Abstract
Strigolactones (SLs) are plant secondary metabolites that were first identified as germination stimulants for the root parasitic weeds witchweeds (Striga spp.) and broomrapes (Orobanche and Phelipanche spp.). In the rhizosphere, SLs also promote root colonization by arbuscular mycorrhizal fungi. In plants, SLs as a novel class of plant hormones regulate various aspects of plant growth and development. Herein I discuss structural diversity of naturally occurring SLs and their distribution in the plant kingdom.
Keywords: strigolactone, germination stimulant, root parasitic plant
Introduction
Strigolactones (SLs) are plant secondary metabolites that are structurally related to strigol (1), which was first identified as a germination stimulant for the root parasitic weed witchweed, Striga lutea.1–3) SLs induce seed germination not only in hemiparasites Striga spp. but also in holoparasites broomrapes (Orobanche and Phelipanche spp.), both of the Orobanchaceae. These root parasitic weeds cause enormous damage to agricultural production all over the world, because appropriate and economically feasible control measures have not yet been developed. Furthermore, these weeds are gradually invading crop land in both developing and developed countries.4,5) Extensive studies on the characterization of SLs produced by various plant species have demonstrated that most land plants produce and release not single but mixtures of SLs.6)
The reason why plants produce and release SLs, which attract root parasitic weeds, has long been sought. In 2005, one SL, 5-deoxystrigol (5DS, 2) was identified as a branching factor for important symbionts the arbuscular mycorrhizal (AM) fungi, with which more than 80% of land plants form symbiotic associations,4,7) demonstrating that plants produce and release SLs to promote symbiotic associations with AM fungi which supply nutrients, especially phosphate. The Brassicaceae and Chenopodiaceae families and some legume plants like white lupin (Lupinus albus) are non-mycotrophic but still produce SLs,8,9) indicating that SLs may have other important functions in plants. In 2008, SLs or their further metabolites were shown to be a novel class of plant hormone inhibiting shoot branching.10,11) These findings attracted researchers to study the biological functions of SLs and they have been shown to be involved in the regulation of growth of lateral roots and root hairs,12,13) secondary growth,14) photomorophogenesis15) and leaf senescence,16,17) and to promote root colonization by root nodule bacteria.18–21)
Approximately 25 natural SLs have been characterized from root exudates of various plant species.22) Structural diversity in these natural SLs is closely related to biosynthesis, metabolism and biological functions of SLs. In this review, I discuss structural diversity of natural SLs and their distribution in the plant kingdom.
1. Natural SLs
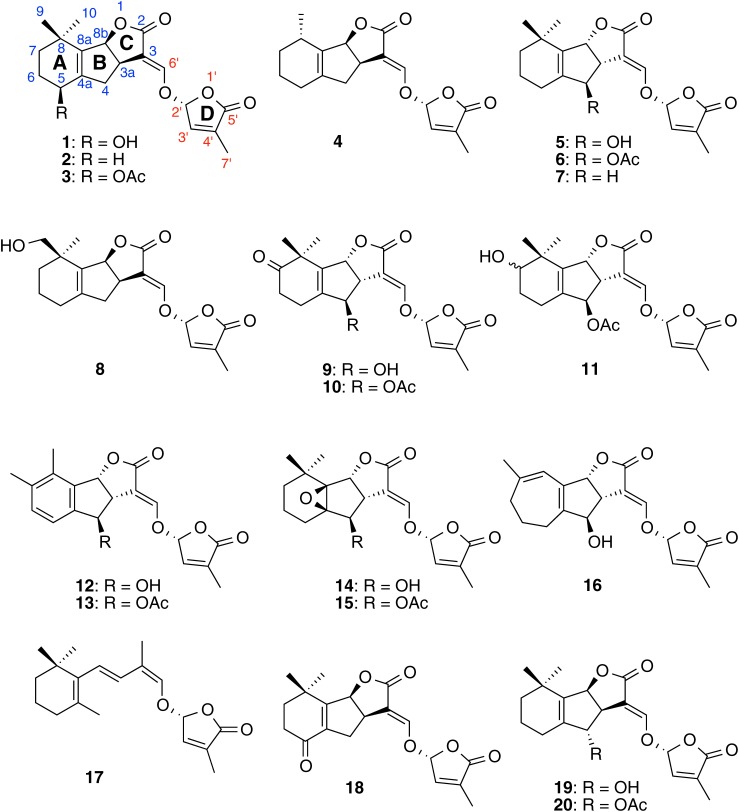
The first reported SLs—strigol and its acetate, strigyl acetate (3)—were isolated from root exudate of cotton (Gossypium hirsutum) a non-host of Striga spp., as germination stimulants for S. lutea.1) Therefore, there were some arguments that SLs were not true host-derived Striga germination stimulants. Then, strigol was isolated from root exudates of maize (Zea mays), sorghum (Sorghum bicolor) and proso-millet (Panicum miliaceum), genuine hosts of Striga.23) We also analyzed root exudates of several maize cultivars, but did not detect strigol.24) Jamil et al. reported that they could not detect any known SLs from root exudates of maize cultivars examined.25) In addition to strigol, two new SLs structurally related to strigol, sorgolactone (4) and alectrol, were purified from root exudates of sorghum26) and cowpea (Vigna unguiculata),27) respectively. Orobanchol, along with alectrol, was characterized as the first Orobanche germination