Abstract
Purpose
Vitamin D receptor (VDR) signaling pathway is implicated in the pathogenesis of breast cancer.
Patients and methods
We selected VDR-associated long noncoding RNAs (lncRNAs) through an in silico analysis of available microarray and RNA-sequencing data and assessed their expression in 75 breast tumor samples and their adjacent noncancerous tissues (ANCTs). We also genotyped two functional polymorphisms within VDR gene in all patients.
Results
VDR, MALAT1, and LINC00511 were significantly upregulated in tumoral tissues compared with ANCTs (fold change [FC] =1.85, P=0.03; FC =1.54, P=0.04; and FC =4.75, P=0.000, respectively). In patients younger than 55 years, significant associations were found between expression levels of both SNHG16 and LINC00511 genes and nuclear grade (P=0.03), expression of LINC00346 and tubule formation (P=0.01), expression of both SNHG16 and SNHG6 genes and family history of cancer (P=0.01 and 0.03, respectively), as well as expression of VDR and progesterone receptor status (P=0.03). We detected significant correlations between expression levels of VDR and SNHG16 in both tumoral tissues and ANCTs. The TT genotype of FokI polymorphism was associated with the higher expression levels of VDR. FokI variants were associated with expression levels of both MALAT1 and SNHG16 in ANCTs (P=0.01 and 0.03, respectively). CdxII variants were associated with expression levels of SNHG16 in ANCTs. A significant correlation was found between FC values of SNHG16 expression and vitamin D levels.
Conclusion
The present study provides further evidence for the contribution of VDR signaling and the related lncRNAs in the pathogenesis of breast cancer and introduces some novel lncRNAs as putative molecules in the interactive functional network of VDR signaling in breast cancer.
Keywords: breast cancer, MALAT1, SNHG16, SNHG6, LINC00346, LINC00511, VDR
Introduction
Breast cancer is a prevalent cancer among women with a huge disease burden, which has been attributed to the lack of suitable biomarkers for the early detection of disease.1 Despite several efforts to find diagnostic and prognostic biomarkers for breast cancer,2–4 the clinical relevance of few biomarkers has been confirmed.5 Vitamin D receptor (VDR) signaling pathway has been regarded as a putative biomarker and therapeutic target in breast cancer.6 Vitamin D has potent antiproliferative, prodifferentiation, and proapoptotic functions in several tissues. Both autocrine VDR signaling and paracrine VDR signaling are important determinants of cell fate in diverse physiological and pathological contexts. In vitro and in vivo studies have provided evidence for the participation of VDR signaling in either chemoprevention or chemotherapy of breast cancer. In contrast, several genetic and epigenetic aberrations have been associated with resistance to VDR functions in cancers.7 VDR binding to calcitriol leads to its translocation into the nucleus and the subsequent interactions with the vitamin D response element (VDRE), which results in the stimulation of the transcription of targeted genes among which are those involved in the carcinogenesis process.2 VDR expression has been documented in several breast cancer cell lines5,6 as well as clinical samples.8,9 Moreover, an association has been found between low VDR expression in breast tumor tissues and aggressive tumor features such as large tumor size, hormone receptor negativity, and high Ki-67 expression.9 Such studies have highlighted the role of VDR signaling in breast cancer. In contrast, VDREs have been detected in noncoding regions of the genome including those encompass the long noncoding RNA (lncRNA) genes.10 These mRNA-like transcripts are involved in several carcinogenesis-associated cellular processes11–13 and are abnormally expressed in breast tumors.4,14,15 Besides, several lncRNAs have been shown to be regulated by VDR signaling in the context of skin cancer.10 Taken together, we hypothesized that VDR expression might be associated with expression levels of certain lncRNAs and clinicopathological features of breast tumors. Consequently, we selected a number of VDR-associated lncRNAs through an in silico analysis of available microarray and RNA-sequencing data and assessed their expression in breast tumor samples compared with the adjacent noncancerous tissues (ANCTs). We also evaluated the associations between expression levels of these genes and tumor features such as stage and grade. In addition, we have genotyped two functional polymorphisms within VDR gene (FokI [rs2228570] and CdxII [rs11568820]) in all patients to evaluate their effects on the expression of VDR in breast samples.
Patients and methods
Patients
This study was approved by the ethical committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.MSP. REC.1396.804). All methods were performed in accordance with the relevant guidelines and regulations. Informed written consent was obtained from all patients. Seventy-five patients with definite diagnosis of invasive ductal carcinoma of breast who were referred to Sina and Farmanieh hospitals entered the study. Tumoral and ANCTs were excised during surgery and transferred in liquid nitrogen to the genetic laboratory for further assessments. Clinicopathological data of patients were collected through questionnaires and the assessment of medical records.
In silico analysis
We used the following online tools for the identification of lncRNAs, which interact with VDR and have functional significance in breast cancer: 1) LncRNA2Target tool (http://www.lncrna2target.org) for the detection of lncRNAs that function the upstream of VDR, 2) cBioPortal for Cancer Genomics (http://cbioportal.org), and 3) the Catalog of Somatic Mutations in Cancer (COSMIC) for the identification of lncRNAs that are targets of genomic alteration in breast cancer tissues.
SNPs genotyping
Genomic DNA was extracted from blood samples of patients using the standard salting out method as previously described.16 The FokI (rs2228570) and CdxII (rs11568820) polymorphisms within VDR gene were genotyped using polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) and tetra-primer amplification refractory mutation system–PCR (ARMS–PCR) techniques, respectively. Table 1 shows the nucleotide sequences of the primers used for SNPs’ genotyping. The PCR program included an initial denaturation at 95°C for 5 minutes followed by 30 cycles at 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds and a final extension step at 72°C for 5 minutes. For FokI (rs2228570) genotyping, the amplified PCR products were digested with FokI restriction endonuclease enzyme (Thermo Fisher Scientific, Waltham, MA, USA) for 15 minutes at 37°C followed by inactivation step at 65°C for 5 minutes.
Table 1.
The nucleotide sequences of primers used for SNPs genotyping
| Locus | Sequence 5′→3′ | PCR product length | Digestion products |
|---|---|---|---|
| FokI | Forward: GGTGGGTGGCACCAAGGAT | 365 bp | CC:336, 29 bp |
| Reverse: CTCCTGTGGCTGTGAGCG | CT: 336, 273, 63, 29 bp | ||
| TT: 273, 63, 29 bp | |||
| CdxII | G-forward: AGGATAGAGAAAATAATAGAAAACATT | Internal control: 297 bp | |
| G-reverse: AACCCATAATAAGAAATAAGTTTTTAC | A allele: 235 bp | ||
| A-forward: TCCTGAGTAAACTAGGTCACAA | G allele: 110 bp | ||
| A-reverse: ACGTTAAGTTCAGAAAGATTAATTC |
Abbreviation: PCR, polymerase chain reaction.
