Abstract
Background
Colorectal cancer (CRC) is one of most common cancers worldwide. Long non-coding RNA SNHG6 has been reported to act as essential regulators in several cancers. However, the functional role and molecular mechanism of SNHG6 in colorectal cancer remain unclear.
Methods
Quantitative real-time polymerase chain reaction (PCR) was performed to evaluate the SNHG6 expression in CRC tissues. Colony formation, transwell assays and in vivo mice models were carried out to assess the effect of SNHG6 on CRC biological functions.
Results
In the present study, we showed that the expression of SNHG6 was significantly upregulated in CRC tissues and cell lines. High expression of SNHG6 was associated with shorter overall survival in CRC patients. Functionally, SNHG6 knockdown significantly inhibited cell proliferation, invasion and migration both in vitro and in vivo. Mechanically, miR-760 was a direct target of SNHG6, and repression of miR-760 could rescue the inhibitory effect of SNHG6 knockdown on CRC progression. In addition, SNHG6 positively regulated FOXC1 expression through sponging miR-760 in CRC cells, thus indicating that SNHG6 exerted an oncogenic role in CRC by acting as a ceRNA of miR-760.
Conclusion
Our results indicate that long non-coding RNA SNHG6 promotes colorectal cancer progression by sequestering miR-760 and activating FOXC1, our findings suggest that SNHG6 may serve as a potential therapeutic target for CRC.
Keywords: lncRNA SNHG6, miR-760, FOXC1, colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the leading causes of death worldwide due to rapid progression and lack of early detection.1 During the past decade, remarkable progress has been made in the treatment of CRC, including surgical resection and chemotherapy. However, the prognosis of CRC remains unsatisfactory, especially in patients diagnosed with distant metastasis.2,3 The liver is the major organ in which the CRC cells prefer to metastasize.4 Unfortunately, resectable liver metastasis accounts for only around 30% of all liver metastasis, the majority needs to be subjected to chemotherapy.5 Thus, investigation of the potential mechanism underlying the progression of CRC is crucial for future CRC treatment.
Recently, genome sequencing has uncovered that actively transcribed DNA sequence accounts for more than 90% of all genome sequence, while only 2% of these transcripts encode proteins, which indicates that most of the transcripts are non-coding RNAs (ncRNAs).6,7 Long ncRNA (lncRNA) is a class of non-protein coding RNA transcripts with length longer than 200 nucleotides.8 Mounting evidence shows that deregulated lncRNAs are involved in various biological processes in cancers, including cell proliferation, apoptosis, chemoresistance and metastasis.9
LncRNA SNHG6 was shown to function as an oncogene in hepatocellular carcinoma and gastric cancer.10,11 Furthermore, in a recent TCGA analysis, SNHG6 was identified as a potential prognostic marker in colon adenocarcinoma without further experimental validation.12 Nevertheless, the functional role and molecular mechanism of SNHG6 in CRC remain unclear.
In the present study, we found that SNHG6 was upregulated in CRC cell lines and patient tissues. Higher expression of SNHG6 was correlated with poor prognosis of CRC patients. SNHG6 knockdown significantly suppressed the proliferation and metastatic ability of CRC cells both in vitro and in vivo by sponging miR-760. Furthermore, forkhead box C1 (FOXC1) was validated as the direct target of miR-760. Collectively, our data reveal the SNHG6/miR-760/FOXC1 signaling axis may serve as a potential therapeutic target for CRC.
Materials and methods
Cell lines
Human CRC cell lines (LOVO, RKO, Ls174t, DLD1, HCT116) and a non-malignant intestinal epithelial cell, IEC, were purchased from the Cell Bank at the Chinese Academy of Sciences. The cells were propagated in Roswell Park Memorial Institute 1640 or DMEM medium supplemented with 10% fetal bovine serum at 37°C, containing 5% CO2.