stimulant from red clover (Trifolium pratense) root exudates.28) This clearly demonstrates that SLs elicit seed germination in both Striga and Orobanche spp. Alectrol was later identified as orobanchyl acetate (6) but was not an isomer of strigol.29,30) 5DS was originally isolated as a branching factor for AM fungi and was then shown to be produced by several plant species.31) However, the retention of 5DS is very similar to that of its isomer 4-deoxyorobanchol (4DO, 7) in both reversed phase and normal phase high performance liquid chromatography, and some plant species reported to produce 5DS may produce 4DO. Sorgomol (8) is an isomer of strigol and orobanchol and was isolated from sorghum root exudates.32) Not only monocots but also dicots including white lupin and Chinese milk vetch (Astragalus sinicus) produce sorgomol.9) The SLs 7-oxoorobanchol (9), 7-oxoorobanchyl acetate (10) and 7-hydroxyorobanchyl acetate (11) were isolated from root exudates of flax (Linum usitatissimum)33) and were detected in root exudates from various plant species including cucumber (Cucumis sativus).34) Solanaceae plants such as tobacco (Nicotiana tabacam) and tomato (Solanum lycopersicum) produce solanacol (12) and solanacyl acetate (13), unique SLs containing a benzene ring.34,35) Fabacol (14) and fabacyl acetate (15), found in root exudate of pea (Pisum sativum), have an epoxide.36) Medicaol (16), a putative didehydro-orobanchol isomer containing a seven-membered A ring, was recently identified from root exudates of barrel medic (Medicago truncatula).37) Root exudates from red clover, tomato and tobacco were found to contain didehydro-orobanchol isomers, which differ from medicaol but with structures still to be elucidated. Putative desmethyl-orobanchyl acetate isomers, desmethyl-7-hydroxyorobanchyl acetate isomers, dihydro-orobanchol isomers and their derivatives have been detected from various plant species (Xie et al., unpublished).
2. Structures and stereochemistry of naturally occurring SLs
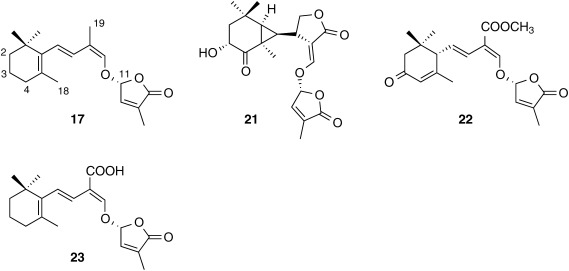
Naturally occurring SLs (natural SLs) are classified into two groups based on their structures. SLs structurally related to strigol are called canonical (or classical) SLs in which the ABC ring moiety, a core structure, connects to a butenolide (D ring) via an enol–ether bridge. By contrast, in non-canonical SLs, the D ring connects to a variety of structures.21) For example, carlactone (CL, 17), a biosynthetic precursor for SLs, contains only the A ring of canonical SLs.38)
Canonical SLs contain three asymmetric carbons (C-3a, C-8b and C-2′) but C-3a should be cis to C-8b as they are at bridge heads of the C ring. Therefore, canonical SLs without substituents on the AB ring (e.g., 5DS and 4DO) consist of four stereoisomers. Since natural CL has an 11-R stereochemistry, which corresponds to the C-2′ in canonical SLs,39) only 5DS with β-oriented C ring and 4DO with α-oriented C ring among four possible stereoisomers are natural SLs. The canonical SLs so far characterized have substituents on the A and/or B rings but no modifications on the CD ring moiety.6,21)
Until the structure of fabacyl acetate was determined in 2009,36) the first reported canonical SL containing an α-oriented C ring, all natural SLs were thought to have a β-oriented C ring and be derived from 5DS. In 2010, total synthesis of solanacol and solanacyl acetate confirmed that these SLs also carry an α-oriented C ring.40) Ueno et al. revised the structures of orobanchol and orobanchyl acetate to have an α-oriented C ring.41) This led to structural revisions of 7-oxoorobanchyl acetate and 7-hydroxyorobanchyl acetate. Consequently, natural canonical SLs can be divided into two types: orobanchol-type with α-oriented C ring and strigol-type with β-oriented C ring.21,35) Strigol- and orobanchol-type SLs seem to be derived from 5DS and 4DO, respectively (Fig. 1).