RNA extraction and quantitative real-time PCR
Expressions of MALAT1 (ENSG00000251562), SNHG16 (ENSG00000163597), SNHG6 (ENSG00000245910), LINC00346 (ENSG00000255874), LINC00511 (ENSG00000227036), and VDR (ENSG00000111424) were assessed in tumoral tissues and ANCTs using the quantitative real-time PCR technique. In brief, total RNA was extracted from all samples using the TRIzol™ Reagent (Thermo Fisher Scientific). RNA samples were treated by DNase (Thermo Fisher Scientific) before cDNA synthesis. Next, cDNA was synthesized by using the RevertAid First Strand cDNA Synthesis Kit (TaKaRa, Otsu, Japan). The relative expression of VDR and mentioned lncRNAs was measured using the SYBR® Premix Ex Taq™ (TaKaRa) and application of B2M expression levels as normalizer. The nucleotide sequences of primers used for expression analysis are shown in Table 2. All experiments were performed in duplicate in the rotor gene 6000 corbett Real-Time PCR System.
Table 2.
The nucleotide sequences of primers used for expression analysis
| Gene | Sequence 5′→3′ | PCR product (bp) |
|---|---|---|
| B2M | F: AGATGAGTATGCCTGCCGTG | 105 |
| R: GCGGCATCTTCAAACCTCCA | ||
| VDR | F: GCCTTTGGGTCTGAAGTGTCT | 94 |
| R: CCATTGCCTCCATCCCTGAA | ||
| MALAT1 | F: GACGGAGGTTGAGATGAAGC | 84 |
| R: ATTCGGGGCTCTGTAGTCCT | ||
| SNHG16 | F: GTCAGCCTCAGTTTCCAAAGC | 104 |
| R: TAAAGACATGGCACTTTGGGTC | ||
| SNHG6 | F: AGGGAGGAAGAAGCGCGAA | 85 |
| R: TCGCAGAGCCCAGCTACG | ||
| LINC00511 | F: TCCCACCAGGAAGTTTAGCAG | 87 |
| R: GCCTCTCAAGAGGTGGTCC | ||
| LINC00346 | F: TGCCCTGGACATTCATGGAC | 150 |
| R: CTGGACAAGCCCACTCTAGC |
Abbreviation: PCR, polymerase chain reaction.
Statistical analysis
Data were presented as mean ± SD. Student’s paired and unpaired t-tests were used for the analysis of differences in gene expression between paired and unpaired samples. The association between clinicopathological data and transcript levels of each gene was assessed using the chi-square test. The expression fold change (FC) was measured using the 2−ΔΔCt method. The pairwise correlation between vitamin D levels and expression of genes or between relative transcripts levels of genes was calculated using the regression model. For all statistical tests, the level of significance was set at P<0.05. We also depicted the receiver operating characteristic (ROC) curve to assess the suitability of gene expression levels for classifying disease status. In order to appraise the probability cutoff of gene expression, the Youden index (J) was applied to maximize the difference between sensitivity (true-positive rate) and 1- specificity (false-positive rate). The precision of each marker for the diagnosis of breast cancer was scored according to the area under curve (AUC) values using the following system: 0.90–1= excellent (A), 0.80–0.90= good (B), 0.70–0.80= fair (C), 0.60–0.70= poor (D), and 0.50–0.60= fail (F).
Results
General demographic data of patients
General demographic data of patients are shown in Table 3.
Table 3.
General demographic data of study participants
| Variables | Values |
|---|---|
| Age (years), mean ± SD (range) | 51.53±12.83 (29–83) |
| Menarche age, mean ± SD (range) | 13.1±1.56 (10–18) |
| Menopause age, mean ± SD (range) | 44.62±14.89 (38–60) |
| First pregnancy age, mean ± SD (range) | 18.97±8.26 (14–34) |
| Breast feeding duration (months), mean ± | 41.48±40.47 (3–240) |
| SD (range) | |
| Serum level of 25-hydroxyvitamin D, mean ± | 22.85±13.98 (6.3–55.8) |
| SD (range) | |
| Positive family history for cancer (%) | 18.7 |
| Cancer stage (%) | |
| I | 25.3 |
| II | 31 |
| III | 33.8 |
| IV | 9.9 |
| Overall grade (%) | |
| I | 16.7 |
| II | 53 |
| III | 30.3 |
| Nuclear grade (%) | |
| I | 10.3 |
| II | 58.8 |
| III | 30.9 |
| Tubule formation (%) | |
| I | 6.5 |
| II | 33.9 |
| III | 59.6 |
| Mitotic rate (%) | |
| I | 40.9 |
| II | 42.9 |
| III | 16.2 |
| Tumor size (%) | |
| <2 cm | 26.1 |
| ≥2 to <5 cm | 71 |
| ≥5 cm | 2.9 |
| ER (%) | |
| Positive | 84.1 |
| Negative | 15.9 |
| PR (%) | |
| Positive | 77.9 |
| Negative | 22.1 |
| Her2/neu expression (%) | |
| Positive | 23.2 |
| Negative | 76.8 |
| Ki67 expression (%) | |
| Positive | 100 |
| Negative | 0 |
Genotyping
Genotyping of CdxII and FokI polymorphisms in breast cancer patients showed AA and CC genotypes as the most prevalent ones among patients, respectively. Table 4 shows the frequency of CdxII and FokI genotypes in breast cancer patients.
Table 4.
Frequency of CdxII and FokI genotypes in breast cancer patients
| CdxII genotypes | Frequency, n (%) | FokI genotypes | Frequency, n (%) |
|---|---|---|---|
| AA | 29 (38.7) | CC | 46 (61.4) |
| AG | 22 (29.3) | CT | 25 (33.3) |
| GG | 24 (32) | TT | 4 (5.3) |
Relative expression of VDR and lncRNAs in tumoral tissues vs ANCTs
VDR, MALAT1, and LINC00511 were significantly upregulated in tumoral tissues compared with ANCTs (FC =1.85, P=0.03; FC =1.54, P=0.04; and FC =4.75, P=0.000, respectively). For MALAT1, such upregulation was more prominent in patients elder than 55 years (FC =2.49, P=0.002). The expression levels of other lncRNAs were not significantly different between tumoral tissues and ANCTs.
Association between relative expression of genes and patients’ clinicopathological data
No significant association was found between expression levels of genes and clinicopathological data of patients except higher expression levels of SNHG16 in patients with positive family history of cancer (any cancer rather than breast cancer) (P=0.01). Then, we assessed associations between these clinicopathological data and relative expression of genes in patients younger than 55 years. In this subgroup of patients, significant associations were found between SNHG16 expression and nuclear grade (P=0.03), LINC00511 expression and nuclear grade (P=0.03), and LINC00346 expression and tubule formation (P=0.01). Higher levels of both SNHG16 and LINC00511 were more frequently detected in tumors with nuclear grade 2. Moreover, higher expression of LINC00511 was more commonly seen in tumors with tubule formation score 2. Besides, higher expression levels of both SNHG16 and SNHG6 genes were associated with the positive family history of cancer (P=0.01 and 0.03, respectively). In addition, a significant positive association was found between the relative expression of VDR and PR status (P=0.03). Table 5 shows the associations between the relative expression of these genes and clinicopathological data in patients younger than 55 years.
Table 5.
