells.
Notes: (A) Flow cytometry analysis of exosomes labeled with PKH67 and incubated with SKBR3 (56.37%), BT-474 (37.21%), and MDA-MB-231 (1.56%) cells. Binding of untargeted exosomes to BT-474 was measured (8.27%) as negative control. Targeted exosomes showed significantly higher binding to HER2+ cells compared to the control; (B-1) merged fluorescent and bright-field microscopy images of MDA-MB231 cell line and PKH-67 as a negative control; (B-2 and 3) merged fluorescent and bright-field microscopy images of targeted exosomes and SKBR3 cells and fluorescent microscopy image. PKH67-Exosomes were observed in green; (C-1–3) fluorescent image of targeted exosomes labeled by PKH-67 and SKBR3 cells and merged image of bright-field and DAPI fluorescent staining; nuclei were stained with DAPI (blue).
Cell treatment and cytotoxicity
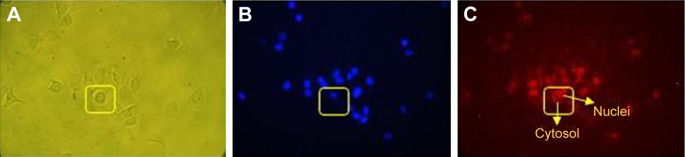
BT-474 cells were treated with 2 µg of exo-DOX, followed by evaluation of binding and uptake after 2 hours using fluorescent microscopy. The results indicated uptake of exo-DOX into the cells and its accumulation in nuclei.
Cytotoxic effect of free DOX and DOX-loaded exosomes was examined on BT-474 and MDA-MB231 cells. IC-50 of DOX for each cell line was determined. Free DOX and exo-DOX considerably reduced cell viability in a dose-dependent manner, and no significant difference was observed (Figure 7).
Figure 7.
Comparison of cytotoxicity of free DOX and exo-DOX.
Notes: (A) BT-474 (HER2+) and MDA-MB-231 (HER2−) cells were used to compare the efficiency of drug delivery of targeted exo-DOX and free DOX at different concentrations. No significant difference was observed. (B) Cytotoxicity of free DOX and targeted exo-DOX on BT-474 cells at a concentration of 1,000 ng after 24 hours. PBS and exosome without DOX were used as control.
Abbreviations: DOX, doxorubicin; PBS, phosphate-buffered saline.
Discussion
Exosomes are small membrane vesicles in the range of 30–100 nm,1 secreted by most cell types, including tumor cells and MSCs. They are also found in biological fluids.1 As the most characterized endogenous vesicles with cell communicative ability,12 exosomes can enter cells through endocytosis, phagocytosis, macropinocytosis, lipid raft-mediated internalization,13 and fusion.14
Exosomes, like liposomes, are surrounded by a bilayer lipid membrane and thus are able to transport both hydrophilic and hydrophobic substances.15 Their membrane is enriched with cholesterol, sphingomyelin, phosphatidylserine, and GM3, compared to that of their parent cells. These lipids are usually not found in liposomes, which is considered an advantages for exosomes. Cholesterol and sphingomyelin can form hydrogen bonds, causing tight packing of sphingomyelin/cholesterol bilayers and reducing water penetrability.16 Furthermore, exosomes exhibit high biocompatibility17 and low immunogenicity and cytotoxicity.9 Hence, exosomes are valuable candidates for next generation of drug delivery systems. Moreover, previous research has proved the potential of exosomes for delivery of curcumin18 and siRNA19 across the blood–brain barrier in mice.
Despite several advantages associated with exosomes, some critical issues, including the source, the drug loading process, limited efficiency of exosome production, and lack of targeting, still remain unaddressed for possible clinical application of these nanoparticles. The small size of exo-somes provides them with the ability to passively target and selectively extravasate into tumor or inflamed tissues owing to the enhanced permeability and retention effect.20 Furthermore, MSC-derived exosomes possess an intrinsic homing ability, causing them to migrate toward tumor and injured tissue, which might be due to acidic pH of tumor environment. Chemokines such as CCR8 and CCL2, TNF-α, and PDGF have been recently identified to play a role in MSC-mediated chemotaxis to tumor environment in vitro.21
Targeted delivery of engineered exosomes has recently been the subject of several studies. Alvarez-Erviti et al19 designed targeted exosomes against neurons by engineering cells expressing LAMP2b fused to neuron-specific rabies viral glycoprotein. Liu et al22 employed modified exosomes expressing rabies viral glycoprotein peptide to deliver µ-opioid receptor siRNA into the brain to reduce morphine addiction. In another study, Ohno et al23 produced targeted exosomes by expressing PDGF receptor fused to the GE11 peptide.
Previously, we produced exosomes targeted with a designed chimeric protein against HER2+ cancer cells.24 These targeted exosomes were subsequently used to deliver siRNA to breast cancer cell lines.8 In the present study, targeted exosomes were used to deliver DOX to HER2+ cancer cells. We aimed to evaluate anticancer effects of DOX-loaded targeted exosomes in a murine tumor model.