Fig. 1. Structures of canonical strigolactones and carlactone.
We identified orobanchol, orobanchyl acetate and 4DO from rice root exudates.35) Orobanchol-type SLs such as 4DO were detected in root exudates of bright yellow tobacco cultivar Tsukuba No. 1. In addition to orobanchol-type, strigol-type SLs were detected in root exudates of burley tobacco cultivar Michinoku No. 1.35) A medicinal plant, dokudami (Houttuynia cordata), was found to produce strigol-type SLs and strigone (18), an oxidized metabolite of strigol, was first detected as a natural SL.42) These results confirm that two biosynthetic pathways lead to two types of canonical SLs and suggest that some plant species produce either strigol- or orobanchol-type SLs as major SLs and the others produce both types of SLs.21,35)
Canonical SLs are synthesized from β-carotene via the biosynthetic intermediate CL.38,39,43) CL contains the A and D rings of canonical SLs but lacks the B and C rings. Oxidations at C-19 and C-18 and the subsequent ring closure appear to convert CL to 5DS or 4DO which are then modified independently.38,43)
Allylic oxidations of 5DS at C-5 and C-4 produce strigol and 4-hydroxy-5DS (19), respectively, and an oxidation at a homoallylic position (C-9) gives sorgomol. Strigol is further oxidized to strigone, and sorgomol may be converted to sorgolactone after oxidation and subsequent decarboxylation. Although we grew a large number of sorghum plants (several different cultivars) and collected root exudates, sorgolactone has never been detected. Strigol and 4-hydroxy-5DS are acetylated to give strigyl acetate and 4-acetoxy-5DS (ent-2′-epi-orobanchyl acetate, 20), respectively. Allylic oxidation of 4DO at C-4 affords orobanchol. The oxidation at C-7 of orobanchol gives 7-hydroxyorobanchol, which is further oxidized to 7-oxoorobanchol. The 7-hydroxyorobanchol may be converted to solanacol via dehydration, oxidation and migration of a methyl group. A seven-membered A ring may be formed before construction of the ABC ring structure. Orobanchol is converted to fabacol by epoxidation. Hydroxy-SLs are acetylated to corresponding acetoxy-SLs. It is intriguing that strigol-type isomers of orobanchol and orobanchyl acetate were detected,35) while either strigol- or orobanchol-type SLs have been detected for other canonical SLs. For example, neither 5-hydroxy-4DO (orobanchol-type isomer of strigol) nor 9-hydroxy-4DO (orobanchol-type isomer of sorgomol) have been detected. Firstly, this is because that all orobanchol-type SLs, except for 4DO, are derived from orobanchol. Secondly, presence of the β-oriented C ring in 5DS may restrict directions and positions of oxidation on the A ring. However, orobanchol, orobanchyl acetate and a didehydro-orobanchol isomer were detected in red clover root exudates but not 4DO,28) indicating that oxidation of 4DO to orobanchol proceeds rapidly. Alternatively, orobanchol may be synthesized not from 4DO but directly from CL or hydroxy-CL in this plant.