Clinical samples
Forty CRC tumor and adjacent normal tissues were obtained from the First Affiliated Hospital of Harbin Medical University from July 2014 to July 2016, with the informed written consent of patients and approval from the ethics committee of Harbin Medical University for the experiments.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from fresh patient tissues after surgery, and from cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA, Carlsbad, CA, USA) or RNeasy Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The probes for miR-760 and endogenous control RNA U6 were purchased from Applied Biosystems (Foster City, CA, USA). The expression of miR-760 and U6 were measured by TaqMan MiRNA assay kit (Applied Biosystems). The expression of SNHG6 and FOXC1 were quantified by using Takara’s SYBR Premix Ex Taq™ II kit (Tli RNaseH Plus) in Applied Biosystems. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. The primers were listed as following: SNHG6: 5′-ATACTTCTGCTTCGTTACCT-3′(F), 5′-CTCATTTTCATCATTTGCT-3′(R); GAPDH: 5′-ACCACAGTCCATGCCATCAC-3′(F), 5′-TCACCACCC TGTTGCTGTA-3′(R); FOXC1: 5′-GGCGAGCAGAGCT ACTACC-3′(F), 5′-TGCGAGTACACGCTCATGG-3′(R). The cDNA amount used in the PCR reaction is 25 ng. Data analysis was performed with ΔΔCt values from each sample calculated by normalizing with the respective control.
Plasmid and cell transfection
To knockdown SNHG6, three different shRNA sequences were cloned into the lentivirus backbone carrying the luciferase. The sequence was as following: sh-SNHG6#1:5′-CTGCGAGGTGCAAGAAAGCCT-3′, sh-SNHG6#2: 5′-GATGAATGTTCTTAGCTGTCA-3′, sh-SHNG6#3: 5′-GCTTCGTTACCTCAAGTGTGG-3′. Full-length SNHG6 was then cloned into the expression vector pcDNA3.1 for overexpression of SNHG6 in CRC cells. Mutated plasmids were introduced using a Quickchange® site-directed muta-genesis kit (Stratagene, La Jolla, CA, USA) according to manufacturer’s instructions. miR-760 mimics, miR-760 inhibitor and negative control miRNA (miR-NC) were purchased from RiboBio (Guangzhou, China). Transfection was performed by using FuGENE®6 (Roche, Nutley, NJ, USA) according to the manufacturer’s instructions.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was assayed using CCK-8 (Beyotime, Hangzhou, China). Briefly, the cells were plated onto 96-well plates in triplicate at approximately 4×103 cells per well and cultured in the medium. The cells were then treated with the CCK-8 reagent, and the absorbance (450 nm) was measured at the indicated time points.
Colony formation assay
Transfected cells were plated in 10 cm dishes and incubated in the growth medium at 37°C for 2 weeks. Two weeks later, the cells were fixed with 4% formaldehyde, and stained with 1% crystal violet. The number of cell colonies was counted and analyzed.
Wound healing assay
Indicated CRC cells were plated on 6-well plates (1×106 cells/well) and then scratches were made using a 100-µL tip. The distance between the two edges of the wound was measured and imaged after 24 hours of wound formation.
Transwell assay
Cell invasion or migration was assessed with or without Matrigel-coated chambers in 24-well plates (BD Biosciences, NY, USA). Briefly, the indicated CRC cells (1×105) were plated in serum-free medium and allowed to translocate toward complete media below the transwell inserted champer. Afterwards, the cells translocating from the membrane to the lower surface were fixed, stained and counted after 24 hours.
Dual luciferase reporter assay
The wild-type SNHG6 (WT-SNHG6) and mutant SNHG6 (Mut-SNHG6) containing the binding sites of miR-760 were established and incorporated into the PGL3 dual-luciferase vector (Promega, Madison, WI, USA). Similarly, the WT and Mut 3′UTR of FOXC1 containing the putative miR-760 binding site was also amplified by PCR and then cloned into pGL3 vector. Subsequently, HEK293 cells were plated into 24-well plates and were co-transfected with indicated plasmids using FuGENE®6. After 48 hours, the cells were harvested and the relative luciferase activities were assessed by Dual-luciferase Reporter Assay System according to the manufacturer’s instructions. All experiments were repeated at least three times.
Western blot assay
The indicated cells were digested and then lysed by radioimmunoprecipitation assay buffer. After protein quantification, 50 µg of the samples were loaded onto 10% polyacrylamide gels, then transferred to polyvinylidene difluoride membranes. Expression of each protein was incubated with primary antibody (FOXC1, Santa Cruz, Dallas, TX, USA [C-18], 1:1,000 diluted; GAPDH Antibody [6C5], 1:2,000 diluted) overnight at 4°C. After incubation with horseradish peroxidase-conjugated secondary antibodies (1:5,000 diluted), membranes were developed using an enhanced chemiluminescence immunoblotting kit (Beyotime).
