Abstract
Purpose
A series of microRNAs (miRNAs) have been identified as associated with the survival of patients with non-small-cell lung cancer (NSCLC). The aim of the present study was to explore whether combination of these experimentally validated individual miRNA biomarkers could be used to further increase their prognostic power in NSCLC.
Patients and methods
Based on previously validated NSCLC prognostic miRNAs, gene signatures that could discriminate high-risk subgroups with poor clinical outcome in four NSCLC miRNA expression datasets (GSE13937, GSE16025, The Cancer Genome Atlas Lung Adenocarcinoma, and TCGA lung squamous cell carcinoma) were developed using the SurvMicro tool. The potential of the miRNA signature established was validated by quantitative real-time PCR analysis of clinical NSCLC samples, and its prognostic power evaluated using the survivalROC method.
Results
We developed two miRNA signatures with prognostic significance for NSCLC, comprising 12- and 7-miRNAs. The 7-miRNA signature (miR-148b, miR-365, miR-32, miR-375, miR-21, miR-125b, and miR-155) was a subset of a 12-miRNA set that retained prognostic power across NSCLC cohorts. Compared with previously established miRNA signatures, our 7-miRNA signature has similar potential, while comprising fewer miRNA components. The prognostic ability of the 7-miRNA signature was validated experimentally in an independent NSCLC cohort using real-time PCR (HR=3.4847, 95% CI=1.3693–8.8680, P=0.0092), and this signature, combined with tumor pathological stage, had superior prognostic ability compared with tumor stage alone.
Conclusion
Our data indicate that the established 7-miRNA signature is simple, robust, and may have greater clinical prognostic utility for patients with NSCLC.
Keywords: lung cancer, gene signature, overall survival, The Cancer Genome Atlas, TCGA, risk classification
Introduction
Non-small cell carcinoma (NSCLC) is the most frequent cause of cancer deaths both in China and globally.1 Despite patients receiving potentially curative treatments, the prognosis of NSCLC remains poor, with a 40% relapse rate within 5 years. As patients with NSCLC and tumors of the same pathological TNM (tumor-lymph node-metastasis) stage exhibit heterogeneous clinical outcomes, the current classification system requires development to improve prognostic models.
MicroRNAs (miRNAs) are a class of small non-coding RNA with potential biological and clinical relevance. With the advent of miRNA expression profiles in multiple types of cancer, including NSCLC, significant efforts have been made to correlate miRNA expression with tumor classification or prognosis.2 A number of single miRNA biomarkers are associated with the clinical outcome of NSCLC,3,4 and recent reports suggest that various signatures comprising multiple miRNA biomarkers may be more powerful in classifying cancer subtypes than single miRNAs.5 Nevertheless, the diagnostic efficiency of these miRNA signatures could not be replicated in additional separate patient cohorts.6
Currently, the majority of miRNA signatures are constructed from miRNA expression profile data; however, whether experimentally validated individual prognostic miRNAs can be used to build a prognostic miRNA signature has not previously been explored. In this study, we established and validated a 7-miRNA prognostic signature for NSCLC from previously validated individual miRNA biomarkers, the discriminatory power of which could be demonstrated in various different patient cohorts. The robust stability and ease of applicability of this miRNA gene set indicate that it has potential clinical utility for prognostic classification and treatment regimen selection in patients with NSCLC.
Material and methods
Selection of NSCLC miRNA biomarkers for miRNA prognostic signature construction
To develop our miRNA signature, 12 validated miRNAs (hsa-mir-126, hsa-mir-100, hsa-mir-148b, hsa-mir-365, hsa-mir-124, hsa-mir-32, hsa-mir-146a, hsa-mir-375, hsa-mir-21, hsa-mir-200c, hsa-mir-125b-1, and hsa-mir-155) were selected from previous publications according to the following criteria: 1) shows statistical significance as prognostic biomarker for NSCLC in meta-analysis; 2) miRNA expression levels measured in formalin-fixed paraffin-embedded (FFPE) or frozen tissues; and 3) miRNA expression levels were detected individually (for example, by real-time PCR).