3. Non-canonical SLs
Canonical SLs described in the previous section are derived from either 5DS or 4DO and contain the core structure, the ABC ring. Several novel germination stimulants that are structural distinct from canonical SLs have been characterized in root exudates from various plant species. Avenaol (21)44) and heliolactone (22)45) are two novel germination stimulants isolated from black oat (Avena strigosa) and sunflower (Helianthus annus), respectively, and are typical non-canonical SLs. Carlactonoic acid (CLA, 23)46) formed from CL by MAX1 oxidation has been detected in root exudates of various plant species including maize, sunflower, spikemoss (Selaginella moellendorfii) and poplar (Populus spp.). In addition to these non-canonical SLs, more than 10 novel germination stimulants structurally related to CL have been suggested. It is intriguing that root exudates from plant species producing these non-canonical SLs as major germination stimulants should exhibit strong germination stimulation activity to seeds of root parasitic plants but do not contain detectable levels of canonical SLs (Fig. 2).
Fig. 2. Structures of non-canonical strigolactones.
4. Distribution of SLs in the plant kingdom
We analyzed SLs produced by various plant species and some results are listed in Table 1.
Table 1. Distribution of strigolactones in the plant kingdom.
| Canonical strigolactones | Non-canonical strigolactones | |||
|---|---|---|---|---|
| Orobanchol-type SL | Strigol-type SL | |||
| Pteridophyte | Selaginella moellendorffii | 4-deoxyorobanchol | CLA | |
| Gymnosperm | Pinus thunbergii | orobanchol, orobanchyl acetate | CLA | |
| Ginkgo biloba | 4-deoxyorobanchol, orobanchol, orobanchyl acetate | |||
| Angiosperm | Oryza sativa | 4-deoxyorobanchol, orobanchol, orobanchyl acetate, 7-oxoorobanchyl acetate | ||
| Pisum sativum | 4-deoxyorobanchol, orobanchol, orobanchyl acetate, fabacol, fabacyl acetate | |||
| Solanum lycopersicum | 4-deoxyorobanchol, orobanchol, solanacol, 7-hydroxyorobanchol | |||
| Cucumis sativus | 4-deoxyorobanchol, orobanchol, orobanchyl acetate, 7-oxoorobanchol, 7-oxoorobanchyl acetate, 7-hydroxyorobanchol, 7-hydroxyorobanchyl acetate | |||
| Nicotiana tabacum | 4-deoxyorobanchol, orobanchol, orobanchyl acetate, solanacol, solanacyl acetate | 5-deoxystrigol, 4-hydroxy-5-deoxystrigol, 4-acetoxy-5-deoxystrigol | ||
| Astragalus sinicus | orobanchyl acetate | 5-deoxystrigol, sorgomol | ||
| Gossypium hirsutum | strigol, strigyl acetate | |||
| Sorghum bicolor | 5-deoxystrigol, strigol, strigyl acetate, sorgomol | |||
| Fragaria×ananassa | 5-deoxystrigol, strigol, strigyl acetate | |||
| Lotus japonicus | 5-deoxystrigol | methyl lotuslactonoate* | ||
| Helianthus annuus | heliolactone, CLA | |||
| Zea mays | 5-deoxystrigol | methyl zealactonoate** CLA | ||
| Populus | 4-deoxyorobanchol | CLA, MeCLA | ||
Strigolactones identified in root exudates are listed. CLA, carlactononic acid; MeCLA, methyl carlactonoate. *, ** Structures of these novel strigolactones will be reported elsewhere.
In angiosperms, rice and cucumber produce orobanchol-type SLs, while cotton and strawberry (Fragaria×ananassa) produce strigol-type SLs. By contrast, Chinese milk vetch is a producer of both types of SLs. Tobacco plants also produce both types of SLs, and the ratio of orobanchol- to strigol-type SLs significantly differ between cultivars. The burley tobacco Michinoku No. 1 produce both types of SLs at similar levels, while amounts of strigol-type SLs are only 1% that of orobanchol-type SLs in root exudates of bright yellow tobacco Tsukuba No. 1. In general, SL production is known to be promoted under phosphorus (P) and nitrogen (N) deficiencies.47–49) It should be noted that in plant species producing both types of SLs, nutrient deficiencies, especially of P and N, more strongly affect production of one type of SL. In the case of tobacco, P deficiency increased production of strigol-type SLs by more than 1000-fold, whereas orobanchol-type SLs were unaffected. Similar results were obtained with Chinese milk vetch, in which P and N deficiencies promoted strigol-type SLs, 5DS and sorgomol, but level of orobanchyl acetate was unaffected.50) These results suggest that production and exudation of strigol- and orobanchol-type SLs are regulated independently.
