Abstract
Background
KIF20A plays an indispensable role in cytokinesis regulation, which is important for tumor proliferation and growth. Recently, the oncogenic role of KIF20A has been well documented in several cancers. However, its clinical role in epithelial ovarian cancer (EOC) remains not reported yet. We investigated its expression and its role in promoting invasion and chemoresistance in EOC cells.
Patients and methods
KIF20A transcription and translation levels were investigated in normal ovarian epithelial cell, ovarian cancer cells, and 10 pairs of fresh EOC tissues and adjacent normal ovarian tissues by real-time quantitative polymerase chain reaction and Western blots. Moreover, KIF20A protein level was also examined by immunohistochemistry in 150 EOC tissues. The correlation between KIF20A expression and clinical variables was analyzed by statistical methods. We also used wound healing assay, transwell assay MTT, and Annexin V/PI to explore KIF20A functions.
Results
KIF20A expression was obviously elevated at both mRNA and protein levels in EOC cell lines and clinical cancer tissues compared with normal ovarian epithelial cell and adjacent normal ovarian tissues. KIF20A protein expression was highly correlated with International Federation of Gynecology and Obstetrics stage (P=0.008), lymph node metastasis (P=0.002), intraperitoneal metastasis (P<0.001), vital status at last follow-up (P<0.001), intraperitoneal recurrence (P=0.030), tumor recurrence (P=0.005), drug resistance (P=0.013), and ascites with tumor cells (P<0.001). KIF20A overexpression was closely related to poorer overall survival and disease progression-free survival. Furthermore, Cox regression analysis revealed that KIF20A can act as an independent hazard indicator for predicting clinical outcomes in EOC patients. Interestingly, KIF20A overexpression promoted invasion and metastasis of EOC cells and also confers resistance to cisplatin.
Conclusion
Our findings indicated that KIF20A overexpression predicts unfavorable clinical outcome, revealing that KIF20A holds a promising potential to serve as a useful prognostic biomarker for EOC patients.
Keywords: epithelial ovarian cancer, KIF20A, clinical prognosis, tumor progression
Introduction
Ovarian cancer was ranked as one of the prevalently lethal gynecologic malignancies, and it was estimated that there is a global incidence of 22,440 new cases per year, with an incidence of 14,080 deaths annually.1 Currently, the standard treatment for ovarian cancer consists of cytoreductive surgery and adjuvant chemotherapy.2 Recent studies have examined platinum-containing chemotherapeutic drugs and targeted therapy, like poly(ADP-ribose) polymerase inhibitors, niraparib, veliparib, and rucaparib. Despite the advancement of new treatment agents, it remains the highest in mortality rate of all gynecological malignancies.3 Moreover, lack of early diagnosis and the appearance of tumor metastasis and drug resistance resulted in barely 20%–40% 5-year survival rate in ovarian cancer patients.4 Most patients are not detected until they progress into advanced stage, ie, International Federation of Gynecology and Obstetrics stage (FIGO) stages III or IV.5 Consequently, there is urgent need to discover new biomarkers. Presently, many serum markers of ovarian cancer, such as CA125, CA199, and CA153, have been used in the clinic for predicting metastasis and prognosis.5,6 Moreover, ETV5, ALX1, and GOLPH3L have been confirmed as potential prognostic and progressive markers in ovarian cancer.7–9 Nevertheless, these indicators are neither remarkably sensitive nor specific for predicting tumor recurrence and progression. Therefore, novel biomarkers that contribute to detect metastasis and chemoresistance and predict tumor progression for patients with ovarian cancer are needed.
KIF20A was localized to the Golgi apparatus and consisted of 890–amino acid. KIF20A, a microtubule-associated motor protein, functions in mitosis, migration, and intracellular transport.10 Importantly, recent studies have shown that KIF20A is a significant downstream target gene of Hedgehog (Hh) signaling, which was related to cancer cell proliferation, invasion, metastasis, and autophagy. In addition, glioma-associated oncogene 2 (GLI2), a principal transcriptional regulator of Hh signaling, combines with KIF20A to form the GLI2–KIF20A axis, which is necessary for the proliferation and progression of human hepatocellular carcinoma cells.11 Some studies have demonstrated a correlation between KIF20A and human cancer of several different organs, involving bladder cancer, breast cancer, gastric cancer, melanoma, and hepatocellular carcinoma.11–15 However, the patterns of KIF20A expression and its clinical significance have not been investigated in epithelial ovarian cancer (EOC). Herein, the goal of our research was to explore the clinical significance of KIF20A expression in EOC.
Materials and methods
Cell lines
The human ovarian cancer cell lines (COV644, COV362, OV90, SKOV3, TOV112D, OVCAR4, A2780, and COV434, TOV21G) and normal ovarian epithelial cells (HOSEpiC) were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). HOSEpiC were cultured in Defined Keratinocyte-SFM (1×) (Invitrogen, Carlsbad, CA, USA). The ovarian cancer cell lines were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) with 10% fetal bovine serum (Gibco, Grand-Island, NY, USA) and 1% penicillin (100 U/mL) and streptomycin (100 µg/mL) at 37°C under 5% CO2 incubator added.
Tissue specimens and patient information
This research was carried out on the paraffin-embedded tissues from 150 EOC patients and 10 freshly paired EOC tissues, which were diagnosed at Sun Yat-sen University Cancer Center between 2001 and 2010. The clinical characteristics of the 150 samples are recorded in Table 1. The median follow-up time for the whole samples was 49.83 months (range from 3 to 178.2 months). The approval from Sun Yat-sen University Cancer Center Institutional Review Ethics Board was obtained (YB2017-037). Written informed consent was obtained from each patient, and the experiments involving human tissue samples were approved by the Institutional Review Ethics Board of Sun Yat-sen University Cancer Center.
Table 1.
Clinicopathological characteristics and tumor expression of KIF20A in epithelial ovarian cancer
| Characteristic | Cases, n (%) |
|---|---|
| Age (years) | |
| ≤53 | 81 (54.0) |
| >53 | 69 (46.0) |
| FIGO stage | |
| I | 25 (18.5) |
| II | 18 (13.3) |
| III | 83 (61.5) |
| IV | 9 (6.7) |
| Histological type | |
| Serous adenocarcinoma | 119 (80.4) |
| Mucoid adenocarcinoma | 24 (16.2) |
| Endometrial adenocarcinoma | 3 (2.0) |
| Clear cell carcinoma | 2 (1.4) |
| Lymph node metastasis | |
| Absent | 71 (56.3) |
| Present | 55 (43.7) |
| Intraperitoneal metastasis | |
| No | 56 (37.3) |
| Yes | 94 (62.7) |
| Intestinal metastasis | |
| No | 68 (48.9) |
| Yes | 71 (51.1) |
| KIF20A expression | |
| Low or none | 60 (40.0) |
| High | 90 (60.0) |
| Vital status at last follow-up | |
| Alive | 69 (46.0) |
| Dead | 81 (54.0) |
| Tumor recurrence | |
| No | 60 (40.3) |
| Yes | 89 (59.7) |
| Intraperitoneal recurrence | |
| No | 69 (69.7) |
| Yes | 30 (30.3) |
| Distant recurrence | |
| No | 35 (60.3) |
| Yes | 23 (39.7) |
| Residual tumor size (cm) | |
| ≤1 | 98 (76.0) |
| >1 | 31 (24.0) |
| Differentiation grade | |
| G1/G2 | 64 (50.8) |
| G3 | 62 (49.2) |
| Neoadjuvant chemotherapy | |
| No | 113 (75.3) |
| Yes | 37 (24.7) |
| Postoperative chemotherapy | |
| No | 7 (4.7) |
| Yes | 143 (95.3) |
| HIPEC | |
| No | 108 (78.3) |
| Yes | 30 (21.7) |
| Ascites with tumor cells | |
| No | 44 (44.9) |
| Yes | 54 (55.1) |
| Cytoreductive surgery | |
| No | 26 (17.3) |
| Yes | 124 (82.7) |
| Drug resistance | |
| No | 30 (52.6) |
| Yes | 27 (47.4) |
| CA125 (U/mL) | |
| ≤35 | 7 (5.2) |
| >35 | 127 (94.8) |
| CA199 (U/mL) | |
| ≤35 | 93 (69.4) |
| >35 | 41 (30.6) |
| CA153 (U/mL) | |
| ≤25 | 27 (22.9) |
| >25 | 91 (77.1) |
| NSE (U/mL) | |
| ≤15.2 | 31 (36.9) |
| >15.2 | 53 (63.1) |
| CEA (U/mL) | |
| ≤5.0 | 90 (79.6) |
| >5.0 | 23 (20.4) |
| β-HCG (U/mL) | |
| ≤3.0 | 54 (80.6) |
| >3.0 | 13 (19.4) |
Abbreviations: β-HCG, β human choriogonadotropin; CEA, carcinoembryonic antigen; FIGO, International Federation of Gynecology and Obstetrics stage; HIPEC, hyperthermic intraperitoneal chemotherapy; NSE, neuron-specific enolase.