Datasets containing NSCLC miRNA expression profiles
Gene-expression profiling data from four miRNA datasets, including two Gene Expression Omnibus (GEO; GSE13937, GSE16025) and two “The Cancer Genome Atlas” (TCGA; TCGA LUAD [adenocarcinomas], TCGA LUSC [squamous cell carcinomas]), were used to develop and cross-validate an miRNA signature.7–9 The details of all datasets are listed in Table S1. A total of 550 NSCLC samples including 259 adenocarcinomas (ADC) and 291 squamous cell carcinomas (SCC) were included in these analyses.
Development of a 12-miRNA signature using four NSCLC datasets
The gene expression profiles of the 12 selected miRNAs from each dataset were quantile-normalized, and then analyzed in relation to overall survival using the univariate Cox regression method. Then, risk scores for the miRNA signature were calculated for each patient based on the sum of the levels of expression of each miRNA adjusted by hazard ratios determined by univariate Cox regression analysis. Next, patients in each dataset were divided into high- and low-risk groups using the median miRNA signature risk score as the cut-off value. This analysis was performed using the SurvMicro online tool (http://bioinformatica.mty.itesm.mx:8080/Biomatec/Survmicro.jsp), as described previously.10 Concordance indices were also calculated as an indicator of the predictive power of the miRNA signature in each dataset.
Development of a 7-miRNA set from the 12-miRNA signature using four NSCLC datasets
We further evaluated which miRNAs in the 12-miRNA set contributed most to the effectiveness of the prognostic signature. Each individual miRNA was removed from the 12-miRNA set sequentially, and prognostic values and concordance indices calculated using the SurvMicro tool. Finally, based on the extraction criterion described above, we found that at least seven miRNA biomarkers were necessary to maintain the prognostic power of the 12-miRNA signature.
NSCLC samples for experimental validation
Frozen NSCLC samples were used to experimentally confirm the prognostic value of the 7-miRNA signature using quantitative real-time PCR. The samples were collected from archived tumors obtained from NSCLC patients by surgery at the First People’s Hospital of Huzhou between July 2009 and October 2013 with the following criteria: 1) sufficient follow-up clinical information was available; 2) the patient had received no chemotherapy or radiotherapy before surgery; and 3) selected samples contained at least 70% tumor cells. A total of 40 NSCLC samples were included in the validation study. This investigation was approved by the ethics committee of the First People’s Hospital of Huzhou, and written informed consent was obtained from all individual participants included in the study.
miRNA quantitative real-time PCR procedure
Quantitative real-time PCR was carried out to determine the expression levels of signature miRNAs relative to the U6 small RNA. Briefly, total RNA was extracted from frozen NSCLC tissues using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription reaction was performed using stem-loop RT primers and a PrimeScript™ RT reagent Kit (Takara Bio, Ohtsu, Japan), according to the manufacturer’s instructions. PCR amplification was carried out on the Roche 480 Lightcycler platform (Roche Diagnostics, Indianapolis, IN, USA) using SYBR Premix Ex Taq (Takara Bio, Ohtsu, Japan). Cycling conditions were 1 cycle of 95°C for 30 seconds, followed by 40 cycles of 95°C for 5 seconds and 60°C for 30 seconds. The primers used for stem-loop qRT-PCR for miRNAs and U6 snRNA are listed in Table S2. Comparative cycle thresholds (Cp) were calculated to determine the expression levels of miRNAs relative to U6 using the 2-ΔCT method.
Survival risk prediction was performed using the supervised principal components method based on expression levels of the seven miRNAs. Ten-fold cross-validation was used to obtain the highest classification accuracy. The SurvivalROC method was used to estimate time-dependent receiver operating characteristic (ROC) curves based on the 7-miRNA signature and tumor stage. All of the above analyses were performed using BRB-Arraytools, according to the developer’s protocols.11
Statistics
Survival was compared between high- and low-risk groups using Kaplan–Meier survival curves and the log-rank test. Multivariate Cox regression analysis was used to investigate whether the miRNA signature was prognostic, independent of other known clinical factors. Sample size for the comparison of survival rates was estimated used a MadCalc (version 15.1, Ostend, Belgium) sampling module. Except where other analysis tools (eg, SurvMicro and BRB-Arraytools) are mentioned, all statistical analyses were performed using MedCalc.