5. Transport of SLs from roots to shoots
Results from reciprocal grafting experiments using wild-type and SL biosynthetic and perception mutants of Arabidopsis, pea and Petunia, reveal that plant hormones inhibiting shoot branching are produced mainly in the roots and transported to shoots.6,10,20,21) The most probable route for SL transport from roots to shoots is xylem and indeed Kohlen et al. detected orobanchol and other SLs in xylem sap from Arabidopsis and tomato.51,52) However, no reports support xylem transport of SLs. We collected large amounts of xylem sap from various plant species including tomato and Arabidopsis but no signals attributable to known canonical or non-canonical SLs were detected by LC–MS/MS analyses.53) Then, deuterated SLs, d1-orobanchol and d6-4DO, were fed to roots of rice plants and the shoots were harvested 2 and 20 hr after SL treatment. The SLs were detected in shoots harvested 20 hr after but not 2 hr after treatment.53) These results strongly suggest that exogenous and endogenous SLs are transported from roots to shoots not through the xylem but through hypodermal passage cells as in Petunia where polar and asymmetric localization of an ABC transporter (PaPDR1) have been shown to mediate shootward SL transport as well as localized exudation into the rhizosphere.54,55) Furthermore, aforementioned species-specific phenomena in SL production also occur in the transport of exogenous SLs from the roots to shoots, indicating that the transport is structure- and stereo-specific.56) For example, in rice plants, which produce orobanchol-type SL, only orobanchol-type SLs are transported from roots to shoots.56) However, strigol applied to roots of SL biosynthetic rice mutant d10 inhibited tiller bud outgrowth,11) indicating that metabolites of SLs or other signaling compounds downstream of SLs—but not SLs themselves—are the true inhibitors of tiller bud outgrowth.
Conclusion
SLs were originally discovered as germination stimulants for root parasitic plants 50 years ago and are now recognized as important signaling compounds not only in the rhizosphere but also in plants. In the rhizosphere, the physicochemical and biological conditions significantly differ from those of bulk soil.57) For example, pH in the rhizosphere is rather acidic and thus alkaline-unstable canonical SLs would endure longer than expected. In addition to canonical SLs, recently discovered non-canonical SLs were shown to be released into the rhizosphere, but their involvement in rhizosphere communications among soil organisms remains largely unknown. Therefore, further studies are needed to understand involvement of canonical and non-canonical SLs in chemical communications between plants and other organisms which have been exposed to these signaling compounds for more than 400 million years. In particular, some soil microorganisms may not only utilize SLs as cues of living plants nearby but also decompose and/or transform them to support their survival. It is therefore important to characterize novel SLs and elucidate their functions and action mechanisms to develop practical applications of SLs in plant production and crop protection. SLs and their agonists and/or antagonists, and biosynthetic inhibitors could be applied to regulate plant architecture, optimize AM symbiosis and control parasitic weeds.
Acknowledgments
This study was conducted at the Weed Science Center and Center for Bioscience Research and Education, Utsunomiya University. This study was supported by KAKENHI; the Program for Promotion of Basic and Applied Research for Innovations in Bio-Oriented Industry; the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry; a special grant UUCOE from Utsunomiya University; and by a grant from JGC-S Scholarship Foundation. The author would like to express his gratitude to Prof. Koichi Yoneyama, Emeritus Prof. Yasutomo Takeuchi (Utsunomiya University), Emeritus Prof. Takao Yokota (Teikyo University) and Prof. Kohki Akiyama (Osaka Prefecture University) for their kind guidance and warm encouragement. The author is also grateful to co-workers and students.
References
- 1).C. E. Cook, L. P. Whichard, B. Turner, M. E. Wall and G. H. Egley: Science 154, 1189–1190 (1966). [DOI] [PubMed] [Google Scholar]
- 2).C. E. Cook, L. P. Whichard, M. E. Wall, G. H. Egley, P. Coggon, P. A. Luhan and A. T. McPhail: J. Am. Chem. Soc. 94, 6198–6199 (1972). [Google Scholar]
- 3).L. G. Butler: “Allelopathy, Organisms, Processes and Applications,” ed. by Inderjit, K. M. M. Dakshini and F. A. Enhelling, American Chemical Society, Washington DC, pp. 158–168, 1995.