As mentioned before, the source of exosomes is of out-most importance in clinical applications.25 Previous studies suggest MSCs as a suitable source for exosomes, with the ability to produce large amounts of these nanoparticles. MSC-derived exosomes are relatively highly tolerated, stable, and possess inherent therapeutic characteristics, including homing ability.25
Extensive research on MSC-derived exosomes indicates controversial effects on tumor growth, which may be due to the source of MSCs25 and variation in tumor-bearing models.26 Therefore, safety of the MSC culture and the applied material and methods for exosome isolation require careful optimization for clinical applications.
For a long time, retroviruses have been considered as the best vehicles for gene delivery. They can integrate target genes into the genome of cells without transferring viral genes into host cells. However, retroviral infection is not optimal for slow-dividing cells. With a doubling time of 35–40 hours, the transduction of human MSCs with retroviruses is not effective.26 Lentiviral vectors can infect both dividing and nondividing cells and are good candidates for gene delivery to MSCs. Amari et al27 used a lentiviral vector to transfer genes into MSCs and reported that lentiviruses had no effect on characterization and immunomodulation properties of these cells. We employed a lentiviral vector for permanent delivery of the LAMP2b-DARPin gene into MSCs and exosomes were isolated from these cells following selection with puromycin.
Isolation of exosomes lacks a standard protocol, while many researchers prefer a combination of differential centrifugation with size exclusion chromatography, sucrose gradient ultracentrifugation, or an antibody-based approach in order to effectively eliminate cellular debris and other subcellular elements during purification.28 Sedimentation through differential centrifugation is the most common exosome purification method. However, this technique has some disadvantages, such as being time-consuming, leading to aggregation of exosomes with cellular proteins and particles in culture media, as well as requiring specialized equipment. Other size exclusion chromatography-based techniques include high-performance liquid chromatography and ultrafiltration. Compared to ultracentrifugation, ultrafiltration requires no specialized equipment and is relatively less time-consuming.29 In the present study, we used column-based cell guidance systems kit for exosomes isolation, along with ultrafiltration for separation of free dye from labeled exosomes, both of which would economize operation cost and time.
DOX is a DNA intercalating agent, currently administered for breast cancer and solid tumor treatment. However, the clinical usage of this drug in free form has been linked to dose-dependent cardiac toxicity.9 Encapsulation of DOX in exosomes can increase limited dosage of DOX and significantly reduce adverse effects on other organs.17 Flavio Rizzolio used exo-DOX to treat ovarian and breast cancer cells and confirmed that exo-DOX was safer and more efficient than free DOX.28
Comparatively simple operation of electroporation generally makes it the first choice for drug loading in exosomes. Adverse effects on the integrity of exosomes and their cargo, along with aggregation of exosomes have been reported for this method. Although, optimization of electroporation parameters and using trehalose pulse media can diminish the unfavorable effects of electroporation.17 In this study, electroporation was used to encapsulate DOX in exosomes and loading efficiency was determined by measuring the auto-fluorescence of DOX.
DOX-targeted exosomes were uptaken by HER2+ cells and showed comparable cytotoxicity effect to that of free DOX (Figure 7), which corroborates with the results of Tian et al.9 Flow cytometry analysis indicated higher binding of targeted exosomes to HER2+ cells relative to control cells. Untargeted exosomes were used as negative control. Our results demonstrated that targeted exosomes bound to SKBR3 (56.37%) and BT-474 (37.21%) cells more efficiently compared with MDA-MB-231 (1.56%) cells. Moreover, untargeted exosomes exhibited lower binding and entrance (8.27%) to HER2+ cells, as negative control (Figure 6A), implying that expression of chimeric protein plays an important role in exosomes binding and uptake, and it is not a result of physical absorption and nonspecific entrance. Nonetheless, the differences in affinity were only detected in lower concentrations. When the dosage of exosomes was increased, no significant variation was observed and it was suggested that physical absorption and nonspecific fusion were the dominant phenomena in higher concentrations. Thus, by using targeted exosomes as delivery vehicles, the clinical dosage can be decreased, without affecting cancer toxicity.
Microscopic images of binding and uptake of exo-DOX illustrated that targeted exosomes specifically bind and enter HER2+ cells and release their content into these cells (Figure 8). This data showed similar to free DOX, exo-DOX can also deliver DOX to the nuclei. MTT assay indicated no significant difference in the cytotoxicity of free DOX and exo-DOX (Figure 7), which supported the results of Toffoli et al,29 Hadla et al2 and Tian et al.9
Figure 8.
Binding and entrance of exo-DOX to BT-474 cells (HER2+ cells).