Results
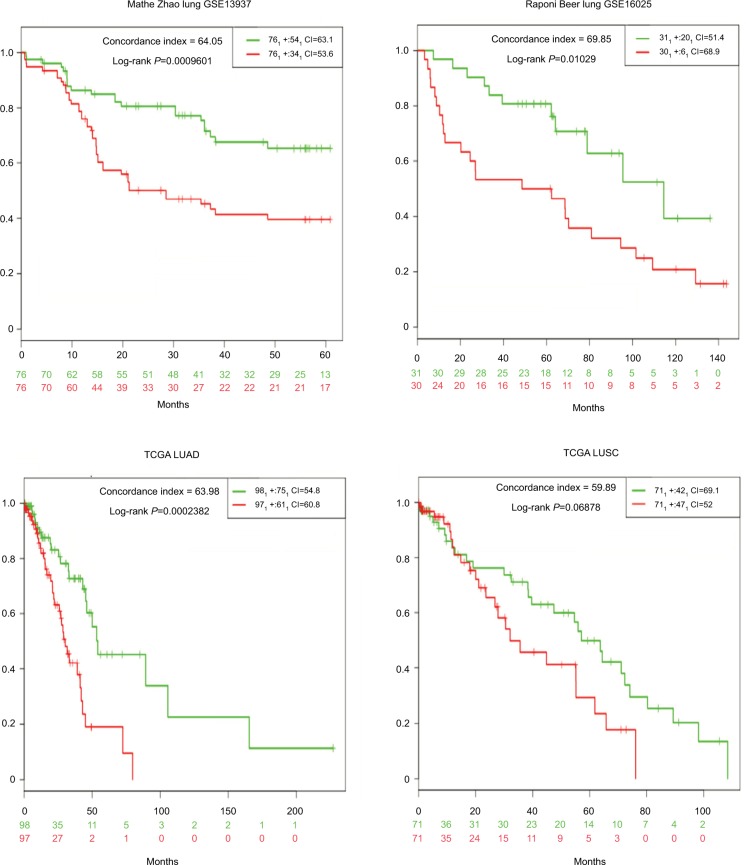
Using the SurvMicro tool, we first calculated 12-miRNA signature risk scores for all tumors in the four NSCLC miRNA datasets separately, ranked patients according to their scores, and separated them into high- or low-risk groups, using the median risk score as the cutoff point. Patients in the high-risk 12-miRNA signature groups had significantly shorter median overall survival than those in the low-risk subgroups in all datasets except for TCGA LUSC, for which the statistical significance was marginal (P=0.071) (Figure 1 and Table 1).
Figure 1.
Kaplan–Meier curves illustrating overall survival according to a 12-miRNA signature.
Notes: High-risk groups had a significantly worse prognosis compared with low-risk groups in the GSE13937, GSE16025, TCGA LUAD, but not the TCGA LUSC, datasets.
Abbreviations: LUSC, lung squamous cell carcinoma; TCGA LUAD, The Cancer Genome Atlas Lung Adenocarcinoma.
Table 1.