The associations between relative expression of these genes and clinicopathological data in patients younger than 55 years
| Clinicopathological Features | VDR up- regulation, n (%) | VDR down- regulation, n (%) | P-value | MALAT1 up- regulation, n (%) | MALAT1 down- regulation, n (%) | P-value | SNHG16 up- regulation, n (%) | SNHG16 down- regulation, n (%) | P-value | SNHG6 up- regulation, n (%) | SNHG6 down- regulation, n (%) | P-value | LINC00511 up- regulation, n (%) | LINC00511 down- regulation, n (%) | P-value | LINC00346 up- regulation, n (%) | LINC00346 down- regulation, n (%) | P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stage | 0.94 | 0.73 | 0.67 | 0.14 | 0.78 | 0.31 | ||||||||||||
| 1 | 5 (50) | 5 (50) | 5 (50) | 5 (50) | 4 (40) | 6 (60) | 1 (14.3) | 6 (85.7) | 7 (70) | 3 (30) | 5 (50) | 5 (50) | ||||||
| 2 | 8 (61.5) | 5 (38.5) | 5 (33.3) | 10 (66.7) | 7 (46.7) | 8 (53.3) | 9 (69.2) | 4 (30.8) | 9 (69.2) | 4 (30.8) | 8 (66.7) | 4 (33.3) | ||||||
| 3 | 9 (64.3) | 5 (35.7) | 8 (53.3) | 7 (46.7) | 9 (64.3) | 5 (35.7) | 8 (53.3) | 7 (46.7) | 12 (80) | 3 (20) | 7 (50) | 7 (50) | ||||||
| 4 | 3 (60) | 2 (40) | 2 (40) | 3 (60) | 2 (40) | 3 (60) | 2 (40) | 3 (60) | 3 (60) | 2 (40) | 4 (100) | 0 (0) | ||||||
| Histological grade | 0.19 | 0.1 | 0.24 | 0.62 | 0.55 | |||||||||||||
| 1 | 5 (71.4) | 2 (28.6) | 6 (75) | 2 (25) | 5 (62.5) | 3 (37.5) | 5 (71.4) | 2 (28.6) | 5 (62.5) | 3 (37.5) | 5 (71.4) | 2 (28.6) | ||||||
| 2 | 14 (70) | 6 (30) | 9 (40.9) | 13 (59.1) | 12 (57.1) | 9 (42.9) | 10 (47.6) | 11 (52.4) | 17 (81) | 4 (19) | 13 (65) | 7 (35) | ||||||
| 3 | 4 (36.4) | 7 (63.6) | 3 (27.3) | 8 (72.7) | 3 (27.3) | 8 (72.7) | 5 (45.5) | 6 (54.5) | 5 (50) | 7 (54.5) | 5 (45.5) | 7 (54.5) | ||||||
| Nuclear grade | 0.31 | 0.14 | 0.03 | 0.79 | 0.03 | 0.27 | ||||||||||||
| 1 | 2 (40) | 3 (60) | 5 (83.3) | 1 (16.7) | 3 (50) | 3 (50) | 2 (50) | 2 (50) | 3 (50) | 3 (50) | 4 (80) | 1 (20) | ||||||
| 2 | 17 (70.8) | 7 (29.2) | 12 (46.2) | 14 (53.8) | 17 (68) | 8 (32.2) | 14 (56) | 11 (44) | 21 (84) | 4 (16.2) | 16 (66.7) | 8 (33.3) | ||||||
| 3 | 5 (50) | 5 (50) | 3 (30) | 7 (70) | 2 (20) | 8 (80) | 4 (40) | 6 (60) | 4 (44.4) | 5 (55.6) | 4 (40) | 6 (60) | ||||||
| Tubule formation | 0.09 | 0.18 | 0.32 | 0.42 | 1 | 0.01 | ||||||||||||
| 1 | 1 (100) | 0 (0) | 2 (100) | 0 (0) | 1 (50) | 1 (50) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 1 (50) | 1 (50) | ||||||
| 2 | 10 (83.3) | 2 (16.7) | 7 (50) | 7 (50) | 9 (69.2) | 4 (30.8) | 8 (61.5) | 5 (38.5) | 9 (69.2) | 4 (30.8) | 11 (97.7) | 1 (8.3) | ||||||
| 3 | 10 (47.6) | 11 (52.4) | 7 (33.3) | 14 (66.7) | 9 (42.9) | 12 (57.1) | 9 (45) | 11 (55) | 13 (65) | 7 (35) | 9 (42.9) | 12 (57.1) | ||||||
| Mitotic rate | 0.24 | 0.16 | 0.43 | 0.18 | 0.1 | 1 | ||||||||||||
| 1 | 9 (81.8) | 2 (18.2) | 8 (66.7) | 4 (33.3) | 8 (66.7) | 4 (33.3) | 9 (75) | 3 (25) | 11 (91.7) | 1 (8.3) | 7 (58.3) | 5 (41.7) | ||||||
| 2 | 8 (53.3) | 7 (46.7) | 5 (29.4) | 12 (70.6) | 8 (50) | 8 (50) | 6 (40) | 9 (60) | 9 (56.3) | 7 (43.8) | 9 (60) | 6 (40) | ||||||
| 3 | 4 (50) | 4 (50) | 3 (37.5) | 5 (62.5) | 3 (37.5) | 5 (62.5) | 4 (50) | 4 (50) | 4 (57.1) | 3 (42.9) | 5 (62.5) | 3 (37.5) | ||||||
| Tumor size | 0.21 | 1 | 0.72 | 1 | 0.71 | 1 | ||||||||||||
| <2 | 5 (55.6) | 4 (44.4) | 5 (50) | 5 (50) | 6 (66.7) | 3 (33.3) | 4 (50) | 4 (50) | 8 (80) | 2 (20) | 6 (66.7) | 3 (33.3) | ||||||
| 2–5 | 19 (65.5) | 10 (34.5) | 14 (45.2) | 17 (54.8) | 15 (45.2) | 16 (51.6) | 15 (53.6) | 13 (46.4) | 20 (69) | 9 (31) | 17 (63) | 10 (37) | ||||||
| >5 | 0 (0) | 2 (100) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | ||||||
| Family history | 0.32 | 0.05 | 0.01 | 0.03 | 1 | 0.44 | ||||||||||||
| + | 5 (55.6) | 4 (44.4) | 7 (77.8) | 2 (22.2) | 8 (88.9) | 1 (11.1) | 6 (85.7) | 1 (14.3) | 7 (77.8) | 2 (22.2) | 7 (77.8) | 2 (22.2) | ||||||
| − | 19 (65.5) | 10 (34.5) | 14 (37.8) | 23 (62.2) | 14 (39.1) | 22 (61.1) | 14 (41.2) | 20 (58.8) | 25 (71.4) | 10 (28.6) | 18 (56.3) | 14 (43.8) | ||||||
| ER status | 0.24 | 1 | 0.28 | 1 | 1 | |||||||||||||
| + | 21 (63.6) | 12 (36.4) | 16 (45.7) | 19 (54.3) | 19 (55.9) | 15 (44.1) | 16 (51.6) | 15 (48.4) | 1 | 25 (71.4) | 10 (28.6) | 19 (61.3) | 12 (38.7) | |||||
| − | 3 (37.5) | 5 (62.5) | 4 (44.4) | 5 (55.6) | 3 (33.3) | 6 (66.7) | 5 (55.6) | 4 (44.4) | 5 (71.4) | 2 (28.6) | 6 (66.7) | 3 (33.3) | ||||||
| PR status | 0.03 | 0.73 | 0.28 | 0.85 | 0.69 | 1 | ||||||||||||
| + | 22 (66.7) | 11 (33.3) | 16 (47.1) | 18 (52.9) | 19 (55.9) | 15 (44.1) | 16 (5.3) | 14 (46.7) | 24 (72.7) | 9 (27.3) | 19 (61.3) | 12 (38.7) | ||||||