Notes: (A) Bright-field microscopy of BT-474 cell; (B) fluorescent microscopy of nuclei stained by DAPI; (C) exo-DOX entered the cells and DOX accumulated in nuclei.
Conclusion
Our results suggested no significant difference in the toxicity of exo-DOX and free DOX on BT-474 and MDA-MB231 cells. This proved that encapsulation has no effect on DOX cytotoxicity, while it can reduce its side effects by decreasing the systemic absorption. Therefore, exosomes derived from transduced MSCs can be used as an effective targeted delivery system to selectively target HER2+ cell lines.
Acknowledgments
We thank Dr Ali Mohammadian for his precious advice and comments. This study is a part of PhD thesis of Hosna Gomari, Tarbiat Modares University, Tehran, Iran.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Li Z, Ma YY, Wang J, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2016;9:139–148. doi: 10.2147/OTT.S95565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urbanelli L, Magini A, Buratta S, et al. Signaling pathways in exosomes biogenesis, secretion and fate. Genes. 2013;4(2):152–170. doi: 10.3390/genes4020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aspe JR, Wall NR. Survivin-T34A: molecular mechanism and therapeutic potential. Onco Targets Ther. 2010;3:247. doi: 10.2147/OTT.S15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9(1):86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salido-Guadarrama I, Romero-Cordoba S, Peralta-Zaragoza O, Hidalgo-Miranda A, Rodríguez-Dorantes M. MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. Onco Targets Ther. 2014;7:1327–1338. doi: 10.2147/OTT.S61562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11(1):108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kooijmans SAA, Fliervoet LAL, van der Meel R, et al. PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time. J Control Release. 2016;224:77–85. doi: 10.1016/j.jconrel.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Limoni SK, Moghadam MF, Moazzeni SM, Gomari H, Salimi F. Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl Biochem Biotechnol. 2018 Jun;:28. doi: 10.1007/s12010-018-2813-4. Epub. [DOI] [PubMed] [Google Scholar]
- 9.Tian Y, Li S, Song J, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 10.Tan CY, Lai RC, Wong W, Dan YY, Lim SK, Ho HK, Hk H. Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res Ther. 2014;5(3):76. doi: 10.1186/scrt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim MS, Haney MJ, Zhao Y, et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12(3):655–664. doi: 10.1016/j.nano.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang T, Martin P, Fogarty B, et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res. 2015;32(6):2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3(1):24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parolini I, Federici C, Raggi C, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284(49):34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haney MJ, Klyachko NL, Zhao Y, et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J Control Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kooijmans SA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomedicine. 2012;7:1525–1541. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles – endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta. 2014;1846(1):75–87. doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Sun D, Zhuang X, Xiang X, et al. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18(9):1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 20.Jiang XC, Gao JQ. Exosomes as novel bio-carriers for gene and drug delivery. Int J Pharm. 2017;521(1–2):167–175. doi: 10.1016/j.ijpharm.2017.02.038. [DOI] [PubMed] [Google Scholar]
- 21.Klopp AH, Spaeth EL, Dembinski JL, et al. Tumor irradiation increases the recruitment of circulating mesenchymal stem cells into the tumor microenvironment. Cancer Res. 2007;67(24):11687–11695. doi: 10.1158/0008-5472.CAN-07-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Li D, Liu Z, et al. Targeted exosome-mediated delivery of opioid receptor Mu siRNA for the treatment of morphine relapse. Sci Rep. 2015;5:17543. doi: 10.1038/srep17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohno S, Takanashi M, Sudo K, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khodashenas Limoni S, Salimi F, Forouzandeh Moghaddam M. Designing pLEX-LAMP-DARPin lentiviral vector for exression of HER2 targeted DARPin on exosome surface. J Mazandaran Univ Med Sci. 2017;27(151):12–23. [Google Scholar]
- 25.Yuan Z, Kolluri KK, Gowers KH, Janes SM. TRAIL delivery by MSC-derived extracellular vesicles is an effective anticancer therapy. J Extracell Vesicles. 2017;6(1):1265291. doi: 10.1080/20013078.2017.1265291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lou G, Song X, Yang F, et al. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8(1):122. doi: 10.1186/s13045-015-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amari A, Ebtekar M, Moazzeni SM, et al. Investigation of immunomodulatory properties of human Wharton’s Jelly-derived mesenchymal stem cells after lentiviral transduction. Cellular Immunology. 2015;293(2):59–66. doi: 10.1016/j.cellimm.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Hadla M, Palazzolo S, Corona G, et al. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine. 2016;11(18):2431–2441. doi: 10.2217/nnm-2016-0154. [DOI] [PubMed] [Google Scholar]
- 29.Toffoli G, Hadla M, Corona G, et al. Exosomal doxorubicin reduces the cardiac toxicity of doxorubicin. Nanomedicine. 2015;10(19):2963–2971. doi: 10.2217/nnm.15.118. [DOI] [PubMed] [Google Scholar]