Xenograft model and liver metastasis model
Six-week-old nude mice were purchased from Experimental Animal Center of Chinese Academy of Sciences (Shanghai, China) housed under standard conditions and cared for according to the institutional guidelines for animal care. For the xenograft model, indicated cells (1×107) were harvested and subcutaneously injected into the posterior flank of mice. Tumor volume was monitored on indicated days by caliper measurements and calculated by the algorithms: tumor volume =1/2*length*width.2 For the liver metastasis model, indicated cells (5×106 cells) carrying the luciferase vector were injected through the splenic vein. Liver metastasis was imaged by bioluminescence. D-luciferin (Xenogen, Hopkinton, MA) was injected intraperitoneally into the mice at 100 mg/kg, and bioluminescence was measured with Berthold NIGHTOWL LB983 imaging machine. Four weeks later, the mice were sacrificed and the tumors were collected. All of the animal experiments were approved by the Institutional Animal Care and Use Committee of Harbin Medical University (Protocol Number: 20160821). All animal work was conducted according to the Guide for the Administration of Affairs Concerning Experimental Animals. All animal experiments were performed in accordance with National Institute of Health guidelines for the use of experimental animals.
Statistical analysis
GraphPad Prism6 software or SPSS were used to assess the statistical significance. Student’s t-test or one-way analysis of variance was applied to determine the significance between groups. The overall survival (OS) and disease-free survival were estimated according to the Kaplan–Meier test and significance was determined by the log-rank test. The statistical correlation between the clinical parameters of CRC patients and different SNHG6 expression group in the CRC cohort was analyzed by chi-squared test or Fisher’s exact chi-squared test. Data are presented as the mean ± SD. All experiments were performed at the least in triplicate. Statistical significance was assessed at *P<0.05, **P<0.01, ***P<0.001; # represents no statistical significance.
Results
lncRNA-SNHG6 is highly expressed in CRC and correlates with poor prognosis
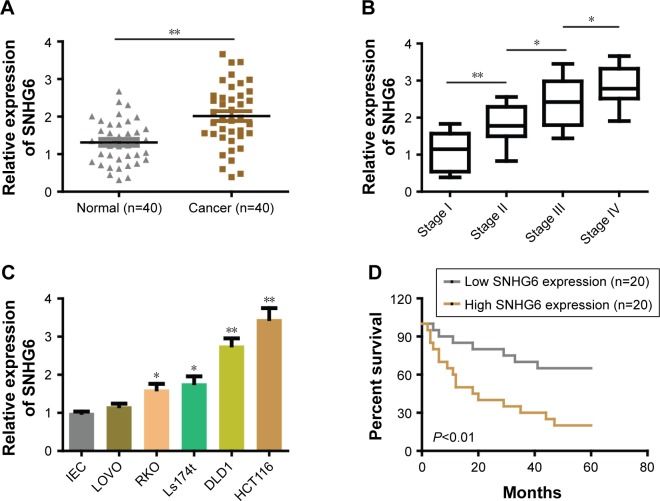
To investigate the role of SNHG6 in CRC, we assessed the expression of SNHG6 in 40 pairs of CRC tissues and adjacent normal tissues by using qRT-PCR. We found that SNHG6 was significantly increased in CRC tissues compared with adjacent normal tissues (Figure 1A). In addition, the expression of SNHG6 significantly correlated with TNM stage (Figure 1B) and lymphatic or distal metastasis by clinicopathological analysis in CRC tissues (Table 1). To further support the above investigation, we detected the levels of SNHG6 in CRC cell lines as well as a non-malignant intestinal epithelial cell line (IEC). Compared with normal epithelial cells, SNHG6 was remarkably upregulated in CRC cells (Figure 1C). Moreover, the Kaplan–Meier analysis indicated that the OS of patients with high SNHG6 expression was significantly shorter compared with low SNHG6 expression (Figure 1D). These results suggest that the SNHG6 level is upregulated in CRC tissues and cell lines, and negatively correlated with the prognosis of CRC patients.
Figure 1.
SNHG6 was overexpressed in CRC tissues and cell lines, and associated with poor prognosis.
Notes: (A) The relative expression of SNHG6 was significantly increased in CRC tissues compared with adjacent normal tissues. (B) The relative expression of SNHG6 in different TNM stages of CRC patients. (C) The relative expression of SNHG6 in CRC cell lines compared with normal IEC. (D) Overall survival of CRC patients by Kaplan–Meier analysis. *P<0.05; **P<0.01.
Abbreviations: CRC, colorectal cancer; IEC, intestinal epithelial cells.
Table 1.