| − | 2 (25) | 6 (75) | 4 (40) | 6 (60) | 3 (33.3) | 6 (66.7) | 5 (50) | 5 (50) | 6 (66.7) | 3 (33.3) | 6 (66.7) | 3 (33.3) | ||||||
| Her2 status | 0.27 | 0.17 | 1 | 0.29 | 0.42 | 0.52 | ||||||||||||
| + | 4 (40) | 6 (60) | 3 (27.3) | 8 (72.7) | 5 (45.5) | 6 (54.5) | 4 (36.4) | 7 (63.6) | 6 (60) | 4 (40) | 6 (54.4) | 5 (45.5) | ||||||
| − | 20 (64.5) | 11 (35.5) | 18 (52.9) | 16 (47.1) | 17 (51.5) | 16 (48.5) | 17 (5.6) | 12 (41.4) | 25 (75.8) | 8 (24.2) | 19 (65.5) | 10 (34.5) |
Notes: The REx of each gene in the tumoral tissue compared with the ANCT of the same patient was assessed. REx values >1 were described as “upregulation”, while REx values ≤1 pointed to “downregulation”. TNM staging was performed based on the AJCC staging guidelines (seventh edition).17 Histological grade was calculated by the combination of scores obtained from nuclear grade, mitotic rate, and tubule formation. Nuclear grade scores were based on the appearance of the nucleus of the cancer cells, with one being the nearest to normal cells and three being the most deviation. Mitotic rate scores 1 and 3 were given to tumors with the slowest and the most rapid rate of mitosis. Tubule formation score represents the percentage of cancer cells that are in tubule formation. Tumor size refers to the greatest dimension of tumor in centimeters.
Abbreviations: AJCC, American Joint Committee on Cancer; ANCT, adjacent noncancerous tissue; REx, relative expression.
Correlation between relative expressions of genes
We assessed correlations between expression levels of genes in both tumoral tissues and ANCTs. The most significant correlations were found between expression levels of VDR and SNHG16 in both tumoral tissues and ANCTs (Table 6).
Table 6.
Coefficients of determination (R2) values between expression levels of genes in both tumoral tissues and ANCTs
| LINC00346 | LINC00511 | SNHG6 | SNHG16 | Malat1 | |
|---|---|---|---|---|---|
| VDR | |||||
| Tumor | 0.06* | 0 | 0.05* | 0.18* | 0.07* |
| ANCT | 0.08* | 0.04 | 0.07 | 0.31* | 0.11* |
| MALAT1 | |||||
| Tumor | 0.03 | 0.08* | 0.01 | 0.01 | |
| ANCT | 0.07* | 0 | 0.03 | 0 | |
| SNHG16 | |||||
| Tumor | 0.04 | 0.06* | 0.08* | ||
| ANCT | 0.11* | 0.03 | 0.12* | ||
| SNHG6 | |||||
| Tumor | 0.05 | 0.12* | |||
| ANCT | 0.05 | 0.07* | |||
| LINC00511 | |||||
| Tumor | 0.05 | ||||
| ANCT | 0 |
Note:
Correlation is significant at P<0.05 level.
Abbreviation: ANCTs, adjacent noncancerous tissues.
Association between polymorphisms’ genotypes and VDR and lncRNAs; relative expressions
FokI polymorphism was associated with the expression of VDR in both tumoral tissues and ANCTs (P=0.01 and 0.000, respectively) (Table 7). The TT and CC genotypes of this polymorphism were associated with the higher and lower expression levels of VDR (Figure 1).
Table 7.
Association between relative expression (delta CT mean ± SD) of genes and VDR polymorphisms
| CdxII | FokI | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Alleles | Expression in ANCTs | P-value | Expression in tumoral tissues | P-value | Alleles | Expression in ANCTs | P-value | Expression in tumoral tissues | P-value |
| VDR | 0.87 | 0.46 | 0.000 | 0.01 | |||||
| AA | 11.01±3.31 | 9.25±2.57 | CC | 12.4±2.72 | 10.21±2.13 | ||||
| AG | 10.91±3.91 | 9.74±2.23 | CT | 8.41±2.05 | 9.29±2.92 | ||||
| GG | 10.53±2.53 | 10.15±2.89 | TT | 7.01±2.51 | 6.74±2.56 | ||||
| MALAT1 | 0.63 | 0.61 | 0.01 | 0.25 | |||||
| AA | –2.6±1.67 | –3.01±2.19 | CC | –2.1±1.81 | –3.37±2.47 | ||||
| AG | –2.87±1.95 | –3.68±2.92 | CT | –3.34±1.49 | –3.25±1.78 | ||||
| GG | –2.6±1.75 | –3.13±2.09 | TT | –2.24±1.6 | –1.3±3.69 | ||||
| SNHG16 | 0.03 | 0.89 | 0.03 | 0.59 | |||||
| AA | 4.61±1.46 | 4.25±2.61 | CC | 3.81±3.28 | 4.12±1.23 | ||||
| AG | 3.73±1.58 | 4.06±1.56 | CT | 3.69±1.51 | 4.5±3.05 | ||||
| GG | 3.62±1.27 | 4.34±1.68 | TT | 4.05±1.6 | 3.53±1.64 | ||||
| SNHG6 | 0.05 | 0.35 | 0.44 | 0.01 | |||||
| AA | 3.06±1.39 | 2.73±1.35 | CC | 3.07±1.52 | 2.85±1.27 | ||||
| AG | 3.45±1.56 | 3.46±2.45 | CT | 2.87±1.41 | 3.59±2.07 | ||||
| GG | 2.37±1.31 | 2.93±1.27 | TT | 2.12±0.85 | 1.17±2.28 | ||||
| LINC00511 | 0.29 | 0.56 | 0.1 | 0.29 | |||||
| AA | 16.24±5.63 | 12.49±4 | CC | 16.42±5.37 | 11.75±3.25 | ||||
| AG | 16.28±4.78 | 11.65±2.96 | CT | 14.65±5.93 | 12.96±3.66 | ||||
| GG | 13.99±6.2 | 12.68±2.82 | TT | 10.93±2.88 | 13.35±2.08 | ||||
| LINC00346 | 0.31 | 0.83 | 0.74 | 0.44 | |||||
| AA | 13.44±3.11 | 12.65±3.42 | CC | 13.75±3.3 | 11.86±3.85 | ||||
| AG | 14.51±3.95 | 12.08±3.21 | CT | 13.92±5.68 | 13.19±4.05 | ||||
| GG | 12.49±5.47 | 11.99±5.65 | TT | 13.55±2.12 | 12.78±8.45 | ||||
Abbreviation: ANCTs, adjacent noncancerous tissues.