Prognostic significance of 12- and 7-miRNA signatures in four independent NSCLC cohorts
| Dataset | 12-miRNA signature
|
7-miRNA signature
|
||||
|---|---|---|---|---|---|---|
| Concordance index | HR (95% CI) | P-value | Concordance index | HR (95% CI) | P-value | |
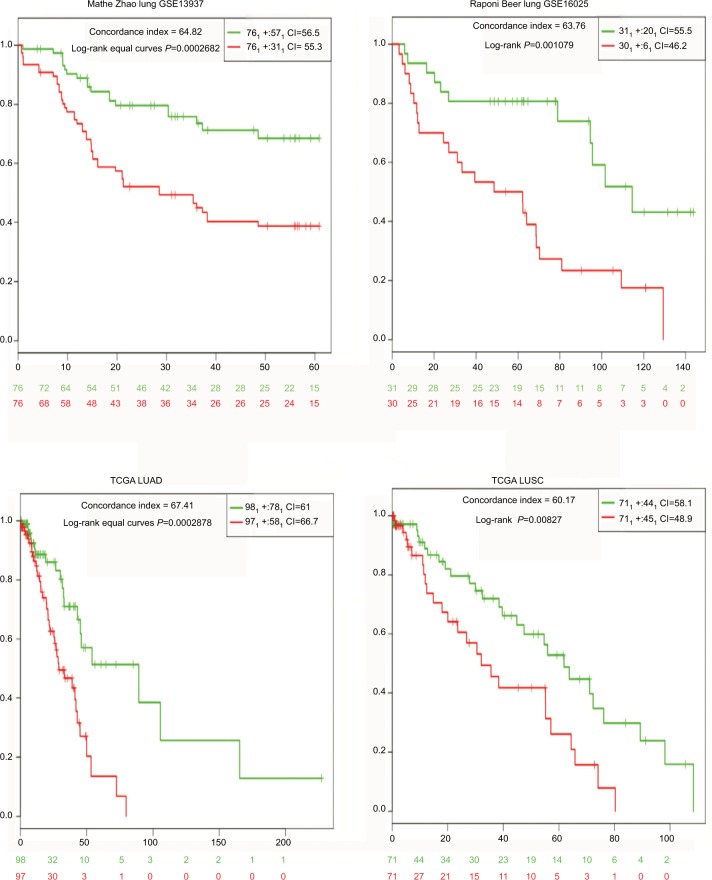
| Mathe Zhao lung GSE13937 | 64.05 | 2.32 (1.38–3.9) | 0.0014 | 64.82 | 2.6 (1.52–4.45) | 0.00027 |
| Raponi Beer lung GSE16025 | 69.85 | 2.49 (1.21–5/13) | 0.013 | 63.76 | 3.13 (1.52–6.42) | 0.0019 |
| TCGA LUAD | 63.98 | 2.78 (1.58–4.9) | 0.0004 | 67.71 | 2.82 (1.57–5.05) | 0.00049 |
| TCGA LUSC | 59.89 | 1.70 (0.95–3.04) | 0.071 | 60.17 | 2.1 (1.2–3.69) | 0.0097 |
Abbreviations: LUSC, lung squamous cell carcinoma; TCGA LUAD, The Cancer Genome Atlas Lung Adenocarcinoma.
Next, we reduced the 12-miRNA set to a 7-miRNA signature (miR-148b, miR-365, miR-32, miR-375, miR-21, miR-125b, miR-155), while retaining the prognostic efficiency in all datasets (Figure 2, Table 1). Moreover, we determined that the concordance indices of the 7-miRNA signature were similar for the different datasets. Hence, the 7-miRNA signature appears to be a more stable prognostic indicator for NSCLC than the 12-miRNA set. We also compared our 7-miRNA signature with two previously established miRNA signatures (Raponi et al,8 20 miRNAs and Yu et al,12 five miRNAs). The data presented in Table 2 demonstrate that our signature has a similar concordance index compared with that of Raponi et al, and a higher index than that of the Yu et al signature.
Figure 2.
Kaplan–Meier curves illustrating overall survival according to a 7-miRNA signature.
Notes: High-risk groups had a significantly worse prognosis compared with low-risk groups in all four datasets.
Table 2.