Relationship between SNHG6 expression and clinicopathologic features of colorectal cancer patients (n=40)
| Variable | Relative SNHG6 expression
|
P-value | |
|---|---|---|---|
| Low (n=20) | High (n=20) | ||
| Age (years) | NS | ||
| <50 | 8 | 11 | |
| >50 | 12 | 9 | |
| Gender | NS | ||
| Male | 14 | 10 | |
| Female | 6 | 10 | |
| Lymphatic metastasis | P<0.001 | ||
| N0/N1 | 15 | 4 | |
| N2/N3 | 5 | 16 | |
| Distal metastasis | P<0.05 | ||
| Negative | 19 | 13 | |
| Positive | 1 | 7 | |
| Tumor stage | P<0.001 | ||
| I/II | 16 | 5 | |
| III/IV | 4 | 15 | |
Notes: Patients were divided into SNHG6 high group (relative fold change was higher than the median) and low group (relative fold change was lower than the median) according to the analysis of quantitative real-time polymerase chain reaction detection. NS, not significant between different groups. Differences among variables were evaluated by chi-squared or Fisher’s exact chi-squared test.
Silencing lncRNA-SNHG6 inhibits the proliferation, colony formation, and metastasis of CRC cells in vitro and in vivo
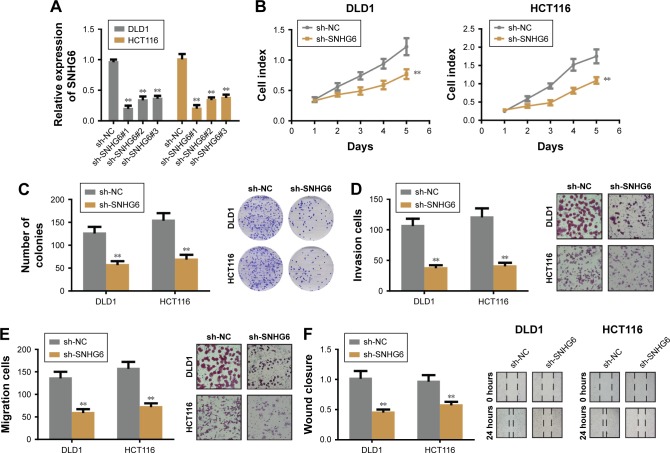
To identify the role of SNHG6 on biological functions of CRC we established SNHG6 stable knockdown cell lines by using three different shRNA and chose the best knockdown efficiency for the following experiments (Figure 2A). CCK-8 assay showed that SNHG6 knockdown significantly inhibited cell growth in HCT116 and DLD1 cells (Figure 2B). Colony formation assay also revealed that SNHG6 silencing attenuated the clonogenic ability of CRC cells compared with negative control (Figure 2C). Next, invasion and migration transwell assays were performed to assess the metastatic ability in CRC cells. As shown in Figure 2D and E, the number of invading cells in the sh-SNHG6 group was greatly reduced compared with that in the control group. Additionally, wound-healing assay showed that the migration abilities of HCT116 and DLD1 cells were obviously decreased by SNHG6 repression (Figure 2F). Conversely, overexpression with SNHG6 promoted the proliferation, colony formation, invasion and migration in LOVO cells (data not shown). To investigate whether SNHG6 inhibited CRC tumorigenic ability in vivo, we subcutaneously injected HCT116-SNHG6 knockdown cells on the dorsal flank of mice. Consistent with the findings in vitro, SNHG6 knockdown remarkably impaired the tumorigenic ability of CRC cells in vivo (Figure 3A). As a result, tumors in the SNHG6 knockdown group showed a lower weight and smaller volume (Figure 3B and C). Next, we used the mice liver metastasis model of CRC by injecting the cells through the splenic vein. Compared with the control group, the metastatic nodule numbers and size were dramatically decreased in the SNHG6 knockdown group (Figure 3D–F). Together, these results demonstrated that SNHG6 knockdown suppressed tumor growth and liver metastasis of CRC both in vitro and in vivo.
Figure 2.
Knockdown of SNHG6 impaired CRC proliferation, invasion and migration in vitro.
Notes: (A) Knockdown of SNHG6 in DLD1 and HCT116 cells by three different shRNA. (B) CCK-8 assays showed that SNHG6 knockdown inhibited cell proliferation in DLD1 and HCT116 cells. (C) Colony formation assay showed that SNHG6 knockdown reduced the number of colonies in DLD1 and HCT116 cells. (D and E) SNHG6 knockdown attenuated cell invasion and migration of DLD1 and HCT116 cells. (F) Wound healing assay indicated that SNHG6 knockdown repressed the migration of DLD1 and HCT116 cells. **P<0.01.