Figure 1.
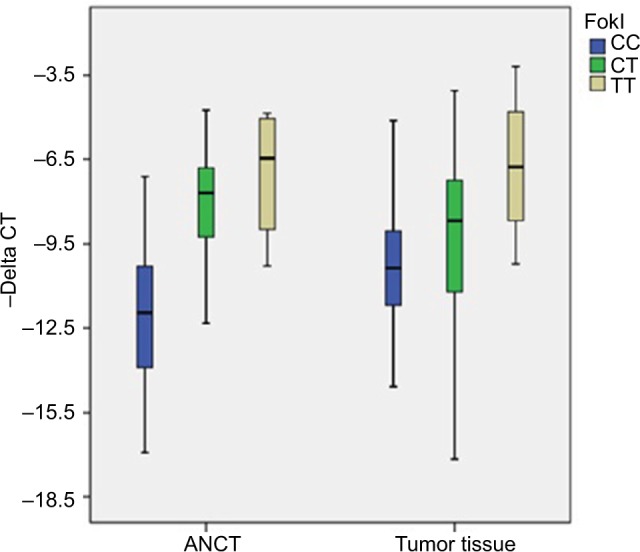
Relative expression of VDR (-delta CT values= CTB2M - CTVDR) in association with different genotypes of FokI polymorphism.
Abbreviations: ANCT, adjacent noncancerous tissue; CT, cycle threshold.
Moreover, FokI variants were associated with expression levels of both MALAT1 and SNHG16 in ANCTs (P=0.01 and 0.03, respectively) but not tumoral tissues. CdxII variants were associated with expression levels of SNHG16 in ANCTs (Table 7). Subsequent association analysis of CdxII genotypes with VDR expression in ER-based subgroups revealed no association in either ER-positive subtypes or ER-negative subtypes (data not shown).
Then, we assessed the associations between VDR polymorphisms and relative expressions of genes in tumoral tissues compared with the corresponding ANCTs and found significant over-representation of CC genotype in patients with the downregulation of both VDR and LINC00511 genes in tumoral tissues compared with ANCTs (Table 8). No association was found between CdxII genotypes and relative expression of genes.
Table 8.
Association between VDR polymorphisms and relative expressions of genes in tumoral tissues compared with the corresponding ANCTs
| CdxII
|
FokI
|
||||||
|---|---|---|---|---|---|---|---|
| Alleles | Upregulated, n (%) | Downregulated, n (%) | P-value | Alleles | Upregulated, n (%) | Downregulated, n (%) | P-value |
| VDR | 0.32 | 0.01 | |||||
| AA | 20 (69) | 9 (31) | CC | 31 (70.5) | 13 (29.5) | ||
| AG | 9 (47.4) | 10 (52.6) | CT | 8 (34.8) | 15 (65.2) | ||
| GG | 13 (59.1) | 9 (40.9) | TT | 3 (75) | 1 (25) | ||
| MALAT1 | 0.8 | 0.07 | 0.07 | ||||
| AA | 14 (48.3) | 15 (51.7) | CC | 29 (63) | 17 (37) | ||
| AG | 12 (57.1) | 9 (42.9) | CT | 9 (36) | 16 (64) | ||
| GG | 12 (52.2) | 11 (47.8) | TT | 2 (50) | 2 (50) | ||
| SNHG16 | 0.59 | 0.29 | |||||
| AA | 16 (55.2) | 13 (44.8) | CC | 24 (52.2) | 22 (47.8) | ||
| AG | 10 (47.6) | 11 (52.4) | CT | 9 (37.5) | 15 (62) | ||
| GG | 9 (40.9) | 13 (59.1) | TT | 2 (75) | 1 (25) | ||
| SNHG6 | 0.55 | 0.16 | |||||
| AA | 15 (55.6) | 12 (44.4) | CC | 23 (59) | 16 (41) | ||
| AG | 10 (52.6) | 9 (47.4) | CT | 8 (33.3) | 16 (66.7) | ||
| GG | 8 (40) | 12 (60) | TT | 2 (50) | 2 (50) | ||
| LINC00511 | 0.88 | 0.01 | |||||
| AA | 21 (72.4) | 8 (27.6) | CC | 36 (83.7) | 7 (16.3) | ||
| AG | 15 (75) | 5 (25) | CT | 15 (60) | 10 (40) | ||
| GG | 15 (68.2) | 7 (31.8) | TT | 1 (25) | 3 (75) | ||
| LINC00346 | 0.71 | 0.63 | |||||
| AA | 18 (64.3) | 10 (35.7) | CC | 27 (61.4) | 17 (38.6) | ||
| AG | 11 (55) | 9 (45) | CT | 15 (65.2) | 8 (34.8) | ||
| GG | 14 (66.7) | 7 (33.3) | TT | 1 (33.3) | 2 (66.7) | ||
Abbreviation: ANCTs, adjacent noncancerous tissues.
Assessment of correlations between serum levels of vitamin D and expression of genes
We assessed the correlations between vitamin D levels and expression of genes in tumoral tissues and ANCTs, as well as relative expression of genes in tumoral vs ANCTs (FC values). A significant correlation was found between FC values of SNHG16 expression and vitamin D levels (P=0.03). Table 9 shows the results of correlation analysis.
Table 9.
The correlations between vitamin D levels and expression of genes were assessed in tumoral tissues and ANCTs separately
| Genes | ANCT
|
Tumor tissue
|
Rex
|
|||
|---|---|---|---|---|---|---|
| R2 | P-value | R2 | P-value | R2 | P-value | |
| VDR | 0.067 | 0.31 | 0.068 | 0.31 | 0.008 | 0.73 |
| MALAT1 | 0.018 | 0.6 | 0.007 | 0.74 | 0.001 | 0.9 |
| SNHG16 | 0.118 | 0.17 | 0.011 | 0.68 | 0.263 | 0.03 |
| SNHG6 | 0.028 | 0.554 | 0.001 | 0.89 | 0.009 | 0.73 |
| LINC00511 | 0.004 | 0.82 | 0.001 | 0.92 | 0.025 | 0.55 |
| LINC00346 | 0.078 | 0.29 | 0.001 | 0.92 | 0.008 | 0.73 |
Notes: Moreover, we calculated REx of each gene in tumoral tissue compared with the ANCT of the same patient and assessed the correlation between REx value of each gene and vitamin D level in each patient.
Abbreviations: ANCTs, adjacent noncancerous tissues; REx, relative expression.
We also compared the serum levels of vitamin D in certain subgroups of patients based on the relative expression of each gene in tumoral tissues vs ANCTs and found no significant difference in vitamin D level between these groups. Figure 2 shows serum vitamin D levels in SNHG16 up-/downregulated and VDR up-/downregulated subjects.
Figure 2.
Serum vitamin D levels in SNHG16 up-/downregulated and VDR up-/downregulated subjects.