- 4).C. Parker: Pest Manag. Sci. 65, 453–459 (2009). [DOI] [PubMed] [Google Scholar]
- 5).C. Parker: Weed Sci. 60, 269–276 (2012). [Google Scholar]
- 6).X. Xie, K. Yoneyama and K. Yoneyama: Annu. Rev. Phytopathol. 48, 93–117 (2010). [DOI] [PubMed] [Google Scholar]
- 7).K. Akiyama, K. Matsuzaki and H. Hayashi: Nature 435, 824–827 (2005). [DOI] [PubMed] [Google Scholar]
- 8).Y. Goldwasser, K. Yoneyama, X. Xie and K. Yoneyama: Plant Growth Regul. 55, 21–28 (2008). [Google Scholar]
- 9).K. Yoneyama, X. Xie, H. Sekimoto, Y. Takeuchi, S. Ogasawara, K. Akiyama, H. Hayashi and K. Yoneyama: New Phytol. 179, 484–494 (2008). [DOI] [PubMed] [Google Scholar]
- 10).V. Gomez-Roldan, S. Fermas, P. B. Brewer, V. Puech-Pagès, E. A. Dun, J.-P. Pillot, F. Letisse, R. Matusova, S. Danoun, J.-C. Portais, H. Bouwmeester, G. Bécard, C. A. Beveridge, C. Rameau and S. F. Rochange: Nature 455, 189–194 (2008). [DOI] [PubMed] [Google Scholar]
- 11).M. Umehara, A. Hanada, S. Yoshida, K. Akiyama, T. Arite, N. Takeda-Kamiya, H. Magome, Y. Kamiya, K. Shirasu, K. Yoneyama, J. Kyozuka and S. Yamaguchi: Nature 455, 195–200 (2008). [DOI] [PubMed] [Google Scholar]
- 12).Y. Kapulnik, P.-M. Delaux, N. Resnick, E. Mayzlish-Gati, S. Wininger, C. Bhattacharya, N. Séjalon-Delmas, J.-P. Combier, G. Bécard, E. Belausov, T. Beeckman, E. Dor, J. Hershenhorn and H. Koltai: Planta 233, 209–216 (2011). [DOI] [PubMed] [Google Scholar]
- 13).C. Ruyter-Spira, W. Kohlen, T. Charnikhova, A. van Zeijl, L. van Bezouwen, N. de Ruijter, C. Cardoso, J. A. Lopez-Raez, R. Matusova, R. Bours, F. Verstappen and H. Bouwmeester: Plant Physiol. 155, 721–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).J. Agusti, S. Herold, M. Schwarz, P. Sanchez, K. Ljung, E. A. Dun, P. B. Brewer, C. A. Beveridge, T. Sieberer, E. M. Sehr and T. Greb: Proc. Natl. Acad. Sci. U.S.A. 108, 20242–20247 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).H. Shen, P. Luong and E. Huq: Plant Physiol. 145, 1471–1483 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Y. Yamada, S. Furusawa, S. Nagasaka, K. Shimomura, S. Yamaguchi and M. Umehara: Planta 240, 399–408 (2014). [DOI] [PubMed] [Google Scholar]
- 17).H. Ueda and M. Kusaba: Plant Physiol. 169, 138–147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).M. J. Soto, M. Fernández-Aparicio, V. Castellanos-Morales, J. M. García-Garrido, J. A. Ocampo, M. J. Delgado and H. Vierheilig: Soil Biol. Biochem. 42, 383–385 (2010). [Google Scholar]
- 19).E. Foo and N. W. Davies: Planta 234, 1073–1081 (2011). [DOI] [PubMed] [Google Scholar]
- 20).Y. Seto, H. Kameoka, S. Yamaguchi and J. Kyozuka: Plant Cell Physiol. 53, 1843–1853 (2012). [DOI] [PubMed] [Google Scholar]
- 21).S. Al-Babili and H. J. Bouwmeester: Annu. Rev. Plant Biol. 66, 161–186 (2015). [DOI] [PubMed] [Google Scholar]
- 22).P. Khetkam, X. Xie, T. Kisugi, H. I. Kim, K. Yoneyama, K. Uchida, T. Yokota, T. Nomura and K. Yoneyama: J. Pestic. Sci. 39, 121–126 (2014). [Google Scholar]