Abbreviations: CRC, colorectal cancer; CCK-8, Cell Counting Kit-8.
Figure 3.
Knockdown of SNHG6 impaired CRC proliferation, invasion and migration in vivo.
Notes: (A) Representative image of xenograft using SNHG6 knockdown and control cells. (B and C) SNHG6 knockdown decreased tumor volume and tumor weight of HCT116 cells. (D–F) Representative bioluminescence images and morphology of liver metastasis in SNHG6 knockdown and control cells. **P<0.01.
Abbreviation: CRC, colorectal cancer.
lncRNA-SNHG6 interacts with miR-760 to regulate CRC cell proliferation, invasion and migration
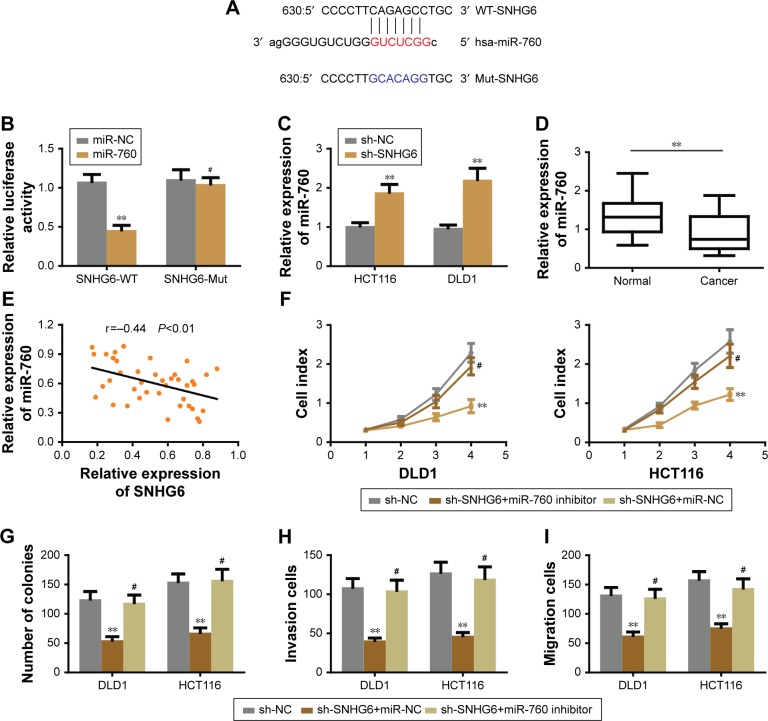
Previous studies have suggested that specific lncRNAs could function as competing endogenous RNAs (ceRNA) by spongactive miRNAs.13 To determine whether SNHG6 could act as a ceRNA in the progression of CRC, we used online bioinformatics software (starbase v2.0 and RegRNA 2.0) to predict the potential lncRNA-miRNA interactions. Based on the prediction, we found that miR-760 was a potential target of SNHG6. Further sequence alignment confirmed that there was a binding site between miR-760 and SNHG6 (Figure 4A). To validate the prediction, we performed the dual luciferase reporter assay to assess the potential binding. As shown in Figure 4B, cells transfected with miR-760 mimic significantly reduced the luciferase activity of WT-SNHG6 expressing cells, while they failed to affect Mut-SNHG6 expressing cells. Then, we proceeded to measure the miR-760 level in CRC cells with SNHG6 silencing. The result showed that, miR-760 was significantly increased after SNHG6 knockdown (Figure 4C). In addition, we checked the expression of miR-760 in the CRC samples and adjacent normal tissues. As shown in Figure 4D, miR-760 was largely decreased in CRC samples compared with normal controls. Furthermore, we analyzed the correlation between miR-760 and SNHG6 by Pearson’s correlation analysis. The result showed that the expression of SNHG6 negatively correlated with miR-760 level in CRC tissues (Figure 4E). Collectively, the above results strongly indicate that SNHG6 directly interacts with miR-760. Next, we sought to determine whether SNHG6-mediated CRC cell proliferation, invasion and migration was miR-760 dependent. We performed the rescue experiments by transfecting miR-760 inhibitor into SNHG6 knockdown cells. The results showed that repression of miR-760 abolished the inhibitory effects of SNHG6 knockdown on CRC cell proliferation (Figure 4F and G), invasion (Figure 4H) and migration (Figure 4I). To sum up, SNHG6 modulates the progression of CRC cells by sponging miR-760.
Figure 4.
SNHG6 directly interacted with miR-760 to regulate CRC cell proliferation, invasion and migration.