Notes: The REx of each gene in the tumoral tissue compared with the ANCT of the same patient was assessed. REx values >1 were described as “up-regulation” while REx values ≤1 pointed to “downregulation”.
Abbreviations: ANCT, adjacent noncancerous tissue; REx, relative expression.
ROC curve analysis
Based on ROC curve analysis results, the accuracy of LINC00511 expression levels for breast cancer diagnosis is fair (Figure 3). Besides, MALAT1 and VDR expression levels were the most specific and sensitive diagnostic markers for breast cancer among the assessed markers, respectively. Table 10 shows the details of ROC curve analysis.
Figure 3.
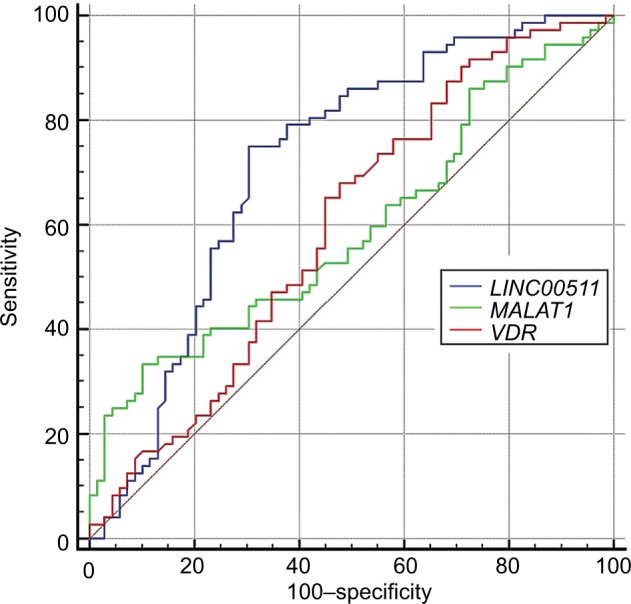
ROC curve for the assessment of LINC00511, MALAT1, and VDR expression levels as a diagnostic marker for breast cancer.
Abbreviation: ROC, receiver operating characteristic.
Table 10.
The results of ROC curve analysis
| Estimate criterion | AUC | Ja | Sensitivity | Specificity | P-valueb | |
|---|---|---|---|---|---|---|
| LINC00511 | >13.14 | 0.716 | 0.43 | 75 | 68.06 | <0.0001 |
| MALAT1 | ≤−4.33 | 0.596 | 0.226 | 32 | 90.67 | 0.03 |
| VDR | ≤12 | 0.597 | 0.19 | 87.84 | 31.94 | 0.04 |
Notes: Estimate criterion: optimal cutoff point for gene expression.
Youden index.
Significance level P (area =0.5).
Abbreviations: AUC, area under curve; ROC, receiver operating characteristic.
Discussion
In the present study, we evaluated expression levels of VDR and five lncRNAs with putative role in VDR signaling pathway in breast tumor samples vs ANCTs. VDR was significantly upregulated in breast tumor tissues compared with ANCTs. Its expression levels were associated with PR status in younger patients but not with other clinicopathological data or vitamin D levels. The observed association between VDR expression and PR status in this subgroup of patients is contrary to the results of Friedrich et al18 and Ditsch et al19 studies. Al-Azhri et al have reported the absence or low expression of VDR in 42% of breast tumors, moderate expression in 32% of breast tumors, and strong expression in the other 25% of samples using the immunohistochemistry (IHC) method. They found inverse association between VDR expression and more aggressive breast cancer behavior but no association between its expression and patients’ survival. Moreover, they detected no association between vitamin D levels and VDR expression.9 The latter finding is in accordance with our result regarding the lack of association between VDR expression in breast tissues and vitamin D levels. The discordance between our results and the mentioned studies can be explained by the difference in expression analysis method (quantitative real-time PCR vs IHC) and the assessment of relative expression in tumoral tissues vs ANCTs in our study but not in their studies.
We also demonstrated a significant overexpression of MALAT1 in tumoral tissues compared with ANCTs. Miao et al have demonstrated MALAT1 overexpression in 85.9% (67/78) of breast cancer tissues compared with ANCTs. They also have shown a significant association between MALAT1 overexpression and lymph metastasis.20 Moreover, Huang et al have reported MALAT1 overexpression in ER-positive breast cancer samples compared with ANCTs. In addition, they have shown associations between its transcript levels and positive ER/PR status.21 However, in the present study, we did not find any association between MALAT1 expression levels and patients’ clinicopathological data. Noticeably, MALAT1 upregulation was more prominent in elderly patients. Such data are in line with the previously reported upregulation of MALAT1 in chronic myelomonocytic leukemia (CMML) and its situation among the highest differentially expressed transcripts in CMML monocytes (CD14+)22 considering the facts that CMML is exceptionally rare in people younger than 50 years and its incidence sharply increases in people elder than 70 years.23 Although the exact mechanism for age-related upregulation of MALAT1 is not clarified, the previously reported higher frequency of somatic mutations in elderly breast cancer patients24 might be implicated in this phenomenon.
In addition, we detected a significant upregulation of LINC00511 in tumoral tissues compared with ANCTs. Besides, its expression was associated with nuclear grade in patients younger than 55 years. LINC00511 has been found to be a highly scored differentially expressed transcript in ER-negative MDA-MB-231 cells compared with the ER-positive T-47D and MCF-7 cells. Its expression has been upregulated in retinoid-related orphan receptor (ROR) γ-depleted MCF-7 and T-47D as well as aggressive basal-like breast cancer subtype.25 In contrast, RORγ function has been shown to be regulated by CYP11A1-derived vitamin D metabolites.26 Consequently, LINC00511 might provide the functional link between VDR signaling pathway and RORγ expression in breast cancer. However, such hypothesis should be evaluated in future studies. Moreover, next-generation sequencing of HER-2-enriched subtype breast cancer has shown LINC00511 as one of the core genes in the dysregulated pathways in this type of breast cancer including MAPK signaling pathway, PI3K–Akt signaling pathway, metabolic pathways, and those regulating cell cycle and actin cytoskeleton.27 Taken together, our results are in line with the previous data regarding the crucial role of this lncRNA in breast cancer pathogenesis. Although the expression levels of other lncRNAs were not significantly different between tumoral tissues and ANCTs, significant associations were found between LINC00346 expression and tubule formation as well as expression levels of both SNHG16 and SNHG6 genes and family history of cancer. Future studies are needed to elaborate the underlying mechanisms of such observations.