- 23).B. A. Siame, Y. Weerasuriya, K. Wood, G. Ejeta and L. G. Butler: J. Agric. Food Chem. 41, 1486–1491 (1993). [Google Scholar]
- 24).K. Yoneyama, R. Arakawa, K. Ishimoto, H. I. Kim, T. Kisugi, X. Xie, T. Nomura, F. Kanampiu, T. Yokota, T. Ezawa and K. Yoneyama: New Phytol. 206, 983–989 (2015). [DOI] [PubMed] [Google Scholar]
- 25).M. Jamil, F. K. Kanampiu, H. Karaya, T. Charnikhova and H. J. Bouwmeester: Field Crops Res. 134, 1–10 (2012). [Google Scholar]
- 26).C. Hauck, S. Müller and H. Schildknecht: J. Plant Physiol. 139, 474–478 (1992). [Google Scholar]
- 27).S. Müller, C. Hauck and H. Schildknecht: J. Plant Growth Regul. 11, 77–84 (1992). [Google Scholar]
- 28).T. Yokota, H. Sakai, K. Okuno, K. Yoneyama and Y. Takeuchi: Phytochemistry 49, 1967–1973 (1998). [Google Scholar]
- 29).X. Xie, K. Yoneyama, D. Kusumoto, Y. Yamada, T. Yokota, Y. Takeuchi and K. Yoneyama: Phytochemistry 69, 427–431 (2008). [DOI] [PubMed] [Google Scholar]
- 30).H. Matsuura, K. Ohashi, H. Sasako, N. Tagawa, Y. Takano, Y. Ioka, K. Nabeta and T. Yoshihara: Plant Growth Regul. 54, 31–36 (2008). [Google Scholar]
- 31).K. Yoneyama, X. Xie, T. Kisugi, T. Nomura, H. Sekimoto, T. Yokota and K. Yoneyama: Plant Growth Regul. 65, 495–504 (2011). [Google Scholar]
- 32).X. Xie, K. Yoneyama, D. Kusumoto, Y. Yamada, Y. Takeuchi, Y. Sugimoto and K. Yoneyama: Tetrahedron Lett. 49, 2066–2068 (2008). [Google Scholar]
- 33).X. Xie, K. Yoneyama, J. Kurita, Y. Harada, Y. Yamada, Y. Takeuchi and K. Yoneyama: Biosci. Biotechnol. Biochem. 73, 1367–1370 (2009). [DOI] [PubMed] [Google Scholar]
- 34).X. Xie, D. Kusumoto, Y. Takeuchi, K. Yoneyama, Y. Yamada and K. Yoneyama: J. Agric. Food Chem. 55, 8067–8072 (2007). [DOI] [PubMed] [Google Scholar]
- 35).X. Xie, K. Yoneyama, T. Kisugi, K. Uchida, S. Ito, K. Akiyama, H. Hayashi, T. Yokota, T. Nomura and K. Yoneyama: Mol. Plant 6, 153–163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).X. Xie, K. Yoneyama, Y. Harada, N. Fusegi, Y. Yamada, S. Ito, T. Yokota, Y. Takeuchi and K. Yoneyama: Phytochemistry 70, 211–215 (2009). [DOI] [PubMed] [Google Scholar]
- 37).T. Tokunaga, H. Hayashi and K. Akiyama: Phytochemistry 111, 91–97 (2015). [DOI] [PubMed] [Google Scholar]
- 38).A. Alder, M. Jamil, M. Marzorati, M. Bruno, M. Vermathen, P. Bigler, S. Ghisla, H. Bouwmeester, P. Beyer and S. Al-Babili: Science 335, 1348–1351 (2012). [DOI] [PubMed] [Google Scholar]
- 39).Y. Seto, A. Sado, K. Asami, A. Hanada, M. Umehara, K. Akiyama and S. Yamaguchi: Proc. Natl. Acad. Sci. U.S.A. 111, 1640–1645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).V. X. Chen, F.-D. Boyer, C. Rameau, P. Retailleau, J.-P. Vors and J.-M. Beau: Chemistry 16, 13941–13945 (2010). [DOI] [PubMed] [Google Scholar]
- 41).K. Ueno, S. Nomura, S. Muranaka, M. Mizutani, H. Takikawa and Y. Sugimoto: J. Agric. Food Chem. 59, 10485–10490 (2011). [DOI] [PubMed] [Google Scholar]