Notes: (A) Bioinformatics analysis of predicted binding sites between SNHG6 and miR-760. (B) Luciferase reporter assay demonstrated that miR-760 mimics significantly decreased the relative luciferase activity of WT-SNHG6 but not Mut-SNHG6. (C) SNHG6 knockdown increased the relative expression of miR-760 in DLD1 and HCT116 cells. (D) The relative expression level of miR-760 in CRC tissues and adjacent normal tissues. (E) Pearson’s correlation analysis of the expression between SNHG6 and miR-760 in CRC tissues. miR-760 inhibition rescued SNHG6 silencing induced cell proliferation (F), colony formation (G), invasion (H) and migration (I) inhibition in HCT116 cells. **P<0.01; #Non-significant.
Abbreviations: CRC, colorectal cancer; Mut, mutant; WT, wild type; NC, negative control.
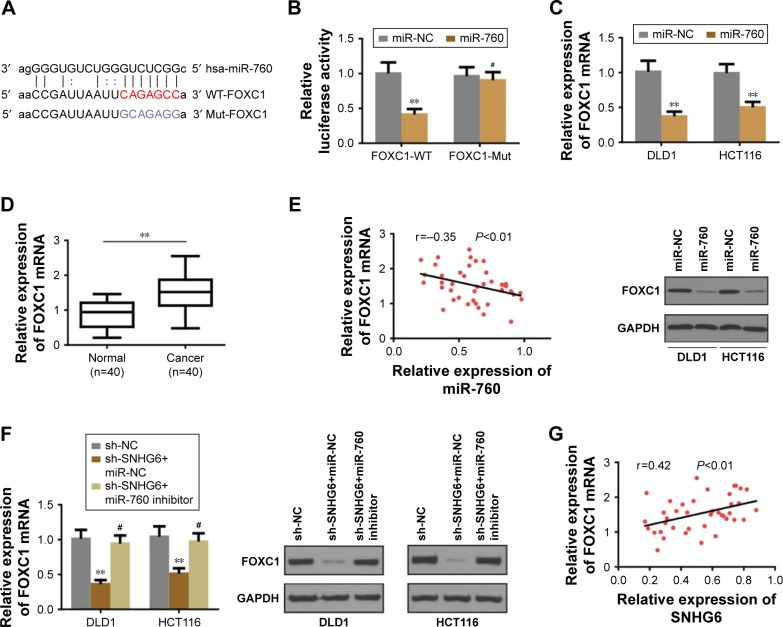
lncRNA-SNHG6 positively regulated FOXC1 expression through inhibiting miR-760
Since miRNAs exert the regulatory effect by directly targeting the 3″-UTR of the downstream target genes we used different online algorithms (TargetScan, miRDB and miRanda) to predict the target gene of miR-760, and found that FOXC1 was a potential target of miR-760 (Figure 5A). To confirm the speculation, the dual luciferase assay was carried out to examine the luciferase activity between miR-760 and FOXC1. As shown in Figure 5B, miR-760 mimics significantly reduced the luciferase activity of WT-FOXC1 3″-UTR but not in the mutated one. Moreover, miR-760 overexpression markedly repressed the protein and mRNA level of FOXC1 (Figure 5C). Conversely, miR-760 inhibition promoted the expression of FOXC1 in protein and mRNA level (data not shown). Further analysis of FOXC1 expression in CRC tissues revealed that FOXC1 was markedly upregulated in CRC tissues (Figure 5D). Pearson’s correlation analysis indicated that miR-760 expression was negatively associated with FOXC1 (Figure 5E). Thus, we concluded that FOXC1 is a direct target of miR-760 in CRC cells. Thereafter, we sought to investigate whether SNHG6 regulated FOXC1 in CRC cells. As expected, the protein and RNA expression of FOXC1 was significantly decreased under SNHG6 knockdown, while miR-760 inhibition could rescue this repressive effect (Figure 5F). Furthermore, Pearson’s correlation analysis indicated that SNHG6 expression was positively associated with FOXC1 in CRC tissues (Figure 5G). Taken together, our data suggest that SNHG6 facilitates the expression of FOXC1 by negatively modulating miR-760.
Figure 5.
FOXC1 was a direct target of miR-760 and positively regulated by SNHG6.