Next, we examined correlations between expression levels of genes in both tumoral tissues and ANCTs. VDR expression in tumoral tissues was correlated with the expression of all assessed lncRNAs except LINC00511. Notably, the most significant correlations were found between expression levels of VDR and SNHG16 in both tumoral tissues and ANCTs. Cai et al have reported frequent overexpression of SNHG16 in breast tumor tissues compared with ANCTs and validated an oncogenic role for this lncRNA in breast cancer. Their in vitro studies indicated that SNHG16 role in cell migration is exerted via miR-98.28 In contrast, Ting et al29 have reported the transcriptional induction of miR-98 by 1α,25-dihydroxy vitamin D3 in LNCaP cells. Taken together, these data suggest the presence of an interactive network between SNHG16, VDR, and miR-98 in the context of cancer. The observed correlation between SNHG16 expression ration and serum vitamin D further supports this hypothesis. Assessment of correlations between transcript levels of these genes also revealed context-dependent correlations. For instance, the correlation between LINC00346 and SNHG16 was seen only in ANCTs, while the correlation between LINC00511 and SNHG16 was only detected in tumoral tissues. Such observation might indicate the presence of context-dependent regulatory mechanisms for these lncRNAs, which need to be assessed in future studies.
We also evaluated VDR expression in tumoral tissues and ANCTs in relation with FokI and CdxII genotypes. FokI polymorphism was associated with the expression of VDR in both tumoral tissues and ANCTs. The TT and CC genotypes of this polymorphism were associated with the higher and lower expression levels of VDR. In addition, we found significant over-representation of CC genotype in patients with the downregulation of VDR in tumoral tissues compared with ANCTs. FokI polymorphism is located in the promoter region 5′ of exon 2. Previous studies have indicated that the C>T change results in the production of a longer protein that is less operative as a transcriptional activator of VDR.30 From our observations in breast cancer patients, it can be deduced that CC genotype can result in a more prominent downregulation of VDR in tumoral tissues compared with ANCTs. Moreover, FokI variants were associated with expression levels of both MALAT1 and SNHG16 in ANCTs, but not tumoral tissues, which provide further evidence for distinct regulation of these lncRNAs in tumoral and nontumoral tissues. CdxII variants were not associated with expression levels of VDR in any sets of tissues, but these variants were associated with expression levels of SNHG16 in ANCTs. Pulito et al31 have found a correlation between VDR mRNA, protein levels, and CdxII genotypes in the ER negative breast cancer cell lines in spite of the lack of such correlations in two ER-positive cell lines. They also reported associations between the AA genotype and low levels of VDR in the majority of breast cancer patients. When they categorized their patients by molecular subtypes, the association between CdxII polymorphism and VDR expression was detected only in Her2-positive and triple-negative subtypes.31 However, we did not find any associations between CdxII polymorphisms and VDR expression in either ER-positive samples or ER-negative samples. Such discordance might be due to low number of ER negative samples (n=11) in our cohort of patients. The observed associations between VDR polymorphisms and the expression levels of certain genes as demonstrated in our study highlight the importance of genotyping of these variants before administration of novel VDR-targeted therapies in cancer patients.
The lncRNAs whose expressions have been assessed in the present study have several mRNA and miRNA targets themselves. For instance, previously published data have shown interaction of MALAT1 with ER,21 miR-129-5p32, DBC1,33 miR-204/ZEB2 axis,34 CD133,35 miR-204,36 CCND1,37 Sox-2,38 miR-1,39 miR-124,40 and VEGFA.41 Moreover, SNHG16 and LINC00511 interact with miR-98/E2F528 and SOX9,25 respectively. So, future studies are needed to assess the dysregulation of all of these targets in the context of breast cancer in an integrative manner.
Finally, we assessed the accuracy of the three dys-regulated genes (LINC00511, MALAT1, and VDR) for the diagnosis of breast cancer using ROC curve analysis but did not find high performance for any of them. However, LINC00511 expression levels had fair accuracy for such purpose. Besides, MALAT1 and VDR expression levels were the most specific and sensitive diagnostic markers for breast cancer among the assessed markers, respectively. Taken together, none of these genes have the ideal features to be used as an individual biomarker. Future studies are needed to assess the accuracy of a panel of these genes in the diagnosis of breast cancer.
Conclusion
The present study provides further support for the contribution of VDR signaling and the related lncRNAs in the pathogenesis of breast cancer and introduces some novel lncRNAs as putative molecules in the interactive functional network of VDR signaling in breast cancer.
Acknowledgments
We thank patients who participated in our study. This article has been extracted from the thesis written by Vahid Kholghi Oskooei in the School of Medicine, Shahid Beheshti University of Medical Sciences (registration no: 46) and is financially supported by “Research Department of the School of Medicine Shahid Beheshti University of Medical Sciences” (grant no 11203).
Footnotes
Author contributions
VKO conducted the experiments and the data analyses. LG contributed to the evaluation of patients. MDO participated in the study design and coordination. SG-F designed and supervised the study and wrote the manuscript. All authors read the manuscript drafts, contributed to the edits, and approved the final manuscript. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ghafouri-Fard S, Shamsi R, Seifi-Alan M, Javaheri M, Tabarestani S. Cancer-testis genes as candidates for immunotherapy in breast cancer. Immunotherapy. 2014;6(2):165–179. doi: 10.2217/imt.13.165. [DOI] [PubMed] [Google Scholar]
- 2.Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007;7(9):684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- 3.Kazemi-Oula G, Ghafouri-Fard S, Mobasheri MB, Geranpayeh L, Modarressi MH. Upregulation of RHOXF2 and ODF4 Expression in Breast Cancer Tissues. Cell J. 2015;17(3):471–497. doi: 10.22074/cellj.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikpayam E, Soudyab M, Tasharrofi B, et al. Expression analysis of long non-coding ATB and its putative target in breast cancer. Breast Dis. 2017;37(1):1–10. doi: 10.3233/BD-160264. [DOI] [PubMed] [Google Scholar]
- 5.Eisman JA, Martin TJ, Macintyre I, Moseley JM. 1,25-dihydroxyvitamin-D-receptor in breast cancer cells. Lancet. 1979;2(8156-8157):1335–1336. doi: 10.1016/s0140-6736(79)92816-2. [DOI] [PubMed] [Google Scholar]
- 6.Evans RM. The steroid and thyroid hormone receptor superfamily. Science (New York, N. Y.) 1988;240(4854):889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorne J, Campbell MJ. The vitamin D receptor in cancer. Proc Nutr Soc. 2008;67(2):115–127. doi: 10.1017/S0029665108006964. [DOI] [PubMed] [Google Scholar]