Plasmids, infection, and transfection
Human KIF20A complementary DNA (cDNA) was polymerase chain reaction (PCR)-amplified by PCR forward: agatctGCCACCATGTCGCAAGGGATCCTTTC; reverse: gaattcTTAGTACTTTTTGCCAAAAG. To silence KIF20A, two siRNAs against KIF20A in pSuper-puro vector were obtained from Ribobio Inc (Guangzhou, Guangdong, China). The siRNA sequences are as follows: si-KIF20A-1 :5′-CTCCGAGATGAAATTTGCA-3′; si-KIF20A-2 :5′-GGTCTGTGGTACGCAAGAA-3′; Transfection was performed using Lipofectamine 3000 reagent (Invitrogen) in line with the manufacturer’s instructions.
RNA extraction reverse transcription and real-time PCR
Total RNA was isolated from 10 freshly paired clinical samples and cell lines by using Trizol reagent (Invitrogen) based on the manufacturer’s instructions. Then, we used RNase-free DNase to inhibit degradation. In line with the manufacturer’s guidelines, we used Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) to synthesize cDNA from RNA (2 µg) obtained from each sample. The qPCR primer sequences were displayed as follows: KIF20A forward primer: 5′-TCAGAGCGCTGCAAAGATCA-3′; KIF20A reverse primer: 5′-CGGTTCTGCTGGTTTTGACG-3′; GAPDH forward primer: 5′-AGAGGCAGGGATGATGTTCTG-3′; and GAPDH reverse primer; 5′-AGAGGCAGGGATGATGTTCTG-3′. We used the 2−ΔΔCt method to detect relative quantitative value, where Ct represents the threshold cycle for each transcript. All experiments were tested in triplicate.
Western blotting
Total protein was obtained from frozen tissues and cultured cells by using a Whole Protein Extraction Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. Western blots analysis was practiced on the basis of standard steps. Briefly, we used ice-cold PBS to wash cells thrice, and then lysed it on ice in 1× SDS lysis buffer with protease inhibitors. Liquid nitrogen was added to fresh clinic tissue samples, and then we grinded it to powder and used sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer to lyse it. Protein concentrations were determined by the Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA). Equal quantities of protein (30 µg) were resolved on 9% SDS polyacrylamide gels, electrotransferred to polyvinylidene fluoride (PVDF) membranes (Immobilon P; Millipore, Bedford, MA, USA). The membranes was immunoblotted with primary antibody against KIF20A (1:200; Sigma-Aldrich, St. Louis, MO, USA; HPA036910) and GAPDH (1:5,000; Sigma-Aldrich, SAB2701826) at 4°C overnight, after the membranes were blocked in 5% skimmed milk for 1 hour. Then, we washed the membranes thrice with Tris-buffered saline/0.1% Tween 20 (TBST) for 10 minutes each time, followed by the secondary antibody (1:3000, anti-rabbit antibody, Abcam, Cambridge, MA, USA, ab6721) for 2 hours at room temperature. Finally, the bound antibodies were visualized by chemiluminescence detection reagent (Amersham Pharmacia Biotech, Piscataway, NJ, USA) in line with the manufacturer’s guideline. Anti-GAPDH antibody was used as a loading control.
Immunohistochemistry
Immunohistochemistry (IHC) analysis was performed to detect KIF20A protein expression in 150 EOC patients. Briefly, all specimens were cut into 4-µm sections and baked at 60°C for 1 hour. We removed paraffin from these sections by using xylene, once in 10 minutes, twice. Then, these sections were treated in different ethanol solutions (100%, 100%, 95%, 90%, and 80%). Then, EDTA antigenic retrieval buffer and microwave were used to retrieve antigenic of the sections. In order to extinguish endogenous peroxidase activity, the samples were processed with 3% hydrogen peroxide in methanol. Then, 1% bovine serum albumin was selected to block nonspecific blinding. Subsequently, rabbit polyclonal anti-KIF20A antibody (1:600; Sigma; HPA036910) was used to incubate the sections overnight at 4°C. PBS served as the negative control. After washing, the clinical sections were processed with prediluted anti-rabbit secondary antibody (Abcam), followed by treatment with streptavidin–horseradish peroxidase complex (Sigma), and then soaked in 3-amino-9-ethylcarbazole. These sections were mounted in Crystal Mount after the processing of counterstaining with 10% Mayer’s hematoxylin and dehydrating. The score of immunostaining was separately evaluated by two pathologists who were blinded to the clinical parameters. Evaluation of KIF20A protein expression and scores of immunostaining were assessed by two observers separately. The staining intensity of KIF20A was classified as follows: 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). The proportion of tumor cells was graded from 0 to 3: 0 (<10%), 1 (10%–50%), 2 (51%–75%), and 3 (>75%). The product of the proportion of tumor cells and staining intensity score was seen as the staining index. According to the methods of this evaluation, we assessed KIF20A expression in ovarian cancer samples by determining the staining intensity (SI), with scores of 0–9. The median SI of all ovarian cancer tissues was 4. Thus, we distinguished high and low KIF20A expressions by the following criteria: SI ≤4 was defined as tumors with low KIF20A expression; SI ≥6 was indicated as tumors with high KIF20A expression.
Transwell assay
Transwell chambers (8-µm pore size, Corning Inc., Corning, NY, USA) coated with Matrigel (BD Biosciences, San Jose, CA, USA) were used to assess the ability of migration and invasion. Briefly, ovarian cancer cells (2×104) were incubated in serum-free medium and then added to the upper chamber of transwell chambers. The lower chamber was added with 10% FBS culture medium. After incubation at 37°C for 24 hours, the cells in the upper chamber were transited into the lower surface. We used 4% paraformaldehyde and stained it with hematoxylin. Finally, we used microscope (Nikon Eclipse 80i) to count the cells (10 random 200 × fields per well). All experiments were conducted three times.
Wound healing assay
The ovarian cancer cells transfected vector KIF20A or KIF20A siRNA were seeded in six-well plates and grown until almost confluence in 24 hours, and then cultured in serum-free medium for 24 hours. We used a 200-µL pipette to scratch an artificial homogeneous wound of cell monolayers, followed by washing with PBS and cultured it in DMEM plus 10% FBS. Inverted microscope was used to measure the average distance between the edge and the center of the straight scratch at time points 0, 12, and 24 hours (with a 10× objective lens).
MTT assays
The sensitivity to cisplatin of EOC cells was measured using MTT assays followed by the manufacturer’s instructions. Briefly, 4×103 cells were put on 96-well plates and cultured at 37°C overnight and treated with different cisplatin concentrations (3, 9, 27, 81, and 243 µm) for 48 hours. We added the MTT solution (20 µL; 5 mg/mL), and the plates were further incubated for 4 hours. At the end point, 200 µL DMSO was added to the plates in order to stabilize the product. The absorbance was measured at 570 nm. The IC50 values were analyzed using the GraphPad Prism® six software.
Annexin V/PI assay
Annexin V Apoptosis Detection Kit (Beyotime) was used to assess apoptosis following the instructions. Briefly, ovarian cancer cells (2×105) were seeded in six-well plates, then treated with cisplatin (5 µM) for 24 hours, and FITC Annexin V and propidium iodide (each at 5 µL/105 cells) were added. After these processes, we incubated cells at room temperature without light for 15 minutes. We used flow cytometer (Beckman Coulter, Brea, CA, USA) to analyze the flow cytometry data.
Statistical analysis
We used the SPSS 20.0 (SPSS Inc., Chicago, IL, USA) to carry out statistical analyses. Chi-squared test and Fisher’s exact test were used to analyze the correlation of KIF20A protein and clinicopathological features. Spearman’s rank correlation coefficients were utilized to assess the bivariate association between the clinical variables. The Kaplan–Meier method and log-rank testing were carried out to plot survival curves of EOC patients with high or low KIF20A expression. Furthermore, the Cox regression model was applied to calculate the relative risk ratios and analyze the survival data. In all experiments, P<0.05 was regarded to be statistically significant.