Comparison of our 7-miRNA signature with two other miRNA signatures in NSCLC
| Dataset | 7-miRNA signature
|
Raponi signature
|
Yu signature
|
|||
|---|---|---|---|---|---|---|
| Concordance index | P-value | Concordance index | P-value | Concordance index | P-value | |
| Mathe Zhao | 64.82 | 0.00027 | 74.76 | 1.063e-06 | 59.75 | 0.052 |
| Raponi Beer | 63.76 | 0.0019 | 77.56 | 0.00016 | 59.68 | 0.1621 |
| TCGA LUAD | 67.71 | 0.00049 | 73.56 | 8.741e-06 | 58.36 | 0.1149 |
| TCGA LUSC | 60.17 | 0.0097 | 63.63 | 0.00086 | 63.04 | 0.1089 |
Abbreviations: LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
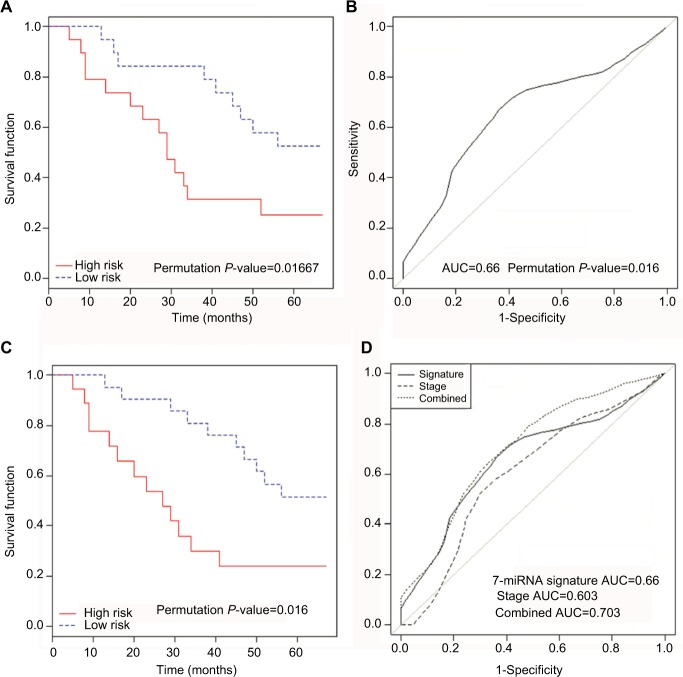
Finally, we experimentally validated the significance of the 7-miRNA signature in 40 clinical NSCLC samples by real-time PCR (power, 0.7; significance level, 0.05). Details of the NSCLC samples used in these experiments are presented in Table 3. This signature demonstrated a similar prognostic ability to discriminate patients at high-risk to that calculated using the four publicly available datasets, with an area under the curve (AUC) of 0.66 (Figure 3A and B). There were no significant associations between the 7-miRNA signature-defined risk subgroups and the clinical characteristics of patients (Table 3). Moreover, multivariate Cox regression analysis revealed that this 7-miRNA signature acted as a prognostic factor independent of tumor size, lymph node metastasis, and tumor grade (Table 4). Most interestingly, a combination of the 7-miRNA signature with tumor stage information provided superior prognostic capacity; as illustrated in Figure 3C and D, patients classified by both miRNA signature and stage exhibited a greater significant difference in overall survival, with a higher AUC (0.703).
Table 3.
Association between miRNA signature risk with clinicopathological characteristics
| Characteristics | 7-miRNA signature
|
P-value | |
|---|---|---|---|
| High-risk (n=20) |
Low-risk (n=20) |
||
| Age (years) | |||
| <60 (n=20) | 7 | 13 | 0.1138 |
| ≥60 (n=20) | 13 | 7 | |
| Gender | |||
| Male (n=36) | 20 | 16 | 0.1138 |
| Female (n=4) | 0 | 4 | |
| Histology | |||
| LUAD (n=17) | 10 | 7 | 0.5524 |
| LUSC (n=23) | 10 | 13 | |
| Tumor size | |||
| T1–2 (n=30) | 14 | 16 | 0.7150 |
| T3–4 (n=10) | 6 | 4 | |
| Lymph node metastasis | |||
| N0 (n=23) | 11 | 12 | 1.000 |
| N1 (n=17) | 9 | 8 | |
| Stage | |||
| I (n=18) | 8 | 10 | |
| II (n=10) | 4 | 6 | 0.3761 |
| III (n=12) | 8 | 4 | |
Abbreviations: LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Figure 3.
Kaplan–Meier and survivalROC plots for the experimental dataset according to the 7-miRNA signature.
Notes: The high-risk groups, as classified using the 7-miRNA signature, had a significantly worse prognosis (A), yielding a significant discriminative value (AUC, 0.66) (B). Combination of the 7-miRNA signature and tumor stage data permitted further discrimination between patients at high risk (C) with a larger AUC value (0.703), indicating superior classification performance (D).
Table 4.