Notes: (A) Bioinformatics analysis of predicted binding sites between miR-760 and FOXC1. (B) Luciferase reporter assay indicated that miR-760 mimics significantly reduced the luciferase activity of FOXC1 in HEK293T cells. (C) MiR-760 overexpression inhibited the mRNA and protein expression levels of FOXC1 in DLD1 and HCT116 cells. (D) The relative expression levels of miR-760 mRNA in CRC tissues. (E) Pearson’s correlation analysis of the expression between miR-760 and FOXC1 in CRC tissues. (F) SNHG6 knockdown inhibited the mRNA and protein expression levels of FOXC1 and rescued by miR-760 inhibition in DLD1 and HCT116 cells. (G) Pearson’s correlation analysis of the expression between SNHG6 and FOXC1 in CRC tissues. **P<0.01; #Non-significant.
Abbreviations: CRC, colorectal cancer; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
Mounting evidence has revealed that lncRNAs could play a critical role in the progression of colorectal cancer.14 In the present study, we found that the expression of SNHG6 was increased in CRC tissues and cell lines compared with normal controls; silencing SNHG6 in CRC cells impaired tumor growth and liver metastasis both in vitro and in vivo. The oncogenic role of SNHG6 is due to its ability to directly sequester miR-760 and subsequently activate FOXC1. Furthermore, the correlation among SNHG6, miR-760 and FOXC1 expression in CRC tissues strengthens the above conclusion. Our findings have explored the potential application of SNHG6/miR-760/FOXC1 pathway in CRC treatment.
miRNAs are a group of small non-coding RNA molecules with 18–23 nucleotides that are involved in various biological processes by binding to the 3′UTRs of mRNA, thus exerting the regulatory effect.15 Based on the previous study, miRNAs could be sequestered by lncRNAs, thus releasing their target genes and functions in CRC. For example, lncRNA CASC2 acts as a ceRNA by sponging miR-18a to inhibit CRC progression.16 LncRNA UCA1 enhances chemoresistance of CRC by inhibiting miR-204–5 p.17 In the current study, we identified miR-760 as the binding non-coding RNA of SNHG6. The role of miR-760 in cancer is context dependent. Liao et al reported that miR-760 promoted ovarian cancer growth by inhibiting PHLPP2,18 while another report revealed that overexpression of miR-760 enhanced the sensitivity of chemotherapy and suppressed metastasis in breast cancer.19,20 In addition, a recent study showed that miR-760 could be a novel biomarker for early detection of CRC.21 However, the exact role and mechanism of miR-760 in CRC still remains ambiguous. In our study, we found that miR-760 was downregulated in CRC. miR-760 inhibition restored SNHG6 silencing induced CRC proliferation, invasion and migration repression. These results suggest that miR-760 functions as a tumor suppressor in CRC.
Long non-coding RNA SNHG6 has been identified as a poor prognostic predictor which is upregulated in colorectal cancer.22 However, no detailed experiments have been performed to elucidate the mechanism with which SNHG6 regulates CRC progression. In this study, we confirmed the upregulation of SNHG6 in CRC tissues and cell lines, which was consistent with previous studies. Meanwhile, we performed functional experiments to show that SNHG6 could promote CRC cell growth, migration and invasion both in vivo and in vitro. Additionally, we found that SNHG6 acted its function by sponging miR-760 and activating FOXC1. To some extent, this mechanism could explain the process of how SNHG6 regulates CRC progression. Nevertheless, long non-coding RNA could play crucial roles in many different ways. For example, some lncRNAs can associate with specific proteins such as transcription factors or RNA binding proteins, and subsequently regulate the function of the proteins.23 Thus, the precise regulatory mechanism that mediates the function of SNHG6 in CRC remains to be further explored.
FOXC1 is one of the members of the forkhead box family of transcription factors.24 Initially, FOXC1 was identified as a crucial transcription factor in the development of organs derived from the neural crest.25 In recent years, FOXC1 has been reported in the regulation of various cancers. For example, FOXC1 was reported to promote metastasis on breast cancer, hepatocellular carcinoma and endometrial cancer.26–28 Overexpression of FOXC1 was associated with poor prognosis of gastric cancer and pancreatic cancer.29,30 However, the role of FOXC1 in CRC is still unclear. Here, we validated that FOXC1 was a direct target of miR-760 in CRC and was highly expressed in CRC tissues. Additional Pearson’s analysis revealed the inverse correlation between miR-760 and FOXC1. SNHG6 upregulated FOXC1 through repressing miR-760.
Conclusion
In summary, we demonstrated that SNHG6 was overexpressed in CRC tissues and cell lines, and associated with poor prognosis of CRC patients. Furthermore, knockdown of SNHG6 expression attenuated cell proliferation, invasion and migration by acting as a ceRNA of miR-760 to regulate FOXC1 expression in CRC cells. Our study highlights that SNHG6 may serve as a potential therapeutic strategy for CRC treatment in the future.