Results
KIF20A expression was upregulated in ovarian cancer cell lines
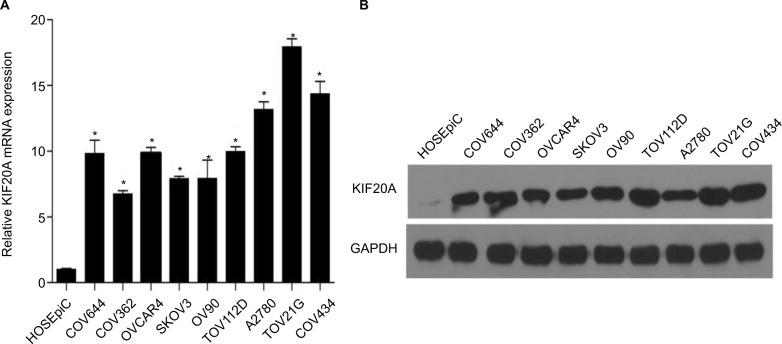
To explore KIF20A expression in ovarian cancer, we detected KIF20A expression in different kinds of ovarian cancer cells (COV644, COV362, OV90, SKOV3, TOV112D, OVCAR4, A2780, COV434) and HOSEpiC at mRNA and protein levels. The results showed that KIF20A was elevated in all of the ovarian cancer cell lines compared with HOSEpiC (P<0.05; Figure 1A, B).
Figure 1.
KIF20A mRNA and protein were overexpressed in EOC cell lines.
Notes: (A, B) Expression of KIF20A mRNA and protein in EOC cell lines (COV644, COV362, OV90, SKOV3, TOV112D, OVCAR4, A2780, COV434, TOV21G) and normal ovarian epithelial cell (HOSEpiC) by real-time PCR (A) and Western blotting (B). Expression levels were normalized to GAPDH expression. Error bars represent standard deviation (SD) of the mean calculated from three parallel experiments. *P<0.05.
Abbreviations: EOC, epithelial ovarian cancer; PCR, polymerase chain reaction.
KIF20A expression in EOC and normal control tissues
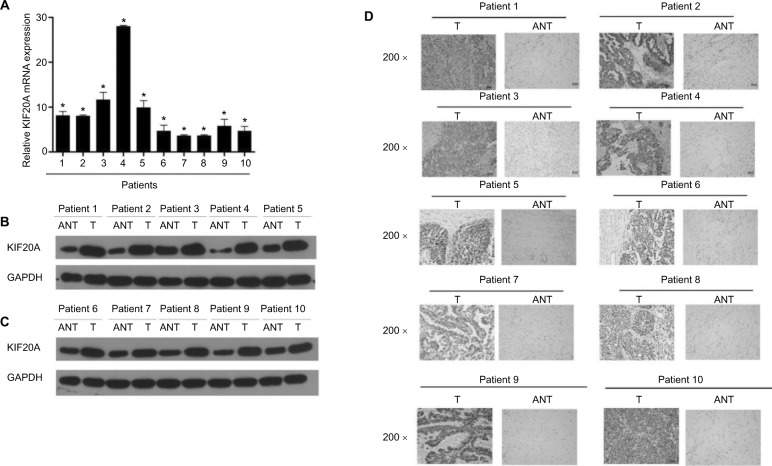
The KIF20A expression of 10 paired fresh EOC tissues and the adjacent noncancerous tissues was examined at transcription and translation levels. As shown in Figure 2A–C, KIF20A expression was significantly upregulated in 10 ovarian cancer tissues compared with the paired noncancerous tissues (P<0.05). Next, we carried out IHC staining to detect the KIF20A protein expression level (Figure 2D and Figure S1A). The results were strongly consistent with the KIF20A protein expression examined by Western blotting.
Figure 2.
Overexpression of KIF20A mRNA and protein in EOC specimens.
Notes: (A) The average T/ANT ratios of KIF20A mRNA expression in the ovarian cancer (T) and adjacent noncancerous tissue sections were quantified using real-time PCR and normalized against GAPDH. Error bars represent standard deviation (SD) of the mean calculated from three parallel experiments. (B, C) Western blotting analyses of KIF20A expression in ten pairs of ovarian cancer tissues (T) and ANT. GAPDH was used as the loading control. (D) Immunohistochemical detection of KIF20A protein in ten pairs of matched ovarian cancer tissues. *P<0.05.
Abbreviations: ANT, adjacent noncancerous tissue; EOC, epithelial ovarian cancer; PCR, polymerase chain reaction.
Association between KIF20A upregulation and EOC clinical features
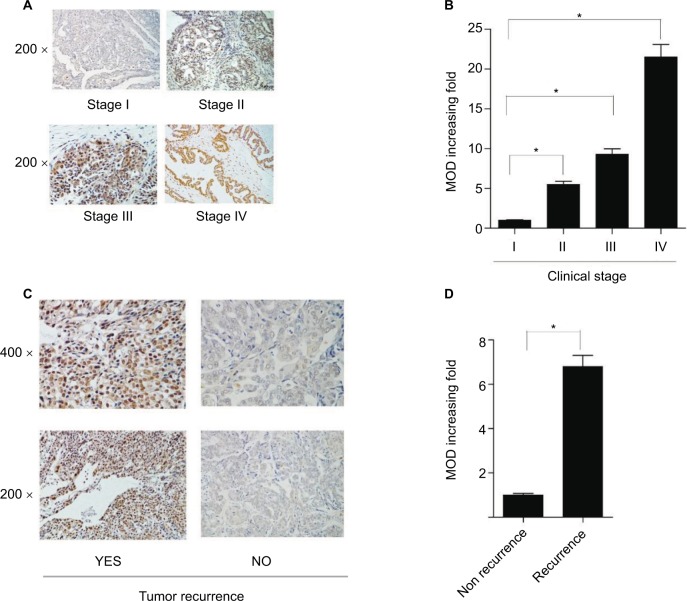
IHC analysis was carried out on 150 EOC samples to assess the value of KIF20A protein upregulation in EOC and then to analyze the correlation between KIF20A expression and the clinicopathological variables. The relationship between KIF20A protein level expression in EOC patients and the clinicopathological variables is presented in Tables 2 and 3. The 150 clinic samples contained 25 stage I cases, 18 stage II cases, 83 stage III cases, and 9 stage IV cases (15 medical records did not have detailed clinical staging information). Of the 150 samples, 90 cases (60%) had a strong KIF20A expression and 60 cases (40%) had a weak or negative KIF20A expression (Table 1). KIF20A was mostly positioned in the nucleus and cytoplasm of the ovarian cancer cells. The expression of KIF20A was low or absent in normal ovarian tissues. The intensity of KIF20A staining increases sequentially from clinical stage I to IV (Figure 3A). As shown in Figure S2A–D, we conducted IHC staining in four histological types of EOC, including serous adenocarcinoma, mucoid adenocarcinoma, endometrial adenocarcinoma, and clear cell adenocarcinoma. Besides, the expression of KIF20A protein was stronger in recurrence group than in the nonrecurrence group (Figure 3). We noted positive KIF20A protein expression in 40% (10/25), 38.89% (7/18), 66.26% (55/83), and 88.89% (8/9) stage I, II, III, and IV tumors, respectively (P<0.05, Table 2). Quantitative IHC analysis suggested that mean optical density (MOD) value of KIF20A was obviously correlated with FIGO stages (Figure 3B) and was higher in recurrence group than in nonrecurrence group (Figure 3D). Moreover, the chi-squared test and Fisher’s exact test were performed to analyze the correlation between KIF20A expression and clinicopathological parameters (Table 2). Spearman’s correlation test indicated that the elevated expression correlated with FIGO stage (P=0.001), intraperitoneal metastasis (P=0.000), lymph node (LN) metastasis (P=0.002), vital status at last follow-up (P<0.001), intraperitoneal recurrence (P=0.030), ascites with tumor cells (P<0.001), and drug resistance (P=0.012). Additionally, patients with KIF20A protein level upregulation were experienced more tumor recurrence (P=0.005); therefore, the vital prognosis of these patients was poor. On the contrary, the correlations between KIF20A expression and age; histological type; intestinal metastasis; residual tumor size; differentiation grade; postoperative chemotherapy; cytoreductive surgery; hyperthermic intraperitoneal chemotherapy (HIPEC); and serum CA125, CA199, carcinoembryonic antigen (CEA), neuron-specific enolase (NSE), and β human choriogonadotropin (β-HCG) levels were not statistically significant (Table 3). These results indicate that KIF20A has an important impact on ovarian cancer recurrence and progression.
Table 2.