- 8.Eisman JA, Suva LJ, Sher E, Pearce PJ, Funder JW, Martin TJ. Frequency of 1,25-dihydroxyvitamin D3 receptor in human breast cancer. Cancer Res. 1981;41(12 Pt 1):5121–5124. [PubMed] [Google Scholar]
- 9.Al-Azhri J, Zhang Y, Bshara W, et al. Tumor Expression of Vitamin D Receptor and Breast Cancer Histopathological Characteristics and Prognosis. Clin Cancer Res. 2017;23(1):97–103. doi: 10.1158/1078-0432.CCR-16-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang YJ, Bikle DD. LncRNA: a new player in 1α, 25(OH)(2) vitamin D(3)/VDR protection against skin cancer formation. Exp Dermatol. 2014;23(3):147–150. doi: 10.1111/exd.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikpayam E, Tasharrofi B, Sarrafzadeh S, Ghafouri-Fard S. The Role of Long Non-Coding RNAs in Ovarian Cancer. Iran Biomed J. 2017;21(1):3–15. doi: 10.6091/.21.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soudyab M, Iranpour M, Ghafouri-Fard S. The Role of Long Non-Coding RNAs in Breast Cancer. Arch Iran Med. 2016;19(7):508–17. [PubMed] [Google Scholar]
- 13.Dianatpour A, Ghafouri-Fard S. The Role of Long Non Coding RNAs in the Repair of DNA Double Strand Breaks. Int J Mol Cell Med. 2017;6(1):1–12. [PMC free article] [PubMed] [Google Scholar]
- 14.Iranpour M, Soudyab M, Geranpayeh L, et al. Expression analysis of four long noncoding RNAs in breast cancer. Tumor Biology. 2016;37(3):2933–2940. doi: 10.1007/s13277-015-4135-2. [DOI] [PubMed] [Google Scholar]
- 15.Khorshidi HR, Taheri M, Noroozi R, Sarrafzadeh S, Sayad A, Ghafouri-Fard S. ANRIL Genetic Variants in Iranian Breast Cancer Patients. Cell J. 2017;19(Suppl 1):72–78. doi: 10.22074/cellj.2017.4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell DW. Rapid Isolation of Mammalian DNA. Cold Spring Harb Protoc. 2006;2006(1) doi: 10.1101/pdb.prot3514. pdb. prot3514. [DOI] [PubMed] [Google Scholar]
- 17.Compton C, Byrd D, Garcia-Aguilar J, et al. AJCC cancer staging manual. Chapter. 2010;14:143–164. [Google Scholar]
- 18.Friedrich M, Villena-Heinsen C, Tilgen W, Schmidt W, Reichrat J, Axt-Fliedner R. Vitamin D receptor (VDR) expression is not a prognostic factor in breast cancer. Anticancer Res. 2002;22(3):1919–1924. [PubMed] [Google Scholar]
- 19.Ditsch N, Toth B, Mayr D, et al. The Association between Vitamin D Receptor Expression and Prolonged Overall Survival in Breast Cancer. Journal of Histochemistry & Cytochemistry. 2012;60(2):121–129. doi: 10.1369/0022155411429155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao Y, Fan R, Chen L, Qian H. Clinical Significance of Long Non-coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Ann Clin Lab Sci. 2016;46(4):418–424. [PubMed] [Google Scholar]
- 21.N-Si Huang, Y-Yun Chi, J-Yan Xue, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget. 2016;7(25):37957–37965. doi: 10.18632/oncotarget.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balasis ME, Merelvede J, Kunigal S, et al. Molecular Characterization of SRSF2 Mutation Identifies a Clinically Relevant Lncrna (MALAT1) in Chronic Myelomonocytic Leukemia (CMML. Am Soc Hematology. 2015 [Google Scholar]
- 23.Guru Murthy GS, Dhakal I, Mehta P. Incidence and survival outcomes of chronic myelomonocytic leukemia in the United States. Leuk Lymphoma. 2017;58(7):1648–1654. doi: 10.1080/10428194.2016.1258700. [DOI] [PubMed] [Google Scholar]
- 24.Azim HA, Nguyen B, Brohée S, Zoppoli G, Sotiriou C. Genomic aberrations in young and elderly breast cancer patients. BMC Med. 2015;13(1):266. doi: 10.1186/s12916-015-0504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tg O, Wang SM, Acharya BR, et al. The Nuclear Receptor, ROR-gamma, Regulates Pathways Necessary for Breast Cancer Metastasis. EBioMedicine. 2016;6:59–72. doi: 10.1016/j.ebiom.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slominski AT, Kim T-K, Takeda Y, et al. RORα and ROR γ are expressed in human skin and serve as receptors for endogenously produced noncalcemic 20-hydroxy- and 20,23-dihydroxyvitamin D. The FASEB Journal. 2014;28(7):2775–2789. doi: 10.1096/fj.13-242040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang F, Lyu S, Dong S, Liu Y, Wang O, Zhang X. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther. 2016;9:761–772. doi: 10.2147/OTT.S97664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485(2):272–278. doi: 10.1016/j.bbrc.2017.02.094. [DOI] [PubMed] [Google Scholar]
- 29.Ting H-J, Messing J, Yasmin-Karim S, Lee Y-F. Identification of microRNA-98 as a Therapeutic Target Inhibiting Prostate Cancer Growth and a Biomarker Induced by Vitamin D. Journal of Biological Chemistry. 2013;288(1):1–9. doi: 10.1074/jbc.M112.395947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerr Whitfield G, Remus LS, Jurutka PW, et al. Functionally relevant polymorphisms in the human nuclear vitamin D receptor gene. Mol Cell Endocrinol. 2001;177(1-2):145–159. doi: 10.1016/s0303-7207(01)00406-3. [DOI] [PubMed] [Google Scholar]
- 31.Pulito C, Terrenato I, di Benedetto A, et al. Cdx2 Polymorphism Affects the Activities of Vitamin D Receptor in Human Breast Cancer Cell Lines and Human Breast Carcinomas. PLoS One. 2015;10(4):e0124894. doi: 10.1371/journal.pone.0124894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother. 2017;95:922–928. doi: 10.1016/j.biopha.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Chen R, Liu Y, Zhuang H, et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017;45(17):9947–9959. doi: 10.1093/nar/gkx600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Zhou Y, Yang Z, et al. MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial–mesenchymal transition in breast cancer. Tumor Biology. 2017;39(7):1010428317690998. doi: 10.1177/1010428317690998. [DOI] [PubMed] [Google Scholar]
- 35.Latorre E, Carelli S, Raimondi I, et al. The Ribonucleic Complex HuR-MALAT1 Represses CD133 Expression and Suppresses Epithelial-Mesenchymal Transition in Breast Cancer. Cancer Res. 2016;76(9):2626–2636. doi: 10.1158/0008-5472.CAN-15-2018. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Yang Z, Wang Y, et al. Long non-coding RNA MALAT1 promote triple-negative breast cancer progression by regulating miR-204 expression. Int J Clin Exp Pathol. 2016;9(2):969–977. [Google Scholar]
- 37.Yang Z, Lu W, Ning L, Hao D, Jian S, Hai-Feng C. Downregulation of long non-coding RNA MALAT1 induces tumor progression of human breast cancer through regulating CCND1 expression. Open Life Sci. 2016;11(1) [Google Scholar]
- 38.Zeng L, Cen Y, Chen J. Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol Lett. 2018;15(2):2117–2122. doi: 10.3892/ol.2017.7557. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Jin C, Bingchuan Yan, Lu Q, Lin Y, Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumor Biology. 2016;37(6):7383–7394. doi: 10.1007/s13277-015-4605-6. [DOI] [PubMed] [Google Scholar]
- 40.Feng T, Shao F, Wu Q, et al. miR-124 downregulation leads to breast cancer progression via LncRNA-MALAT1 regulation and CDK4/E2F1 signal activation. Oncotarget. 2016;7(13):16205–16216. doi: 10.18632/oncotarget.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruszko M, Milano E, Forcato M, et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep. 2017;18(8):1331–1351. doi: 10.15252/embr.201643370. [DOI] [PMC free article] [PubMed] [Google Scholar]