Acknowledgments
We acknowledge Dr Qingbin Ding for the help in the collection of clinical samples after surgery.
This study was supported by Natural Science Foundation of Heilongjiang Province (Grant No ZD201713).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967–976. doi: 10.2147/CIA.S109285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hugen N, van de Velde CJ, de Wilt JH, Nagtegaal ID. Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol. 2014;25(3):651–657. doi: 10.1093/annonc/mdt591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244(2):254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung U, Gönen M, Allen PJ, et al. Colorectal cancer liver metastases and concurrent extrahepatic disease treated with resection. Ann Surg. 2017;265(1):158–165. doi: 10.1097/SLA.0000000000001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X, Sun M, Liu H, Yao Y, Song Y. Long-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Chang L, Yuan Y, Li C, et al. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 2016;383(2):183–194. doi: 10.1016/j.canlet.2016.09.034. [DOI] [PubMed] [Google Scholar]
- 11.Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is Associated with Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation and EMT through Epigenetically Silencing p27 and Sponging miR-101-3p. Cell Physiol Biochem. 2017;42(3):999–1012. doi: 10.1159/000478682. [DOI] [PubMed] [Google Scholar]
- 12.Xue W, Li J, Wang F, Han P, Liu Y, Cui B. A long non-coding RNA expression signature to predict survival of patients with colon adenocarcinoma. Oncotarget. 2017;8(60):101298–101308. doi: 10.18632/oncotarget.21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361(1):13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 16.Huang G, Wu X, Li S, Xu X, Zhu H, Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci Rep. 2016;6:26524. doi: 10.1038/srep26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bian Z, Jin L, Zhang J, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao Y, Deng Y, Liu J, et al. MiR-760 overexpression promotes proliferation in ovarian cancer by downregulation of PHLPP2 expression. Gynecol Oncol. 2016;143(3):655–663. doi: 10.1016/j.ygyno.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Hu SH, Wang CH, Huang ZJ, et al. miR-760 mediates chemoresistance through inhibition of epithelial mesenchymal transition in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20(23):5002–5008. [PubMed] [Google Scholar]
- 20.Han ML, Wang F, Gu YT, Yt G, et al. MicroR-760 suppresses cancer stem cell subpopulation and breast cancer cell proliferation and metastasis: By down-regulating NANOG. Biomed Pharmacother. 2016;80:304–310. doi: 10.1016/j.biopha.2016.03.024. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Huang Z, Ni S, et al. Plasma miR-601 and miR-760 are Novel Biomarkers for the Early Detection of Colorectal Cancer. PLoS One. 2012;7(9):e44398. doi: 10.1371/journal.pone.0044398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Bian Z, Yao S, et al. Up-regulated expression of SNHG6 predicts poor prognosis in colorectal cancer. Pathol Res Pract. 2018;214(5):784–789. doi: 10.1016/j.prp.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Huang Z, Ni S, et al. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One. 2012;7(9):e44398. doi: 10.1371/journal.pone.0044398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 25.Kidson SH, Kume T, Deng K, Winfrey V, Hogan BL. The forkhead/winged-helix gene, Mf1, is necessary for the normal development of the cornea and formation of the anterior chamber in the mouse eye. Dev Biol. 1999;211(2):306–322. doi: 10.1006/dbio.1999.9314. [DOI] [PubMed] [Google Scholar]
- 26.Jin Y, Han B, Chen J, et al. FOXC1 is a critical mediator of EGFR function in human basal-like breast cancer. Ann Surg Oncol. 2014;21(Suppl 4):758–766. doi: 10.1245/s10434-014-3980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia L, Huang W, Tian D, et al. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2013;57(2):610–624. doi: 10.1002/hep.26029. [DOI] [PubMed] [Google Scholar]
- 28.Chung TK, Lau TS, Cheung TH, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130(5):1036–1045. doi: 10.1002/ijc.26060. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Shao QS, Yao HB, Jin Y, Ma YY, Jia LH. Overexpression of FOXC1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2014;64(7):963–970. doi: 10.1111/his.12347. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Gu F, Liu CY, Wang RJ, Li J, Xu JY, Jy X. High level of FOXC1 expression is associated with poor prognosis in pancreatic ductal adenocarcinoma. Tumour Biol. 2013;34(2):853–858. doi: 10.1007/s13277-012-0617-7. [DOI] [PubMed] [Google Scholar]