Correlation between KIF20A expression and the clinicopathological features of epithelial ovarian cancer
| Characteristic | Total | KIF20A expression
|
Chi- square test P-value | Fisher’s exact test P-value | ||
|---|---|---|---|---|---|---|
| None/Weak | Moderate/Strong | |||||
| Age (years) | ≤53 | 81 | 33 (22.0) | 48 (32.0) | 0.841 | 0.869 |
| >53 | 69 | 27 (18.0) | 42 (28) | |||
| Histological type | Serous adenocarcinoma | 119 | 46 (31.1) | 73 (49.3) | 0.114 | – |
| Mucoid adenocarcinoma | 24 | 10 (6.7) | 14 (9.5) | |||
| Endometrial adenocarcinoma | 3 | 3 (2.0) | 0 (0) | |||
| Clear cell carcinoma | 2 | 0 (0) | 2 (1.3) | |||
| FIGO stage | I | 25 | 15 (11.1) | 10 (6.8) | 0.008 | – |
| II | 18 | 11 (8.1) | 7 (5.2) | |||
| III | 83 | 28 (20.7) | 55 (40.7) | |||
| IV | 9 | 1 (0.7) | 8 (5.9) | |||
| Lymph node metastasis | Absent | 71 | 37 (29.4) | 34 (27.0) | 0.002 | 0.003 |
| Present | 55 | 14 (11.1) | 41 (32.5) | |||
| Intraperitoneal metastasis | No | 56 | 35 (23.3) | 21 (14.0) | <0.001 | <0.001 |
| Yes | 94 | 25 (16.7) | 69 (46.0) | |||
| Intestinal metastasis | No | 68 | 30 (21.6) | 38 (27.3) | 0.283 | 0.303 |
| Yes | 71 | 25 (18.0) | 46 (33.1) | |||
| Vital status at last follow-up | Alive | 69 | 45 (30.0) | 24 (16.0) | <0.001 | <0.001 |
| Dead | 81 | 15 (10.0) | 66 (44.0) | |||
| Intraperitoneal recurrence | No | 69 | 37 (37.3) | 32 (32.3) | 0.030 | 0.048 |
| Yes | 30 | 9 (9.1) | 21 (21.2) | |||
| Distant recurrence | No | 35 | 11 (19.0) | 24 (41.4) | 0.546 | 0.583 |
| Yes | 23 | 9 (15.5) | 14 (24.1) | |||
| Tumor recurrence | No | 60 | 32 (21.5) | 28 (18.8) | 0.005 | 0.006 |
| Yes | 89 | 27 (18.8) | 62 (29.0) | |||
| Residual tumor size (cm) | ≤1 | 98 | 42 (32.6) | 56 (43.4) | 0.467 | 0.534 |
| >1 | 31 | 11 (8.5) | 20 (15.5) | |||
| Differentiation grade | G1/G2 | 64 | 25 (19.8) | 39 (31.0) | 0.743 | 0.856 |
| G3 | 62 | 26 (20.6) | 36 (28.6) | |||
| Neoadjuvant chemotherapy | No | 113 | 50 (33.3) | 63 (42.0) | 0.063 | 0.082 |
| Yes | 37 | 10 (6.7) | 27 (18.0) | |||
| Postoperative chemotherapy | No | 7 | 3 (2.0) | 4 (2.7) | 0.724 | 1.000 |
| Yes | 143 | 58 (38.7) | 85 (56.7) | |||
| Drug resistance | No | 30 | 15 (26.3) | 15 (26.3) | 0.013 | 0.025 |
| Yes | 27 | 5 (8.8) | 22 (38.6) | |||
| HIPEC | No | 108 | 46 (33.3) | 62 (44.9) | 0.213 | 0.292 |
| Yes | 30 | 9 (6.5) | 21 (15.2) | |||
| Ascites with tumor cells | No | 44 | 27 (27.6) | 14 (14.3) | <0.001 | <0.001 |
| Yes | 54 | 13 (13.3) | 41 (41.8) | |||
| Cytoreductive surgery | No | 26 | 10 (6.7) | 16 (10.7) | 0.860 | 1.000 |
| Yes | 124 | 50 (33.3) | 74 (49.3) | |||
| CA125 (U/mL) | ≤35 | 7 | 2 (1.5) | 5 (3.7) | 0.568 | 0.706 |
| >35 | 127 | 50 (37.3) | 77 (57.5) | |||
| CA199 (U/mL) | ≤35 | 93 | 33 (24.6) | 60 (44.8) | 0.147 | 0.180 |
| >35 | 41 | 20 (14.9) | 21 (15.7) | |||
| CA153 (U/mL) | ≤25 | 27 | 13 (11.0) | 14 (11.9) | 0.223 | 0.262 |
| >25 | 91 | 32 (27.1) | 59 (50.0) | |||
| NSE (U/mL) | ≤15.2 | 31 | 10 (11.9) | 21 (25.0) | 0.313 | 0.361 |
| >15.2 | 53 | 23 (27.4) | 30 (35.7) | |||
| CEA (U/mL) | ≤5.0 | 90 | 39 (34.5) | 51 (45.1) | 0.132 | 0.157 |
| >5.0 | 23 | 6 (5.3) | 17 (15.0) | |||
| β-HCG (U/mL) | ≤3.0 | 54 | 21 (31.3) | 33 (49.3) | 0.977 | 1.000 |
| >3.0 | 13 | 5 (7.5) | 8 (11.9) | |||
Abbreviations: β-HCG, β human choriogonadotropin; CEA, carcinoembryonic antigen; FIGO, International Federation of Gynecology and Obstetrics stage; HIPEC, hyperthermic intraperitoneal chemotherapy; NSE, neuron-specific enolase.
Table 3.
Correlation between KIF20A expression and the clinicopathological characteristics of epithelial ovarian cancer
| Variable | KIF20A expression
|
|
|---|---|---|
| Spearman’s correlation coefficient | P-value | |
| Age | 0.016 | 0.842 |
| Histological type | –0.54 | 0.518 |
| FIGO stage | 0.291 | 0.001 |
| Intraperitoneal metastasis | 0.354 | <0.001 |
| Lymph node metastasis | 0.269 | 0.002 |
| Intestinal metastasis | 0.091 | 0.286 |
| Vital status at last follow-up | 0.475 | <0.001 |
| Tumor recurrence | 0.231 | 0.005 |
| Intraperitoneal recurrence | 0.218 | 0.030 |
| Distant recurrence | –0.079 | 0.554 |
| Residual tumor size (cm) | 0.064 | 0.471 |
| Differentiation grade | –0.029 | 0.745 |
| Neoadjuvant chemotherapy | 0.152 | 0.064 |
| Postoperative chemotherapy | –0.029 | 0.726 |
| HIPEC | 0.106 | 0.216 |
| Ascites with tumor cells | 0.377 | <0.001 |
| Cytoreductive surgery | –0.014 | 0.861 |
| Drug resistance | 0.329 | 0.012 |
| CA125 (U/mL) | –0.049 | 0.572 |
| CA199 (U/mL) | –0.125 | 0.149 |
| CA153 (U/mL) | 0.112 | 0.226 |
| NSE (U/mL) | –0.110 | 0.319 |
| CEA (U/mL) | 0.142 | 0.134 |
| β-HCG (U/mL) | 0.003 | 0.978 |
Abbreviations: β-HCG, β human choriogonadotropin; CEA, carcinoembryonic antigen; FIGO, International Federation of Gynecology and Obstetrics stage; HIPEC, hyperthermic intraperitoneal chemotherapy; NSE, neuron-specific enolase.
Figure 3.
KIF20A protein expression in paraffin-embedded tissues.
Notes: (A) Representative images from immunohistochemical staining of KIF20A protein expression in ovarian cancer tissues at four clinical stages. (B) The average MOD of KIF20A staining in four clinical stages groups *P<0.05 (C). Representative images of KIF20A expression in recurrence group and nonrecurrence group. (D) The statistical analyses of the average MOD of KIF20A staining in the recurrence group and nonrecurrence group *P<0.05.
Abbreviation: MOD, mean optical density.