Multivariate Cox regression analysis of 7-miRNA signature risk score and clinicopathological factors with overall survival
| Prognostic factors | P-value | Hazard ratio | 95% CI |
|---|---|---|---|
| Grade | 0.7911 | 1.1601 | 0.3889–3.4606 |
| Lymph node metastasis | 0.0103 | 3.2550 | 1.3281–7.9774 |
| Tumor size | 0.5791 | 0.7213 | 0.2287–2.2747 |
| 7-miRNA signature risk score | 0.0092 | 3.4847 | 1.3693–8.8680 |
Discussion
Omics-based technologies are important in establishing the utility of non-coding RNAs for evaluation of patients with cancer.13,14 In addition to the diagnostic and prognostic significance of single miRNA biomarkers or signatures, some driver miRNAs have key modulatory effects during lung cancer development and a number of miRNA signatures are associated with therapeutic responses.15,16 In this study, we developed a new 7-miRNA signature for the prediction of clinical outcome in NSCLC patients. Unlike previously reported signatures, all of the miRNAs included in this signature have confirmed prognostic value for NSCLC. The prognostic power of this signature was well replicated across four published data sets and one experimental sample set and across different platforms (RNA-seq, microarray, and real-time PCR). More meaningfully, this signature can provide additional prognostic information beyond current stage classification systems, which is a high priority for NSCLC management.
Numerous individual miRNAs have been reported as associated with the survival of patients with NSCLC; however, their predictive power in different study cohorts is heterogeneous. For example, individually, some of the seven miRNAs included in our signature failed to demonstrate significant association with survival prognosis in the four datas-ets used in this study (data not shown). Therefore, the model we built can overcome the limitations of single mi RNAs and performed well in survival prediction for patients with NSCLC. Compared with other miRNA signatures, our model has similar power; however, it includes fewer miRNAs and can be determined by real-time PCR, increasing its potential for development into a clinically applicable procedure.
According to the results of previous studies, among the miRNAs included in our model, miR-21, miR-125, and miR-155 are associated with increased risk, while miR-148b, miR-365, miR-32, and miR-375 are protective.13–22 As none of the seven miRNAs in the signature can be deleted while maintaining prognostic power, there may be no significant interaction among the individual miRNAs. All the miRNAs included in the signature have functional implications for the development of lung cancers. Determination of the regulatory mechanisms involved and the interactions of these seven miRNAs will be helpful to increase our understanding of the molecular pathogenesis of NSCLC. Comprehensive functional analyses of these seven miRNAs indicate that their target genes are highly enriched in insulin and mTOR signaling pathways. Whether or not these miRNAs can collaboratively influence the biological behaviors of lung cancer cells also warrants further exploration.
Conclusion
In conclusion, using a list of confirmed prognostic mi RNAs, we developed a 7-miRNA signature as a simple and robust tool for prediction of the prognosis for patients with NSCLC. Our findings also contribute to the understanding of the roles of miRNA interactions in NSCLC and provide a basis to develop more robust and applicable strategies to improve current classification systems for patients with lung cancer.
Supplementary materials
Table S1.
NSCLC miRNA expression profiles datasets used in our analysis
| Dataset | Platform | Samples | Ref |
|---|---|---|---|
| Mathe Zhao Lung GSE13937 | Microarray | 152 (64 ADC, 88 SCC) | 1 |
| Raponi Beer Lung GSE16025 | Microarray | 61 SCC | 2 |
| TCGA Lung Adenocarcinoma | miRNA-Seq | 195 ADC | 3 |
| TCGA Lung Squamous Cell Carcinoma | miRNA-Seq | 142 SCC | 3 |
Abbreviations: TCGA, The Cancer Genome Atlas.
Table S2.