- 42).T. Kisugi, X. Xie, H. I. Kim, K. Yoneyama, A. Sado, K. Akiyama, H. Hayashi, K. Uchida, T. Yokota, T. Nomura and K. Yoneyama: Phytochemistry 87, 60–64 (2013). [DOI] [PubMed] [Google Scholar]
- 43).Y. Zhang, A. D. J. van Dijk, A. Scaffidi, G. R. Flematti, M. Hofmann, T. Charnikhova, F. Verstappen, J. Hpeworth, S. van der Krol, O. Leyser, S. M. Smith, B. Zwanenburg, S. Al-Babili, C. Ruyter-Spira and H. J. Bouwmeester: Nat. Chem. Biol. 10, 1028–1033 (2014). [DOI] [PubMed] [Google Scholar]
- 44).H. I. Kim, T. Kisugi, P. Khetkam, X. Xie, K. Yoneyama, K. Uchida, T. Yokota, T. Nomura, C. S. P. McErlean and K. Yoneyama: Phytochemistry 103, 85–88 (2014). [DOI] [PubMed] [Google Scholar]
- 45).K. Ueno, T. Furumoto, S. Umeda, M. Mizutani, H. Takikawa, R. Batchvarova and Y. Sugimoto: Phytochemistry 108, 122–128 (2014). [DOI] [PubMed] [Google Scholar]
- 46).S. Abe, A. Sado, K. Tanaka, T. Kisugi, K. Asami, S. Ota, H. I. Kim, K. Yoneyama, X. Xie, T. Ohnishi, Y. Seto, S. Yamaguchi, K. Akiyama, K. Yoneyama and T. Nomura: Proc. Natl. Acad. Sci. U.S.A. 111, 18084–18089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).K. Yoneyama, K. Yoneyama, Y. Takeuchi and H. Sekimoto: Planta 225, 1031–1038 (2007). [DOI] [PubMed] [Google Scholar]
- 48).K. Yoneyama, X. Xie, D. Kusumoto, H. Sekimoto, Y. Sugimoto, Y. Takeuchi and K. Yoneyama: Planta 227, 125–132 (2007). [DOI] [PubMed] [Google Scholar]
- 49).J. A. López-Ráez, T. Charnikhova, V. Gómez-Roldán, R. Matusova, W. Kohlen, R. De Vos, F. Verstappen, V. Puech-Pages, G. Bécard, P. Mulder and H. Bouwmeester: New Phytol. 178, 863–874 (2008). [DOI] [PubMed] [Google Scholar]
- 50).K. Yoneyama, X. Xie, H. I. Kim, T. Kisugi, T. Nomura, H. Sekimoto, T. Yokota and K. Yoneyama: Planta 235, 1197–1207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).W. Kohlen, T. Charnikhova, Q. Liu, R. Bours, M. A. Domagalska, S. Beguerie, F. Verstappen, O. Leyser, H. Bouwmeester and C. Ruyter-Spira: Plant Physiol. 155, 974–987 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).W. Kohlen, T. Charnikhova, M. Lammers, T. Pollina, P. Toth, I. Haider, M. J. Pozo, R. A. de Maagd, C. Ruyter-Spira, H. J. Bouwmeester and J. A. Lopez-Raez: New Phytol. 196, 535–547 (2012). [DOI] [PubMed] [Google Scholar]
- 53).X. Xie, K. Yoneyama, T. Kisugi, T. Nomura, K. Akiyama, T. Asami and K. Yoneyama: J. Pestic. Sci. 40, 214–216 (2015). [Google Scholar]
- 54).T. Kretzschmar, W. Kohlen, J. Sasse, L. Borghi, M. Schlegel, J. B. Bachelier, D. Reinhardt, R. Bours, H. J. Bouwmeester and E. Martinoia: Nature 483, 341–344 (2012). [DOI] [PubMed] [Google Scholar]
- 55).J. Sasse, S. Simon, C. Gübeli, G.-W. Liu, X. Cheng, J. Friml, H. Bouwmeester, E. Martinoia and L. Borghi: Curr. Biol. 25, 647–655 (2015). [DOI] [PubMed] [Google Scholar]
- 56).X. Xie, K. Yoneyama, T. Kisugi, T. Nomura, K. Akiyama, T. Asami and K. Yoneyama: J. Pestic. Sci. 41, 55–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).C. Bertin, X. Yang and L. A. Weston: Plant Soil 256, 67–83 (2003). [Google Scholar]