High KIF20A expression was obviously related with poor prognosis in EOC
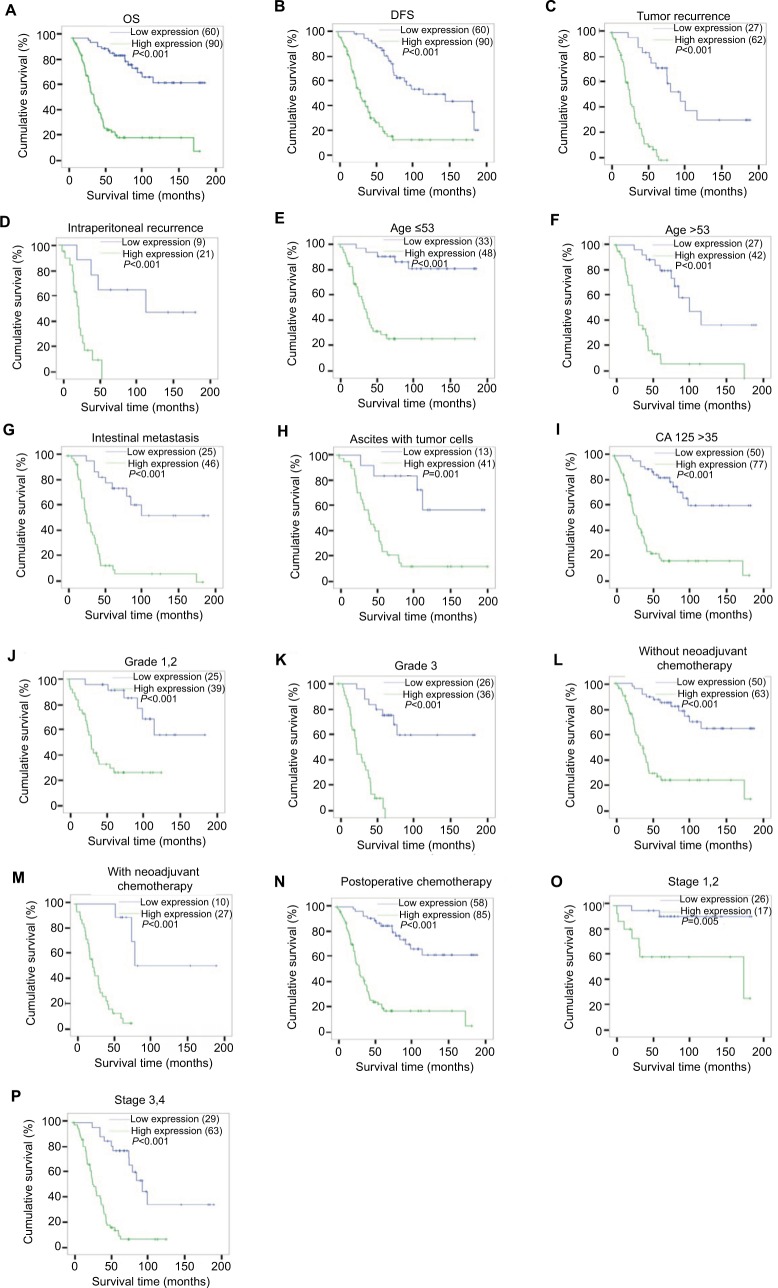
Survival analysis pointed that patients with KIF20A overexpression had a significantly shorter overall survival (OS) and disease-free survival than those with low KIF20A expression (Figure 4A, B, log-rank, P<0.001). In the cohort, patients with elevated KIF20A expression had an obviously lower cumulative 5-year survival rate compared with those with low KIF20A expression (27.4% vs 84.0%, respectively; P<0.05). We also examined the prognostic value of KIF20A expression in subgroups stratified by tumor recurrence (Figure 4C); intraperitoneal recurrence (Figure 4D); age (Figure 4E, F); intestinal metastasis (Figure 4G); ascites with tumor cells (Figure 4H); serum biomarker levels CA125 (Figure 4I); differentiation grade (Figure 4J, K); neoadjuvant chemotherapy (Figure 4L, M); postoperative chemotherapy (Figure 4N); FIGO stages (Figure 4O, P); Univariate Cox analysis showed that KIF20A protein level (P<0.001), age (P=0.025), CA153 levels (P=0.003), CA199 levels (P=0.021), intraperitoneal metastasis (P<0.001), intestinal metastasis (P<0.001), ascites with tumor cells (P<0.001), neoadjuvant chemotherapy (P=0.001), LN metastasis (P<0.001), and tumor recurrence (P<0.001) could serve as important prognostic factors (Table 4). Furthermore, multivariate Cox regression analysis showed that only KIF20A protein expression level (P=0.017) and tumor recurrence (P=0.009) were statistically significant (Table 4).
Figure 4.
Kaplan–Meier curves of univariate analysis data (log-rank test).
Notes: Survival curves for the patients in select patient subgroups (log-rank test). The OS and DFS of the patients with low KIF20A expression vs high KIF20A expression (A, B). OS rates for patients with tumor recurrence (C), intraperitoneal recurrence (D), aged ≤53 years (E), aged >53 years (F), intestinal metastasis (G), ascites with tumor cells (H), CA125 >35 U/mL (I) at differentiation grades 1 and 2 (J), at differentiation grade 3 (K), without neoadjuvant chemotherapy (L), with neoadjuvant chemotherapy (M), postoperative chemotherapy (N), at stages 1, 2 (O), and at stages 3, 4 (P).
Abbreviations: DFS, disease-free survival; OS, overall survival.
Table 4.
Cox regression univariate and multivariate analyses of prognostic factors in epithelial ovarian cancer
| Variable | Patients (n) | Univariate analysis
|
Multivariate analysis
|
|||
|---|---|---|---|---|---|---|
| P-value | Regression coefficient (SE) | P-value | Relative risk | 95% CI | ||
| KIF20A | ||||||
| Low expression | 60 | <0.001 | 1.902 (0.293) | 0.017 | 4.784 | 1.318–17.369 |
| High expression | 90 | |||||
| Intraperitoneal metastasis | ||||||
| No | 56 | <0.001 | 1.880 (0.329) | 0.209 | 2.885 | 0.552–15.074 |
| Yes | 94 | |||||
| Age (years) | ||||||
| ≤53 | 81 | 0.025 | 0.503 (0.224) | 0.582 | 0.804 | 0.369–1.751 |
| >53 | 69 | |||||
| Intestinal metastasis | ||||||
| No | 68 | 0.017 | 0.566 (0.236) | 0.35 | 0.686 | 0.312–1.512 |
| Yes | 71 | |||||
| Neoadjuvant chemotherapy | ||||||
| No | 113 | 0.001 | 0.785 (0.253) | 0.882 | 1.063 | 0.472–2.397 |
| Yes | 37 | |||||
| CA153 (U/mL) | ||||||
| ≤25 | 27 | 0.003 | 1.300 (0.430) | 0.122 | 2.92 | 0.75–11.372 |
| >25 | 91 | |||||
| CA199 (U/mL) | ||||||
| ≤35 | 93 | 0.021 | –0.655 (0.285) | 0.828 | 1.104 | 0.452–2.694 |
| >35 | 41 | |||||
| Ascites with tumor cells (+) | ||||||
| No | 44 | 0.001 | 1.005 (0.311) | 0.811 | 0.875 | 0.294–2.605 |
| Yes | 54 | |||||
| Lymph node metastasis | ||||||
| No | 71 | <0.001 | 0.923 (0.251) | 0.484 | 1.357 | 0.578–3.188 |
| Yes | 55 | |||||
| Tumor recurrence | ||||||
| No | 60 | <0.001 | 1.585 (0.290) | 0.009 | 6.961 | 1.614–30.019 |
| Yes | 89 | |||||
Overexpression of KIF20A facilitates the invasion and metastasis abilities of EOC cells
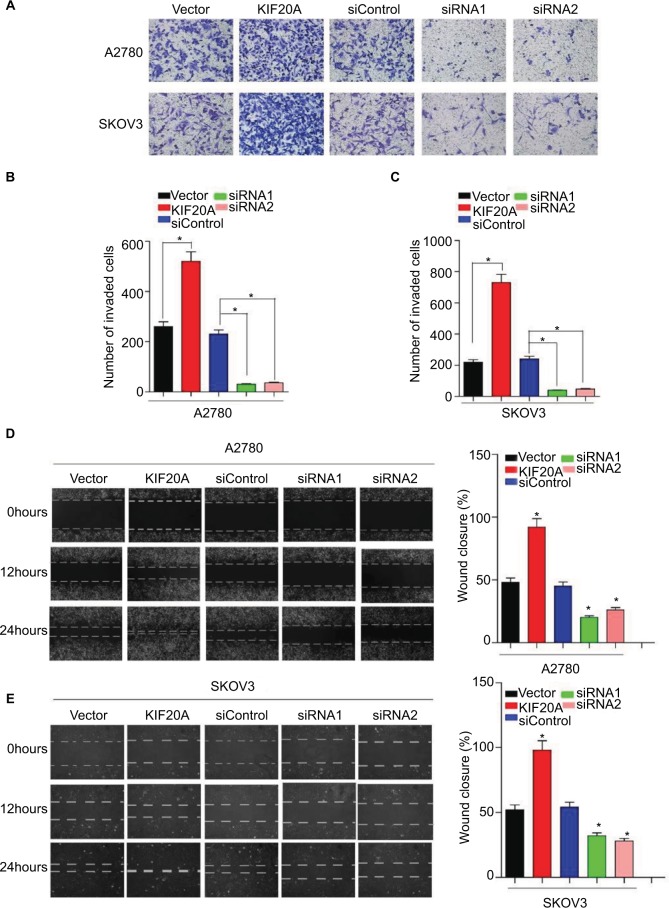
To further explore the function of KIF20A in the invasion and metastasis of EOC, we transiently transfected KIF20A cDNA and KIF20A-siRNA to A2780 cells and SKOV3 cells for 24 hours. Western blot assay was used to evaluate the efficiency (Figure S1). As shown in Figure 5A–C, overexpression of KIF20A enhanced invasion ability of A2780 and SKOV3 cells, whereas silence of KIF20A remarkably reduced the invasive cell numbers. Furthermore, by using the wound healing assay, we found that the KIF20A-transduced EOC cells exhibit an increase in migration ability, while the KIF20A-silenced cells showed an inhibition in their migration ability (Figure 5D, E). These data show that overexpression of KIF20A facilitates EOC cells’ invasion and migration abilities compared with vector control cells.
Figure 5.
Upregulation of KIF20A enhances the invasive and metastatic abilities of EOC cells, while knockdown of KIF20A represses the invasive and metastatic abilities.
Notes: (A–C) The invasive and metastatic abilities of EOC cells were assessed by transwell assay. Each bar represents the mean±SD of three parallel experiments. (D, E) Representative images of wound healing assay; mobility of cells was evaluated by wound closure (%) at 0, 12, 24 hours *P<0.05.
Abbreviation: EOC, epithelial ovarian cancer.
KIF20A confers resistance to cisplatin in EOC cells
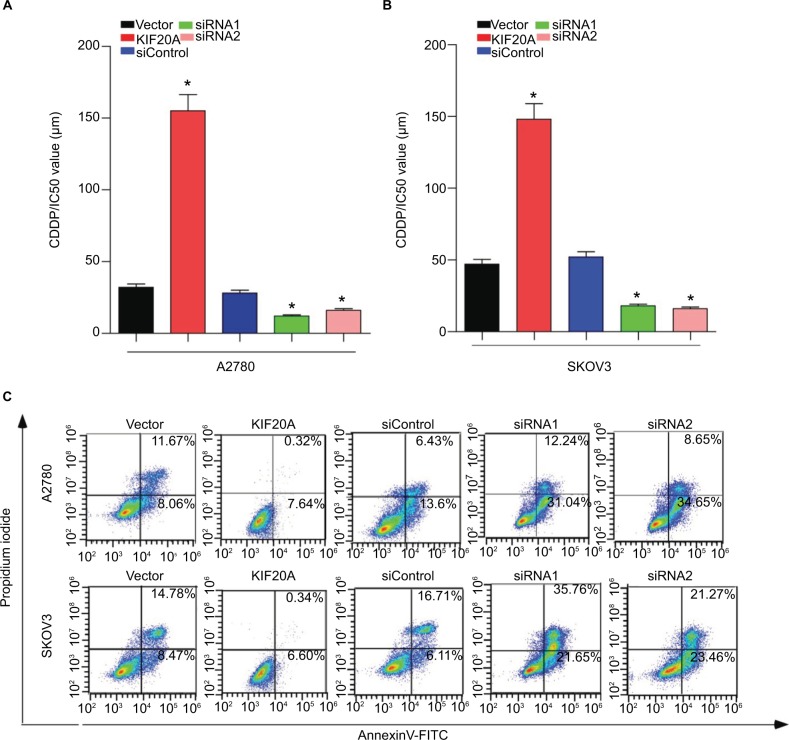
To further verify that KIF20A mediates ovarian cancer cells’ chemoresistance, MTT/IC50 assay and Annexin V-FITC/PI assay were performed on the indicated cells. MTT/IC50 assay indicated that the IC50 value for cisplatin was obviously raised in KIF20A-overexpression ovarian cancer cells, while it was decreased in KIF20A-silenced cells (Figure 6A, B). Consistently, we also found that the proportion of apoptotic cells in KIF20A-overexpression group treated with cisplatin was much lower compared with the control group, yet much higher in the KIF20A-silenced group (Figure 6C). Collectively, these results strongly suggest that KIF20A confers chemoresistance in EOC cells.
Figure 6.
KIF20A confers cisplatin resistance in EOC cells.
Notes: (A, B) MTT/IC50 assay evaluates the viability of KIF20A-overexpressed and KIF20A-silenced EOC cells after cisplatin treatment for 48 hours. (C) Apoptosis assay analyzes the percentage of apoptosis cells for KIF20A-overexpressed and KIF20A-silenced EOC cells after cisplatin treatment (5 µM) for 24 hours *P<0.05.
Abbreviation: EOC, epithelial ovarian cancer.
Discussion
Similar to our study, recent studies have reported that KIF20A is upregulated in several cancers.11–15 Besides, significant evidence indicated that high expression of KIF20A promotes cell metastasis and proliferation in various types of cancers, indicating that overexpression of KIF20A is related to tumor progression.14,15,17,18
As far as we know, this is the first exploration that KIF20A overexpression is closely related to unfavorable prognosis and clinical characteristics, especially tumor recurrence, in EOC. We found that the upregulation of KIF20A occurred at both mRNA and protein levels in EOC tissues in comparison with normal ovarian tissues. High KIF20A expression was closely correlated with FIGO stage, intraperitoneal metastasis, LN metastasis, vital status at last follow-up, tumor recurrence, intraperitoneal recurrence, ascites with tumor cells, and drug resistance. Patients with upregulated KIF20A expression were more likely to experience tumor recurrence during treatment, which is an important cause of poor prognosis of EOC. Univariate and multivariate regression analyses revealed that KIF20A expression can serve as an independent prognostic marker in EOC. Our results show that high KIF20A protein expression correlates with tumor progression and may be viewed as a valuable prognostic factor of clinical outcomes in EOC.
There are many factors related to this poor prognosis in ovarian cancer, of which an important one is tumor progression. Drug resistance is an important reason for tumor progression. In most cases, ovarian cancer is one of the most treatable solid tumors at the early stage of treatment. However, ovarian cancer cells may develop drug chemoresistance during treatment, which causes further treatments to be ineffective and induce tumor progression. Ultimately, there is minimal improvement in OS of ovarian cancer patients during the past 30 years.19 Even in the era of precision treatment, predicting the chemotherapy response and tumor progression of patients with EOC is extremely difficult. A platinum-based drug and a taxane were regarded as the first-line chemotherapy for advanced EOC patients.20 Paclitaxel belongs to the family of antimitotic anticancer agents and blocks mitosis by stabilizing microtubules, leading to the blockage of cell division and therefore cell survival.16 KIF20A is a microtubule-associated motor protein, and it plays the role of intracellular organelle transport and cell division.17 Paclitaxel primarily disturbs mitotic spindle dynamics and triggers the mitotic checkpoint to induce extended G2/M arrest that can incur cell death via the intrinsic (mitochondrial) apoptotic pathway.19 It is not difficult to surmise that high KIF20A expression correlates with chemoresistance. Additionally, in breast cancer, fork-head box M1-induced upregulation of KIF20A at transcriptional level confers drug resistance.20,22 Consistent with these reports, our findings provided evidence that overexpression of KIF20A strongly reduced the proportion of apoptosis cells, while knockdown of KIF20A obviously increased apoptosis cells compared with the control cells. Moreover, there are numerous mechanisms of drug resistance, involving the elevation of efflux mediated by P-glycoprotein coded by the ATP binding cassette subfamily B member 1 (MDR1/ABCB1) gene, spindle assembly checkpoint defects, mitotic slippage, activation of prosurvival signaling, and apoptosis evasion by modulating the activity of p53 and BCL2 family proteins.21 However, the precise mechanisms by which KIF20A affects chemoresistance require further investigation.
From the intraperitoneal metastasis subgroup data, KIF20A expression was nearly three times that in the nonintraperitoneal metastasis subgroup. Intraperitoneal metastasis may be another independent prognostic factor associated with poorer outcomes in EOC.20 In the majority of ovarian cancer cases, patients are always at an advanced stage, in which metastatic disease is already present, and detected, thus resulting in unfavorable prognosis. Unlike most solid tumors, EOC metastasis often occurs through the shedding of cancer cells from the ovarian tumor directly into the peritoneal cavity.23 In addition, intraperitoneal metastasis may have an important role in the clinical staging of EOC according to FIGO 2014. Therefore, it is important to detect intraperitoneal metastasis at the early stages in EOC.24 However, to date, there is lack of sensitive molecular bio-markers for detecting early intraperitoneal metastasis.28 Our findings show that KIF20A expression correlates strongly with intraperitoneal metastasis in EOC. Additionally, Cox regression analyses showed that intraperitoneal metastasis is an important factor that shortens the OS of EOC. KIF20A expression in noncancerous ovarian tissue and primary ovarian cancer without intraperitoneal metastasis group was very low; however, ovarian cancer with intraperitoneal metastasis had remarkably high KIF20A expression. This suggests that KIF20A overexpression is related to intraperitoneal metastasis and poor outcome in EOC. Therefore, KIF20A can been seen as a biomarker for detecting intraperitoneal metastasis at the early stages of EOC and may contribute to improving survival of EOC patients.24,28
LN metastasis is another valuable prognostic factor of EOC25 and is also an important factor for doctors to select suitable treatments. Clinically, the 5-year survival rate of EOC is >90% when the tumor is limited to the ovary; however, it decreases to only 30% when the metastasis has happened.1 Patients with negative LNs have longer OS than patients with positive LNs.26 Furthermore, detecting LN metastasis preoperatively can help doctors to determine whether there is a need for LN dissection. It contributes to the reduction of postoperative complications and to improving patients’ quality of life.27,29 In our study, using univariate Cox regression analyses, we found that LN metastasis was a significant prognostic factor (P<0.001; Table 4). Therefore, detecting LN metastasis at the early stages may improve OS of EOC patients. However, currently, there are no markers for predicting LN status with high sensitivity and specificity.30 Our findings show an obvious correlation between KIF20A overexpression with LN metastasis. Therefore, we suggest that KIF20A may be considered as a valuable biomarker for detecting LN metastasis in ovarian cancer. Thus, during further studies, a larger EOC cohort is needed to verify our results on LN metastasis, and the mechanism of KIF20A promoting LN metastasis of EOC should also be investigated.
Also, wound healing assay and transwell assay were used to evaluate the ability of migration and invasion of EOC cells. Our research shows that upregulation of KIF20A strongly enhanced migration and invasion abilities in EOC cells, but KIF20A-silenced cells obviously reduced migration and invasion. Angiogenesis is an important factor in the growth and invasion of malignant tumor. Vascular endothelial growth factor A (VEGF-A) belonging to VEGF family, plays an important role in angiogenesis, which is necessary for cell invasion and migration.31,32 The association between upregulation of VEGF-A and invasion has been reported in several cancers, including ovarian cancer.33–36 Exertier et al reported that KIF20A/MKLP2 combined with VEGF-A stimulates angiogenesis in vivo, and mitosis-independent vascular outgrowth was potently impaired by KIF20A inhibitor.37 Collectively, we suggest that overexpression of KIF20A may enhance the ability of migration and invasion by regulating VEGF. Nevertheless, we should conduct further exploration to validate it.
Cancer immunotherapy has emerged recently as a promising means of treating cancer; consequently, researching and developing peptide vaccines targeting tumor-specific antigens is important.38,39 Several methods of EOC immunotherapy have been studied at present.39 Moreover, antitumor immune responses by the immune system has been considered as a promising cancer treatment. Previous studies have shown that KIF20A has been used in immunotherapy for various cancers. Asahara et al performed an immunotherapy treatment that utilized KIF20A-66 cancer vaccine reagent, a human leukocyte antigen-A24-restricted epitope peptide obtained from KIF20A, which is significantly transactivated in pancreatic cancer, to treat pancreatic cancer. Patients who were vaccinated with KIF20A-66 had a better clinical outcome than the control group, and further clinical research using this peptide also achieved high potential benefit for patients in advanced stage of pancreatic cancer.38 In immunotherapy of lung cancer and cholangiocellular carcinoma, a Phase I/II clinical trial is currently being carried out using KIF20A-derived short peptides.40 Excitingly, Tomita et al reported that KIF20A long peptides (LP) stimulate KIF20A-specific cytotoxic T lymphocytes in vitro and in vivo.40 KIF20A-specific T-helper type 1 (TH1) cell responses were investigated in head and neck cancer patients and then they received this immunotherapy (8/16, 50%). This finding reveals that the induction of KIF20A-specific TH1 cells in response to KIF20A-LP vaccination may promote the clinical response to chemotherapy or other standard therapies, such as radiotherapy or surgery.41–43 Therefore, based on these previous studies, we believe that conducting research on KIF20A peptide-based immunotherapy of EOC is worthwhile. KIF20A may be a potential cancer therapeutic target to improve survival in EOC.
There are some limitations in our research. Firstly, our work was a retrospective study. Secondly, we performed migration, invasion, and chemoresistance related experiments in vitro, while we lacked some experiments in vivo to confirm our idea. But, in future experiments we will further conduct related in vivo experiments to verify our hypothesis and investigate the molecule mechanism.
Conclusions
In conclusion, our study provides convincing data that KIF20A is overexpressed in the majority of EOC tissues, and KIF20A expression could serve as an independent prognostic indicator for EOC patients. However, further examination of the molecular mechanisms behind the KIF20A-mediated potential tumorigenic effects in ovarian cancer is needed.
Supplementary materials
(A) Immunohistochemical detection of KiF20a protein in 10 pairs of matched ovarian cancer tissues (×400). (B) The silence efficiency and overexpression efficiency of endogenous KIF20A expression on protein levels were detected by Western blotting analysis.
Representative immunohistochemical images of four histological types of epithelial ovarian cancer tissues (×200).
Notes: (A) Serous adenocarcinoma, (B) mucoid adenocarcinoma, (C) endometrial adenocarcinoma, and (D) clear cell carcinoma.
Acknowledgments
Our research was funded by Natural Science Foundation of China (No. 81672579).
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, Zhang B, Xing G, et al. Neoadjuvant chemotherapy versus primary debulking surgery in advanced epithelial ovarian cancer: A meta-analysis of peri-operative outcome. PLoS One. 2017;12(10):e0186725. doi: 10.1371/journal.pone.0186725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh C. Targeted therapy for ovarian cancer: the rapidly evolving landscape of PARP inhibitor use. Minerva Ginecol. 2018;70(2):150–170. doi: 10.23736/S0026-4784.17.04152-1. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton CA, Miller A, Casablanca Y, et al. Clinicopathologic characteristics associated with long-term survival in advanced epithelial ovarian cancer: an NRG Oncology/Gynecologic Oncology Group ancillary data study. Gynecol Oncol. 2018;148(2):275–280. doi: 10.1016/j.ygyno.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mcguire WP, Markman M. Primary ovarian cancer chemotherapy: current standards of care. Br J Cancer. 2003;89(Suppl 3(Suppl 3)):S3–S8. doi: 10.1038/sj.bjc.6601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch HT, Casey MJ, Lynch J, White TE, Godwin AK. Genetics and ovarian carcinoma. Semin Oncol. 1998;25(3):265–280. [PubMed] [Google Scholar]
- 7.Llauradó M, Abal M, Castellví J, et al. ETV5 transcription factor is overexpressed in ovarian cancer and regulates cell adhesion in ovarian cancer cells. Int J Cancer. 2012;130(7):1532–1543. doi: 10.1002/ijc.26148. [DOI] [PubMed] [Google Scholar]
- 8.Yuan H, Kajiyama H, Ito S, et al. ALX1 induces snail expression to promote epithelial-to-mesenchymal transition and invasion of ovarian cancer cells. Cancer Res. 2013;73(5):1581–1590. doi: 10.1158/0008-5472.CAN-12-2377. [DOI] [PubMed] [Google Scholar]
- 9.He S, Niu G, Shang J, et al. The oncogenic Golgi phosphoprotein 3 like overexpression is associated with cisplatin resistance in ovarian carcinoma and activating the NF-κB signaling pathway. J Exp Clin Cancer Res. 2017;36(1):137. doi: 10.1186/s13046-017-0607-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Lai F, Fernald AA, Zhao N, Le Beau MM. cDNA cloning, expression pattern, genomic structure and chromosomal location of RAB6KIFL, a human kinesin-like gene. Gene. 2000;248(1–2):117–125. doi: 10.1016/s0378-1119(00)00135-9. [DOI] [PubMed] [Google Scholar]
- 11.Shi C, Huang D, Lu N, et al. Aberrantly activated Gli2-KIF20A axis is crucial for growth of hepatocellular carcinoma and predicts poor prognosis. Oncotarget. 2016;7(18):26206–26219. doi: 10.18632/oncotarget.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho JR, Chapeaublanc E, Kirkwood L, et al. Deregulation of Rab and Rab effector genes in bladder cancer. PLoS One. 2012;7(6):e39469. doi: 10.1371/journal.pone.0039469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claerhout S, Lim JY, Choi W, et al. Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PLoS One. 2011;6(9):e24662. doi: 10.1371/journal.pone.0024662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita J, Fukushima S, Jinnin M, et al. Kinesin family member 20A is a novel melanoma-associated antigen. Acta Derm Venereol. 2012;92(6):593–597. doi: 10.2340/00015555-1416. [DOI] [PubMed] [Google Scholar]
- 15.Zou JX, Duan Z, Wang J, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res. 2014;12(4):539–549. doi: 10.1158/1541-7786.MCR-13-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nestal de Moraes G, Ji Z, Fan LY, et al. SUMOylation modulates FOXK2-mediated paclitaxel sensitivity in breast cancer cells. Oncogenesis. 2018;7(3):29. doi: 10.1038/s41389-018-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stangel D, Erkan M, Buchholz M, et al. KIF20A inhibition reduces migration and invasion of pancreatic cancer cells. J Surg Res. 2015;197(1):91–100. doi: 10.1016/j.jss.2015.03.070. [DOI] [PubMed] [Google Scholar]
- 18.Gasnereau I, Boissan M, Margall-Ducos G, et al. KIF20A mRNA and its product MKlp2 are increased during hepatocyte proliferation and hepatocarcinogenesis. Am J Pathol. 2012;180(1):131–140. doi: 10.1016/j.ajpath.2011.09.040. [DOI] [PubMed] [Google Scholar]
- 19.Januchowski R, Sterzynska K, Zawierucha P, et al. Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget. 2017;8(30):49944–49958. doi: 10.18632/oncotarget.18278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennessy BT, Coleman RL, Markman M. Ovarian cancer. Lancet. 2009;374(9698):1371–1382. doi: 10.1016/S0140-6736(09)61338-6. [DOI] [PubMed] [Google Scholar]
- 21.Wei W, Birrer MJ. Spleen tyrosine kinase confers paclitaxel resistance in ovarian cancer. Cancer Cell. 2015;28(1):7–9. doi: 10.1016/j.ccell.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Khongkow P, Gomes AR, Gong C, et al. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2016;35(8):990–1002. doi: 10.1038/onc.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klymenko Y, Kim O, Loughran E, et al. Cadherin composition and multicellular aggregate invasion in organotypic models of epithelial ovarian cancer intraperitoneal metastasis. Oncogene. 2017;36(42):5840–5851. doi: 10.1038/onc.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye Y, Yin M, Huang B, Wang Y, Li X, Lou G. CLIC1 a novel biomarker of intraperitoneal metastasis in serous epithelial ovarian cancer. Tumour Biol. 2015;36(6):4175–4179. doi: 10.1007/s13277-015-3052-8. [DOI] [PubMed] [Google Scholar]
- 25.Peng F, Zhong Y, Liu Y, et al. SPARC suppresses lymph node metastasis by regulating the expression of VEGFs in ovarian carcinoma. Int J Oncol. 2017;51(6):1920–1928. doi: 10.3892/ijo.2017.4168. [DOI] [PubMed] [Google Scholar]
- 26.di Re F, Baiocchi G, Fontanelli R, et al. Systematic pelvic and paraaortic lymphadenectomy for advanced ovarian cancer: prognostic significance of node metastases. Gynecol Oncol. 1996;62(3):360–365. doi: 10.1006/gyno.1996.0249. [DOI] [PubMed] [Google Scholar]
- 27.Concin N, Hefler L, van Bavel J, et al. Biological markers in pT1 and pT2 ovarian cancer with lymph node metastases. Gynecol Oncol. 2003;89(1):9–15. doi: 10.1016/s0090-8258(02)00147-6. [DOI] [PubMed] [Google Scholar]
- 28.Elsnerova K, Bartakova A, Tihlarik J, et al. Gene expression profiling reveals novel candidate markers of ovarian carcinoma intraperitoneal metastasis. J Cancer. 2017;8(17):3598–3606. doi: 10.7150/jca.20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayhan A, Gultekin M, Taskiran C, et al. Lymphatic metastasis in epithelial ovarian carcinoma with respect to clinicopathological variables. Gynecol Oncol. 2005;97(2):400–404. doi: 10.1016/j.ygyno.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 30.Noordhuis MG, Fehrmann RS, Wisman GB, et al. Involvement of the TGF-beta and beta-catenin pathways in pelvic lymph node metastasis in early-stage cervical cancer. Clin Cancer Res. 2011;17(6):1317–1330. doi: 10.1158/1078-0432.CCR-10-2320. [DOI] [PubMed] [Google Scholar]
- 31.Zhang D, Li B, Shi J, et al. Suppression of tumor growth and metastasis by simultaneously blocking vascular endothelial growth factor (VEGF)-A and VEGF-C with a receptor-immunoglobulin fusion protein. Cancer Res. 2010;70(6):2495–2503. doi: 10.1158/0008-5472.CAN-09-3488. [DOI] [PubMed] [Google Scholar]
- 32.Feng J, Zhang Y, Xing D. Low-power laser irradiation (LPLI) promotes VEGF expression and vascular endothelial cell proliferation through the activation of ERK/Sp1 pathway. Cell Signal. 2012;24(6):1116–1125. doi: 10.1016/j.cellsig.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Gu F, Niu C, et al. VEGF111b, a C-terminal splice variant of VEGF-A and induced by mitomycin C, inhibits ovarian cancer growth. J Transl Med. 2015;13:164. doi: 10.1186/s12967-015-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Xiong Z, Wei T, et al. Nuclear factor 90 promotes angiogenesis by regulating HIF-1α/VEGF-A expression through the PI3K/Akt signaling pathway in human cervical cancer. Cell Death Dis. 2018;9(3):276. doi: 10.1038/s41419-018-0334-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geng J, Li X, Zhou Z, Wu CL, Dai M, Bai X. EZH2 promotes tumor progression via regulating VEGF-A/AKT signaling in non-small cell lung cancer. Cancer Lett. 2015;359(2):275–287. doi: 10.1016/j.canlet.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, Xiao H, Wang ZH, et al. miR-29a suppresses growth and invasion of gastric cancer cells in vitro by targeting VEGF-A. BMB Rep. 2014;47(1):39–44. doi: 10.5483/BMBRep.2014.47.1.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Exertier P, Javerzat S, Wang B, et al. Impaired angiogenesis and tumor development by inhibition of the mitotic kinesin Eg5. Oncotarget. 2013;4(12):2302–2316. doi: 10.18632/oncotarget.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asahara S, Takeda K, Yamao K, Maguchi H, Yamaue H. Phase I/II clinical trial using HLA-A24-restricted peptide vaccine derived from KIF20A for patients with advanced pancreatic cancer. J Transl Med. 2013;11:291. doi: 10.1186/1479-5876-11-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alipour S, Zoghi S, Khalili N, Hirbod-Mobarakeh A, Emens LA, Rezaei N. Specific immunotherapy in ovarian cancer: a systematic review. Immunotherapy. 2016;8(10):1193–1204. doi: 10.2217/imt-2016-0034. [DOI] [PubMed] [Google Scholar]
- 40.Tomita Y, Yuno A, Tsukamoto H, et al. Identification of promiscuous KIF20A long peptides bearing both CD4+ and CD8+ T-cell epitopes: KIF20A-specific CD4+ T-cell immunity in patients with malignant tumor. Clin Cancer Res. 2013;19(16):4508–4520. doi: 10.1158/1078-0432.CCR-13-0197. [DOI] [PubMed] [Google Scholar]
- 41.Ding ZC, Zhou G. Cytotoxic chemotherapy and CD4+ effector T cells: an emerging alliance for durable antitumor effects. Clin Dev Immunol. 2012;2012:890178–12. doi: 10.1155/2012/890178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lesterhuis WJ, Haanen JB, Punt CJ. Cancer immunotherapy-revisited. Nat Rev Drug Discov. 2011;10(8):591–600. doi: 10.1038/nrd3500. [DOI] [PubMed] [Google Scholar]
- 43.Walter S, Weinschenk T, Stenzl A, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med. 2012;18(8):1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Immunohistochemical detection of KiF20a protein in 10 pairs of matched ovarian cancer tissues (×400). (B) The silence efficiency and overexpression efficiency of endogenous KIF20A expression on protein levels were detected by Western blotting analysis.
Representative immunohistochemical images of four histological types of epithelial ovarian cancer tissues (×200).
Notes: (A) Serous adenocarcinoma, (B) mucoid adenocarcinoma, (C) endometrial adenocarcinoma, and (D) clear cell carcinoma.