Primers for the signature miRNAs assays by real-time PCR
| miRNA | Primer | Primer sequence |
|---|---|---|
| hsa-miR-32 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGACTGGATACGACTGCAACTT-3′ |
| Forward | 5′-GTGCAGGGTCCGAGGTATT-3′ | |
| Reverse | 5′-GCCGCTATTGCACATTACTAAGTT-3′ | |
| hsa-miR-21 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA-3′ |
| Forward | 5′-GCCCGCTAGCTTATCAGACTGATG-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-miR-125b | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACAAG-3′ |
| Forward | 5′-GCGGCTCCCTGAGACCCTAAC-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-miR-155 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCCTA-3′ |
| Forward | 5′-CTCGTGGTAATGCTAATTGTGA-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-mir-148b | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCTGA-3′ |
| Forward | 5′-GGCGCAAGTTCTGTTATACAC-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-miR-365 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATAAGG-3′ |
| Forward | 5′-CGTAATGCCCCTAAAAAT-3′ | |
| Reverse | 5′-AGCCGTTTGTTCGTTCGGCT-3′ | |
| hsa-miR-367 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACGC-3′ |
| Forward | 5′-AGCCGTTTGTTCGTTCGGCT-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| U6 snRNA | RT | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
| Forward | 5′-GCTTCGGCAGCACATATACTAAAAT-3′ | |
| Reverse | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
References
- 1.Mathé EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15(19):6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69(14):5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81501830), the Zhejiang Natural Science Foundation (LY16H160040), the Medicine and Health Project of Zhejiang Province (2015RCB028), the Zhejiang Province Science and Technology Department of Public Welfare Project (2017C33178, 2017C33187), and the Huzhou Science and Technology Fund (2014GZ11).
Footnotes
Author contributions
All authors contributed towards data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zhou C. Lung cancer molecular epidemiology in China: recent trends. Transl Lung Cancer Res. 2014;3(5):270–279. doi: 10.3978/j.issn.2218-6751.2014.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrovic N, Ergün S, Isenovic ER. Levels of microRNA heterogeneity in cancer biology. Mol Diagn Ther. 2017;21(5):511–523. doi: 10.1007/s40291-017-0285-9. [DOI] [PubMed] [Google Scholar]
- 3.Zhan B, Lu D, Luo P, Wang B. Prognostic value of expression of microRNAs in non-small cell lung cancer: a systematic review and meta-analysis. Clin Lab. 2016;62(11):2203–2211. doi: 10.7754/Clin.Lab.2016.160426. [DOI] [PubMed] [Google Scholar]
- 4.Yang M, Shen H, Qiu C, et al. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer. 2013;49(3):604–615. doi: 10.1016/j.ejca.2012.09.031. [DOI] [PubMed] [Google Scholar]
- 5.Kwan JY, Psarianos P, Bruce JP, Yip KW, Liu FF. The complexity of microRNAs in human cancer. J Radiat Res. 2016;57(Suppl 1):i106–i111. doi: 10.1093/jrr/rrw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh R, Ramasubramanian B, Kanji S, Chakraborty AR, Haque SJ, Chakravarti A. Circulating microRNAs in cancer: Hope or hype? Cancer Lett. 2016;381(1):113–121. doi: 10.1016/j.canlet.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Mathé EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esopha-gus: associations with survival. Clin Cancer Res. 2009;15(19):6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69(14):5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguirre-Gamboa R, Trevino V. SurvMicro: assessment of miRNA-based prognostic signatures for cancer clinical outcomes by multivariate survival analysis. Bioinformatics. 2014;30(11):1630–1632. doi: 10.1093/bioinformatics/btu087. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y, Simon R. BRB-ArrayTools Data Archive for human cancer gene expression: a unique and efficient data sharing resource. Cancer Inform. 2008;6:9–15. doi: 10.4137/cin.s448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 12.He B, Lu P, Guan L, et al. Identifying key regulating miRNAs in hepatocellular carcinomas by an omics’ method. Oncotarget. 2017;8(61):103919–103930. doi: 10.18632/oncotarget.21865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavridis K, Gueugnon F, Petit-Courty A, et al. The oncomiR miR-197 is a novel prognostic indicator for non-small cell lung cancer patients. Br J Cancer. 2015;112(9):1527–1535. doi: 10.1038/bjc.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu QG, Zhang SM, Ding XX, He B, Zhang HQ. Driver genes in non-small cell lung cancer: characteristics, detection methods, and targeted therapies. Oncotarget. 2017;8(34):57680–57692. doi: 10.18632/oncotarget.17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berghmans T, Ameye L, Willems L, et al. Identification of microRNA-based signatures for response and survival for non-small cell lung cancer treated with cisplatinvinorelbine A ELCWP prospective study. Lung Cancer. 2013;82(2):340–345. doi: 10.1016/j.lungcan.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 16.Zhu W, Xu B. MicroRNA-21 identified as predictor of cancer outcome: a meta-analysis. PLoS One. 2014;9(8):e103373. doi: 10.1371/journal.pone.0103373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X, Zhang S, Ma X. Prognostic value of microRNA-125 in various human malignant neoplasms: a meta-analysis. Clin Lab. 2015;61(11):1667–1674. doi: 10.7754/clin.lab.2015.150408. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Li J, Tong L, et al. The prognostic value of miR-21 and miR-155 in non-small-cell lung cancer: a meta-analysis. Jpn J Clin Oncol. 2013;43(8):813–820. doi: 10.1093/jjco/hyt084. [DOI] [PubMed] [Google Scholar]
- 19.Wang R, Ye F, Zhen Q, et al. MicroRNA-148b is a potential prognostic biomarker and predictor of response to radiotherapy in non-small-cell lung cancer. J Physiol Biochem. 2016;72(2):337–343. doi: 10.1007/s13105-016-0485-5. [DOI] [PubMed] [Google Scholar]
- 20.Sun R, Liu Z, Ma G, et al. Associations of deregulation of mir-365 and its target mRNA TTF-1 and survival in patients with NSCLC. Int J Clin Exp Pathol. 2015;8(3):2392–2399. [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Y, Wang YL, Yao WJ, et al. Expression of miR-32 in human non-small cell lung cancer and its correlation with tumor progression and patient survival. Int J Clin Exp Pathol. 2015;8(1):824–829. [PMC free article] [PubMed] [Google Scholar]
- 22.Chen LJ, Li XY, Zhao YQ, et al. Down-regulated microRNA-375 expression as a predictive biomarker in non-small cell lung cancer brain metastasis and its prognostic significance. Pathol Res Pract. 2017;213(8):882–888. doi: 10.1016/j.prp.2017.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
NSCLC miRNA expression profiles datasets used in our analysis
| Dataset | Platform | Samples | Ref |
|---|---|---|---|
| Mathe Zhao Lung GSE13937 | Microarray | 152 (64 ADC, 88 SCC) | 1 |
| Raponi Beer Lung GSE16025 | Microarray | 61 SCC | 2 |
| TCGA Lung Adenocarcinoma | miRNA-Seq | 195 ADC | 3 |
| TCGA Lung Squamous Cell Carcinoma | miRNA-Seq | 142 SCC | 3 |
Abbreviations: TCGA, The Cancer Genome Atlas.
Table S2.
Primers for the signature miRNAs assays by real-time PCR
| miRNA | Primer | Primer sequence |
|---|---|---|
| hsa-miR-32 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGACTGGATACGACTGCAACTT-3′ |
| Forward | 5′-GTGCAGGGTCCGAGGTATT-3′ | |
| Reverse | 5′-GCCGCTATTGCACATTACTAAGTT-3′ | |
| hsa-miR-21 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA-3′ |
| Forward | 5′-GCCCGCTAGCTTATCAGACTGATG-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-miR-125b | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACAAG-3′ |
| Forward | 5′-GCGGCTCCCTGAGACCCTAAC-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-miR-155 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCCTA-3′ |
| Forward | 5′-CTCGTGGTAATGCTAATTGTGA-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-mir-148b | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCTGA-3′ |
| Forward | 5′-GGCGCAAGTTCTGTTATACAC-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-miR-365 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATAAGG-3′ |
| Forward | 5′-CGTAATGCCCCTAAAAAT-3′ | |
| Reverse | 5′-AGCCGTTTGTTCGTTCGGCT-3′ | |
| hsa-miR-367 | RT | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACGC-3′ |
| Forward | 5′-AGCCGTTTGTTCGTTCGGCT-3′ | |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| U6 snRNA | RT | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
| Forward | 5′-GCTTCGGCAGCACATATACTAAAAT-3′ | |
| Reverse | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |