Abstract
Background
Liver cancer is a type of malignant tumor with high morbidity and mortality in People’s Republic of China. Its occurrence and development involve the variation and expression changes of multiple genes, and the pathogenesis and related regulatory networks are complex.
Purpose
In the present research, we investigate the involvement of NEAT1_2 and SFPQ in cisplatin resistance in liver cancer. The effects of LncRNA NEAT1 and SFPQ expression on the chemotherapeutic resistance of liver cancer cells were analyzed.
Methods
The expression level of NEAT1_2 and SFPQ mRNA in tissue specimens or cell lines were examined by RT-qPCR and western blotting. CCK-8 assay was performed to evaluate cell viability. Cell proliferation was performed using the EdU cell proliferation assay.
Results
Our data showed that increase NEAT1_2 and SFPQ expressions in liver cancer specimens were associated with the development of cisplatin resistance; high SFPQ expression level impaired patients’ survival from liver cancer. Gain-and loss-of function assay using NEAT1_2 knock-in and knock-out cells constructed using CRISPER/Cas9 system revealed that NEAT1_2 is essential for liver cancer cell survival and mediates cisplatin resistance in liver cancer cells at least partially through SFPQ. Artificial change in NEAT1_2 expression level didn’t significantly influence SFPQ transcription or translation level.
Conclusion
Our data revealed NEAT1_2—SFPQ axis as a novel cisplatin resistance mechanism in liver cancer cells in vitro.
Keywords: NEAT1, NEAT1_1, NEAT1_2, SFPQ, liver cancer, cisplatin resistance
Introduction
Liver cancer is the second leading cause of cancer related death in People’s Republic of China.1 Chemotherapy of different approaches using platinum drugs is seemingly an optimal strategy for treating late stage liver cancer,2,3 but primary or acquired platinum drug resistance remains a major obstacle in clinic practice. Alteration in the nucleotide excision repair (NER) pathway plays an important part in the development of platinum drug resistance, and the expression level of NER-related proteins has been proposed as potential biomarker for poor response to platinum-based chemotherapy.4,5 Briefly, platinum drugs exhibit anti-cancer effects by inducing DNA damage via forming a DNA adduct with inter- or intra-strand crosslinks between guanine bases, thus distorting the DNA helix, inhibiting DNA replication, eliciting DNA damage response/repair mechanisms in cells, and eventually activating the apoptosis program when the damage is unrepairable. The DNA damage response process in living cells includes cell cycle arrest, damage repair, and cell cycle restart. DNA helix distortion is caused by platinum drug-induced DNA damage triggering the activation of NER system,6 which removes the DNA lesions caused by platinum drugs via sensing and excision of the lesion followed by DNA re-synthesis and ligation.7 In platinum drug sensitive cells, the prolonged DNA damage repair process leads to P53-dependent and independent apoptosis, while those cells with hyper-activated NER machinery or overexpressed NER proteins often show resistance to platinum drug-induced cytotoxity.8–12
NEAT1 is an lncRNA that includes two transcript subtypes, NEAT1_1 and NEAT1_2. NEAT1_1 and NEAT1_2 are 3.7 kb and 23 kb in length, respectively. NEAT1_1 is part of NEAT1_2. NEAT1_1 and NEAT1_2 have a 3.7 kb overlap at the 5′ end. One of our preliminary studies screened transcript abundance changes associated with cisplatin resistance and suggested that a nuclear associated lncRNA, NEAT1_2, might be involved in the development of platinum drug resistance in liver cancer cells.17 The cancer-promoting role of NEAT1 in different cancer types, including liver cancer, with its considerable prognostic value, has been revealed by different studies,13–17 the molecular mechanism of which, however, remains largely unresolved. Despite the increasing focus on NEAT1 regulating microRNA expression, a recent report by Adriaens et al raised the possibility that NEAT1_2 encourages platinum drug resistance by reducing the accumulation of DNA damage and replication stress-induced cell death.18
As a key component in paraspeckle, a subnuclear structure involved in regulating gene expression, stress response, and cell cycle,19,20 NEAT1_2 interacts with three out of five discovered paraspeckle proteins, namely PSPC1, SFPQ, and NONO, all of which belong to the DBHS protein family and are fundamentally and dynamically dimerized.21,22 The involvement of PSPC1 and NONO in platinum drug resistance has been reported,23,24 but the role of SFPQ in this pathophysiological development remains undetermined. By constructing NEAT1_2 knock-out cell lines with or without SFPQ RNA interference, we verified the involvement of both NEAT1_2 and SFPQ in the development of cisplatin resistance in liver cancer cells. We hope the results of the present research could provide some new insight into the molecular mechanism of platinum drug resistance.
Materials and methods
Liver cancer patients and tissue specimens
This research was approved by the medical ethics committee of the First Affiliated Hospital of Zhengzhou University. Five patients diagnosed with primary liver cancer on the first visit were enrolled in the naïve group, compared to five patients enrolled in the cisplatin-resistant group who bore cisplatin-resistant primary liver cancers. Written informed consent was obtained from each patient before enrollment. All patients’ primary liver cancers were diagnosed following a practical standard in reference to American Association for the Study of Liver Diseases criteria. Patients’ liver cancer pathologic tissue specimens and non-pathologic adjacent tissue specimens were obtained by aspiration biopsy for therapeutic purposes. Determination of cisplatin-resistant patients: in patients with liver cancer after the first and second cycles of cisplatin chemotherapy, the tumor was relieved; however, in the third and fourth cycles, tumor remission was not obvious and progressed, with some side effects, such as nausea, vomiting, liver area pain, low fever, hair loss, hepatomegaly; some patients had ascites in the abdomen. The CT results showed that the cancer did not shrink, and the blood AFP increased again.
RT-qPCR and Western blotting
RT-qPCR of NEAT1_2 and SFPQ mRNA in tissue specimens or cell lines were performed using a custom-made RT-qPCR kit (GeneCopoeia, Maryland, USA), following the manufacturer’s instructions. The semi-quantitative 2−ΔΔCt method was employed for qPCR data analysis, and expression levels of NEAT1_1, NEAT1_2 or SFPQ mRNA in each sample were normalized to that of GAPDH mRNA before further analysis. Western blotting of SFPQ in different cell cultures was performed using a rabbit anti-human SFPQ monoclonal antibody (ab177149), rabbit anti-human GAPDH monoclonal antibody (ab128915) and horseradish peroxidase-conjugated goat anti-rabbit second antibody (ab205718, Abcam, Cambridge, UK), following the manufacturer’s instructions. Western blotting results were further analyzed by normalizing the gray scale of the SFPQ band in each sample to that of GAPDH using ImageJ software before statistical analysis.
Cell culture and preparation
The six liver cancer cells used in this research were previously purchased from the American Type Culture Collection and maintained in liquid nitrogen before use. Cells were cultured in DMEM medium supplemented with 10% FBS, 100 U/mL penicillin and 100 µg/mL streptomycin in a humidified sterile cell culture incubator with 37°C, 5% CO2, 100% humidity atmosphere. NEAT1 knock-out in QGY-7703 liver cancer cells at log-phase was performed using a custom-made Cas9-sgRNA NEAT1 knock-out kit (GeneCo-poeia), following the manufacturer’s instructions. Briefly, vectors loaded with Cas9 gene and sgRNA targeting the promoter sequence of NEAT1 were transfected into QGY-7703 cells, before single cell clones were isolated by serial dilutions. NEAT1 knock-out clones were selected using the IndelCheck™ kit provided with the NEAT1 knock-out kit, following the manufacturer’s instructions. NEAT1 knock-in in HUH-7 cells was performed using a custom-made AAVS1 safe harbor gene knock-in kit (GeneCopoeia), following the manufacturer’s instructions. Briefly, AAVS1 sgRNA/Cas9 expression vector and NEAT1 donor vector were co-transfected into HUH-7 cells. Positive knock-in cells were selected by neomycin screening. Transient SFPQ gene silencing was performed by shRNA targeting, using a custom-made SFPQ knockdown kit (GeneCopoeia), following the manufacturer’s instructions. Gene editing of NEAT1 or SFPQ in each cell line was verified by RT-PCR before being subjected to this research (data not shown).
Cell function assays
The CCK-8 assay was performed to evaluate cell viability, using CCK-8 reagent purchased from Beyotime (Shanghai, People’s Republic of China), following the manufacturer’s instructions. OD450 of each cell culture after addition of CCK-8 reagent was measured at different time points with a microplate reader. Annexin V/propidium iodide double staining was used for apoptosis assay, using a dead cell apoptosis kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. Cell staining of fluorescent conjugated Annexin V and propidium iodide was measured by flow cytometry. The cell proliferation assay was performed using an EdU cell proliferation detection kit (Sigma-Aldrich, St Louis, MO, USA), following the manufacturer’s instructions. Fluorescent EdU and DAPI double staining of each cell culture were evaluated microscopically. Cell proliferation rate was calculated by comparing EdU positively stained cells over all DAPI positively stained cells.
Statistical analysis
All statistical analyses and visualization in this research were performed using GraphPad Prism 7 software. Unless indicated otherwise, all data is presented as mean ± SD; Student’s t-test was used to test for significance between groups, and a P<0.05 was considered statistically significant.
Results
High NEAT1_2 and SFPQ expression correlates with cisplatin resistance and liver cancer development
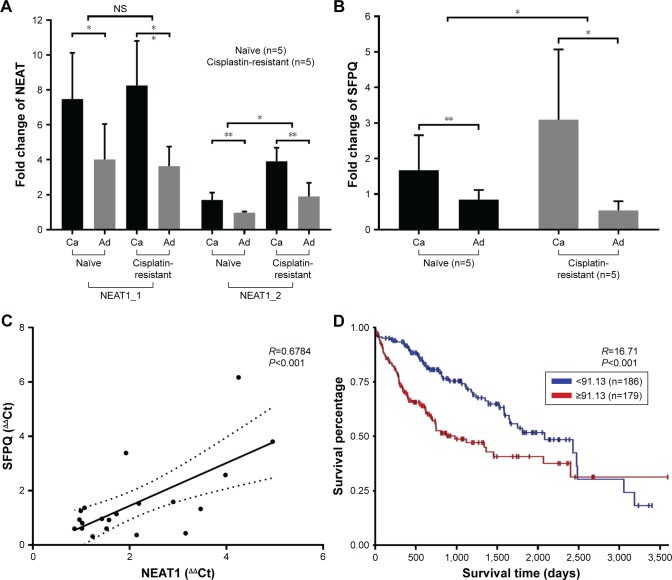
We first evaluated NEAT1_1, NEAT1_2 and SFPQ expression levels in paired liver cancer tissue specimens and non-cancerous adjacent tissue specimens obtained from five liver cancer patients who were diagnosed on the first visit (naïve) or five patients who had developed cisplatin resistance. NEAT1_1 expression increased in cancerous tissue specimens comparing to non-cancerous adjacent tissue specimens but was 2- to 3-fold lower than that of NEAT1_2, while no significant difference between specimens from first naïve patients and cisplatin resistance patients was observed (data not shown), which implied that NEAT1_1 or its upregulation might not be imperative for the development of cisplatin resistance in liver cancer. Transcriptional levels of both NEAT1_2 and SFPQ were also significantly increased in liver cancer tissue specimens, compared to non-cancerous adjacent tissue specimens, but were further upregulated in cisplatin-resistant patients’ tissue specimens (Figure 1A and B). Pearson’s correlation coefficient curve analysis showed a significant correlation between NEAT1_2 and SFPQ expression levels in all obtained tissue specimens (Figure 1C), while Kaplan–Meier curve analysis of SFPQ expression levels over liver cancer patients’ survival using TCGA public data suggested that high SFPQ expression correlates with unfavorable prognosis. The result was statistically significant, P=0.0003 (Figure 1D). These data preliminarily confirmed the relevance of NEAT1_2 and SFPQ to liver cancer development and cisplatin resistance.
Figure 1.
NEAT1_2 or SFPQ expression level change correlates with liver cancer development and cisplatin resistance.
Notes: (A and B) Comparison of transcription levels of NEAT1_2 and SFPQ in different tissue specimens; data presented as fold change comparing to naïve-adjacent group after normalization to GAPDH; one-way ANOVA was employed for statistical significance tests between naïve and cisplatin resistance groups. (C) Correlation of NEAT1_2 and SFPQ transcription levels among all obtained tissue specimens. (D) Kaplan–Meier plot of survival of liver cancer patients with different SFPQ transcription levels. Naïve refers to patients diagnosed on the first visit; cisplatin-resistance refers to patients with cisplatin-resistant primary liver cancer. *P<0.05; **P<0.01.
Abbreviations: Ca, cancerous (liver cancer tissue specimens); Ad, adjacent (adjacent non-cancerous tissue specimens); NS, normal saline.
NEAT1 promotes the proliferation of liver cancer cells in vitro
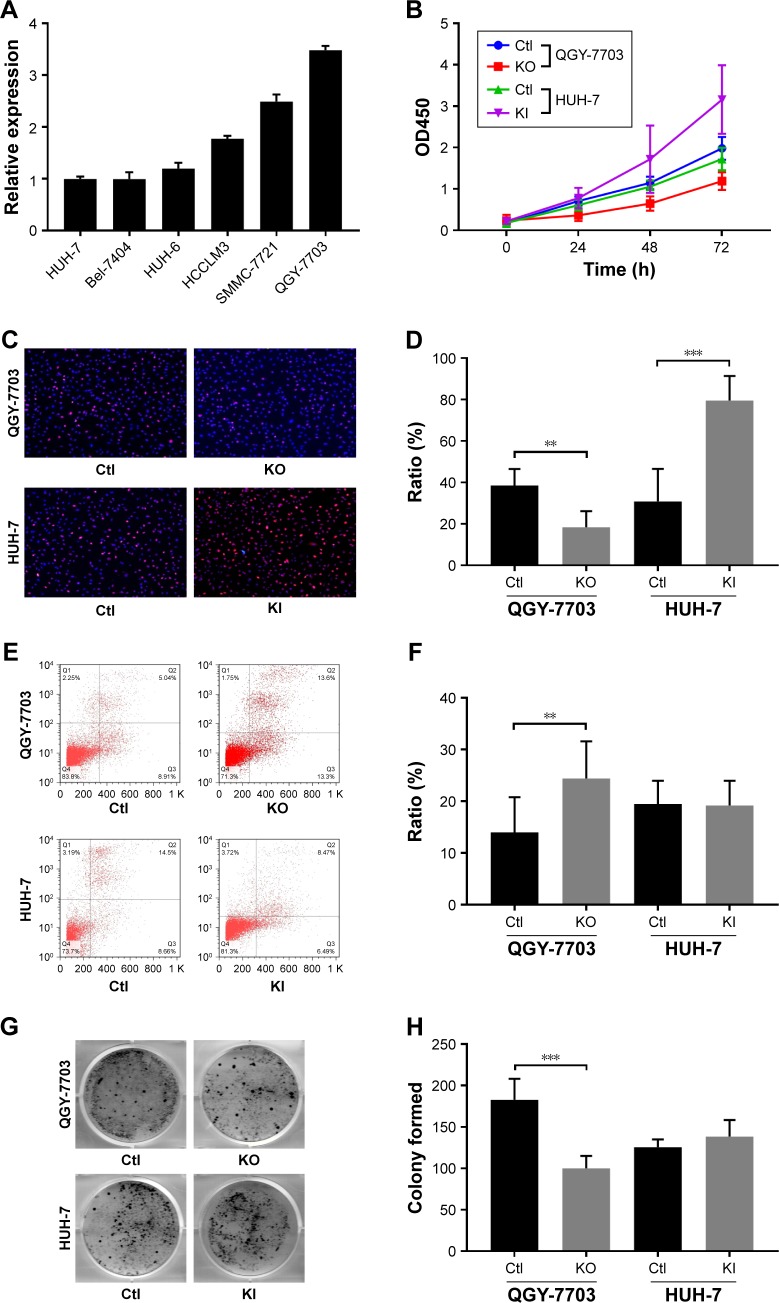
NEAT1_2 expression in six different liver cancer cell lines was first evaluated by RT-qPCR, the result of which showed that QGY-7703 had the highest NEAT1_2 expression level, while HUH-7 showed the lowest among the six liver cancer cell lines (Figure 2A). Therefore, QGY-7703 cells were chosen for NEAT1 knock-out and HUH-7 for NEAT1 knock-in before cell viability, proliferation, apoptosis, and colony formation of these two cell lines with different NEAT1_2 expression levels were evaluated. Our data showed that NEAT1 knock-out significantly decreased cell viability and cell proliferation rate of QGY-7703 cells, while NEAT1 knock-in in HUH-7 cells oppositely influenced its cell viability and proliferation rate (Figure 2B–D). NEAT1 knock-out also significantly increased cell apoptosis rate and decreased colony formation ability of QGY-7703 cells, but NEAT1_2 knock-in displayed no significant impact on that of HUH-7 cells (Figure 2E–H). These data suggested that NEAT1 might promote the proliferation of liver cancer cells and might be involved in their self-maintenance in vitro.
Figure 2.
NEAT1_2 facilitates liver cancer cell activity in vitro.
Notes: (A) NEAT1_2 expression levels in six different liver cancer cell lines, revealed by RT-qPCR. (B) Cell viability of QGY-7703 or HUH-7 cells with different NEAT1 gene modifications. (C and D) Representation and statistics of cell proliferation in different cell groups. (E and F) Representation and statistics of cell apoptosis in different cell groups. (G and H) Representation and statistics of colony formation in different cell groups. **P<0.01; ***P<0.001.
Abbreviations: Ctl, knock-out or knock-in control (wild type); KO, NEAT1 knock-out; KI, NEAT1 knock-in.
NEAT1_2 mediates cisplatin resistance in liver cancer cells partially through SFPQ
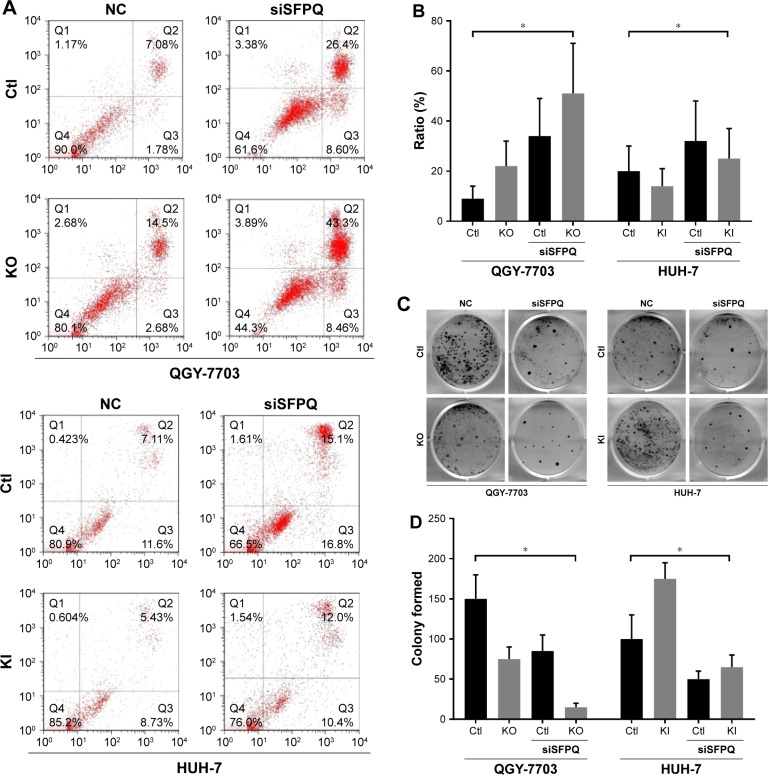
Because the enzymatic activity of any isoform of NEAT1 has not been reported, we hypothesize that NEAT1_2 does not mediate cisplatin resistance alone but interacts with other mediators. SFPQ or other paraspeckle proteins interacting with NEAT1_2 have already been demonstrated to facilitate DNA damage repair. We therefore investigated the cisplatin resistance of QGY-7703 and HUH-7 liver cancer cells with different NEAT1_2 or SFPQ expression levels. NEAT1_2 knock-out increased vulnerability of QGY-7703 cells to cisplatin cytotoxicity, which was synergized by SFPQ knockdown, while NEAT1_2 overexpression by knock-in enhanced cisplatin resistance of HUH-7 cells, which was attenuated by SFPQ knockdown (Figure 3). Knocking out NEAT1_2 and knocking down SFPQ simultaneously promotes apoptosis in QGY-7703 liver cancer cells; over-expression of NEAT1_2 and knockdown SFPQ inhibits apoptosis in HUH-7 liver cancer cells (Figure 3A and B). Inhibition of NEAT1_2 and SFPQ expression can inhibit cell colony formation in QGY-7703 liver cancer cells; NEAT1_2 knock-in and SFPQ knockdown can promote cell colony formation in HUH-7 liver cancer cells (Figure 3C and D). These data suggest that NEAT1_2 is essential for cisplatin resistance of liver cancer cells in vitro, and SFPQ is possibly part of the NEAT1_2 mediated cisplatin resistance mechanism. Our data further showed that NEAT1_2 knock-out or knock-in did not significantly change SFPQ transcriptional or translational levels in QGY-7703 or HUH-7 cells, revealed by RT-qPCR (Figure 4C) and Western blotting (Figure 4A and B), suggesting that NEAT1 facilitates cisplatin resistance in liver cancer cells but not by directly increasing increasing SFPQ expression.
Figure 3.
NEAT1_2 or SFPQ expression is essential for cisplatin resistance in liver cancer cells in vitro.
Notes: (A and B) Representation and statistics of cell apoptosis in QGY-7703 or HUH-7 cells with different NEAT1 gene modifications with the presence of 0.25 µg/mL cisplatin in culture media. (C and D) Representation and statistics of colony formation in different cell groups with same cisplatin treatment as in (A or B). *P<0.05.
Abbreviations: Ctl, knock-out or knock-in control (wild type); KO, NEAT1 knock-out; KI, NEAT1 knock-in; NC, non-specific control; siSFPQ, SFPQ knock-down by shRNA targeting.
Figure 4.
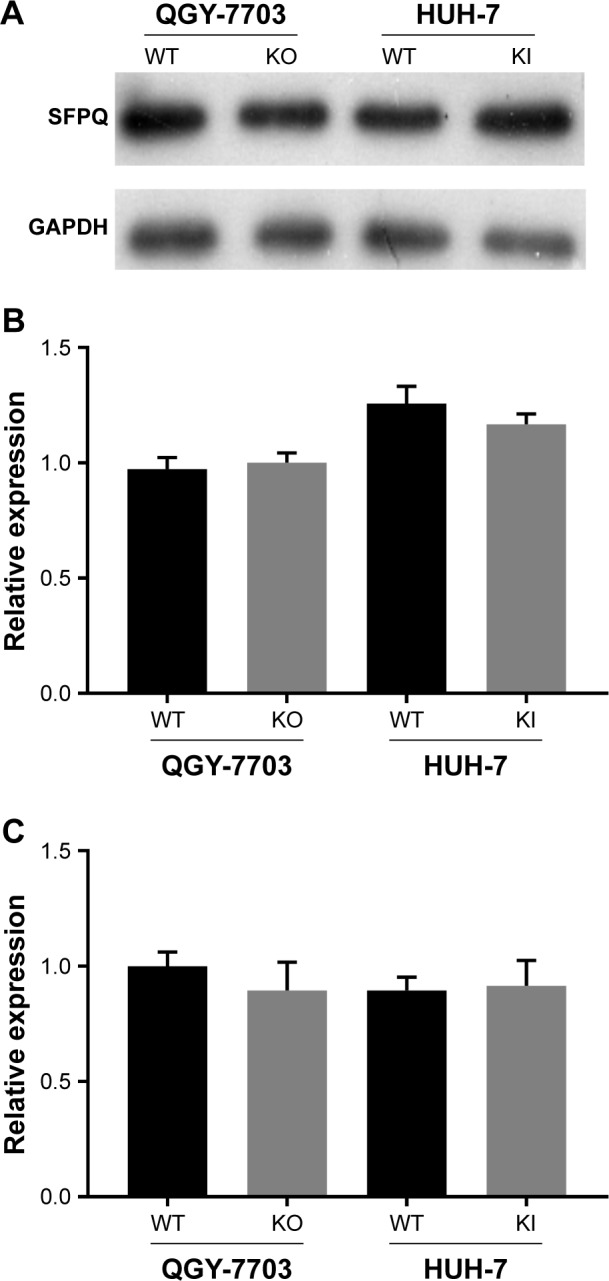
Artificial NEAT1 gene modification did not affect SFPQ transcription or translation.
Notes: (A and B) Representation and statistics of SFPQ protein expression in QGY-7703 or HUH-7 cells with different NEAT1 gene modifications. (C) Statistics of SFPQ mRNA expression in different cell groups.
Abbreviations: WT, unmodified cells; KO, NEAT1 knock-out; KI, NEAT1 knock-in.
Discussion
Chemo-resistance is a major obstacle in clinical liver cancer management. Platinum-based drugs are commonly used in standard chemotherapy, and decreasing the development of platinum drug resistance may significantly improve therapeutic outcome and patients’ survival from liver cancer. In the present research, our data clearly demonstrated that NEAT1_2 is involved in liver cancer cell chemo-resistance in vitro, possibly in part through interacting with SFPQ. We speculate that NEAT1_2 interacts with SFPQ because Imamura et al found that NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli.25
The 3.7-kb NEAT1_1 and 23-kb NEAT1_2, previously named MENε and MENβ, are the two major isoforms of non-coding RNA NEAT1. Their transcriptions are governed by the same promoter but different post-transcriptional 3′-end processing.26,27 It has been well demonstrated that NEAT1_2 but not NEAT1_1 is fundamental for the assembly and maintenance of paraspeckle, a subnuclear structure not essential for normal cell growth or mammalian development but formed in response to DNA damage and that paraspeckle promotes oncogenesis, tumor progression and chemo-resistance.18,19,24–28 In the present research, we first investigated NEAT1_1 and NEAT1_2 expression levels in different tissue specimens obtained from liver cancer patients that were diagnosed on the first visit to our department or burdened with cisplatin-resistant liver cancer. Our data clearly demonstrated that NEAT1_2 but not NEAT1_1 is the dominant NEAT1 isoform for oncogenesis and cisplatin resistance of liver cancer, which was further confirmed by the gain-of-function and loss-of-function assay in vitro. Targeting NEAT1_2 but not NEAT1_1 on its specific 3′ region by shRNA achieved similar but more transient results (data not shown) compared to the Cas9/sgRNA-based genome editing method we used for the present research.
Cancer cells are known for their genome instability and high mutagenesis rate, which is probably due to the continuous cellular stress caused by altered cell proliferation and metabolism machineries, and a strengthened genome repair mechanism is therefore required for preventing cell death caused by the accumulation of DNA damage. Targeting the DNA damage repair pathway has been proposed as a promising strategy against chemo-resistance of different types of cancers.29–31 SFPQ, NONO, and PSPC1 are three characteristic paraspeckle proteins that have been proposed to facilitate DNA damage repair and the consequential chemo- or radio-resistance in cancer cells via seemingly different mechanisms.22–24,32,33 In the present research, our data suggested that NEAT1_2 and SFPQ synergistically promote cisplatin resistance in liver cancer cells in vitro, but change in NEAT1 expression did not affect SFPQ mRNA transcription or protein translation. We therefore speculated that NEAT1_2 supports cisplatin resistance in liver cancer cells possibly by functioning as a scaffold for adaptor and effector paraspeckle proteins required for DNA damage repair.
Paraspeckle is a complex protein aggregate. Naganuma et al identified 36 novel paraspeckle proteins, many of which carry RNA binding motifs.26 Paraspeckle has been linked to tumor progression in different aspects by several different studies. While paraspeckle is seemingly unnecessary for homeostasis, the molecular mechanisms of its cancer promoting role remain unresolved. NEAT1_2 is for now the only RNA molecule identified in paraspeckle and is vital for its integrity; thus, targeting this lncRNA may be a possible therapeutic strategy for tumor management with low collateral damage, especially for advanced tumors with metastasis or chemo-resistance.
Acknowledgments
This research was funded by the 2016 Henan Science and Technology Innovation Outstanding Talent Project (No 164200510010).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 2.Chang L, Wang Y, Zhang J, Guo T. The best strategy for HCC patients at each BCLC stage: a network meta-analysis of observational studies. Oncotarget. 2017;8(12):20418–20427. doi: 10.18632/oncotarget.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: Chemoembolization improves survival. Hepatology. 2003;37(2):429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- 4.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res. 2008;14(5):1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 5.Bowden NA. Nucleotide excision repair: why is it not used to predict response to platinum-based chemotherapy? Cancer Lett. 2014;346(2):163–171. doi: 10.1016/j.canlet.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Chu G. Cellular responses to cisplatin. The roles of DNA-binding proteins and DNA repair. J Biol Chem. 1994;269(2):787–790. [PubMed] [Google Scholar]
- 7.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10(11):756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 8.Portich JP, dos Santos RP, Kersting N, et al. DNA damage response in patients with pediatric Acute Lymphoid Leukemia during induction therapy. Leuk Res. 2017;54:59–65. doi: 10.1016/j.leukres.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Qiu ZQ, Zhao K. Expression of ERCC1, RRM1 and LRP in non-small cell lung cancers and their influence on chemotherapeutic efficacy of gemcitabine concomitant with nedaplatin. Asian Pac J Cancer Prev. 2014;15(17):7303–7307. doi: 10.7314/apjcp.2014.15.17.7303. [DOI] [PubMed] [Google Scholar]
- 10.Hirakawa M, Sato Y, Ohnuma H, et al. A phase II study of neoadjuvant combination chemotherapy with docetaxel, cisplatin, and S-1 for locally advanced resectable gastric cancer: nucleotide excision repair (NER) as potential chemoresistance marker. Cancer Chemother Pharmacol. 2013;71(3):789–797. doi: 10.1007/s00280-013-2073-5. [DOI] [PubMed] [Google Scholar]
- 11.Brevik A, Gaivão I, Medin T, et al. Supplementation of a western diet with golden kiwifruits (Actinidia chinensis var.‘Hort 16A’:) effects on biomarkers of oxidation damage and antioxidant protection. Nutr J. 2011;10:54. doi: 10.1186/1475-2891-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Booton R, Ward T, Heighway J, et al. Xeroderma pigmentosum group D haplotype predicts for response, survival, and toxicity after platinum-based chemotherapy in advanced nonsmall cell lung cancer. Cancer. 2006;106(11):2421–2427. doi: 10.1002/cncr.21885. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Lun L, Li H, et al. The Value of lncRNA NEAT1 as a Prognostic Factor for Survival of Cancer Outcome: A Meta-Analysis. Sci Rep. 2017;7(1):13080. doi: 10.1038/s41598-017-10001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, Li Z, Li Y, et al. Long non-coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: a systematic review and meta-analysis. Oncotarget. 2017;8(2):2672–2680. doi: 10.18632/oncotarget.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo PK, Wolfson B, Zhou Q, Pk L, Cellular ZQ. Cellular, physiological and pathological aspects of the long non-coding RNA NEAT1. Front Biol. 2016;11(6):413–426. doi: 10.1007/s11515-016-1433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen T, Wang H, Yang P, He ZY, Zy H. Prognostic role of long non-coding RNA NEAT1 in various carcinomas: a meta-analysis. Onco Targets Ther. 2017;10:993–1000. doi: 10.2147/OTT.S128588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Chang Q, Yang F, et al. Long non-coding RNA NEAT1 over-expression is associated with unfavorable prognosis in patients with hepatocellular carcinoma after hepatectomy: A Chinese population-based study. Eur J Surg Oncol. 2017;43(9):1697–1703. doi: 10.1016/j.ejso.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Adriaens C, Standaert L, Barra J, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22(8):861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 19.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10(3):456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki YT, Hirose T. How to build a paraspeckle. Genome Biol. 2009;10(7):227. doi: 10.1186/gb-2009-10-7-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2(7):a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knott GJ, Bond CS, Fox AH. The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 2016;44(9):3989–4004. doi: 10.1093/nar/gkw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaafar L, Li Z, Li S, Dynan WS. SFPQ•NONO and XLF function separately and together to promote DNA double-strand break repair via canonical nonhomologous end joining. Nucleic Acids Res. 2017;45(4):1848–1859. doi: 10.1093/nar/gkw1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao X, Kong L, Lu X, et al. Paraspeckle protein 1 (PSPC1) is involved in the cisplatin induced DNA damage response – role in G1/S checkpoint. PLoS One. 2014;9(5):e97174. doi: 10.1371/journal.pone.0097174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. Embo J. 2012;31(20):4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193(1):31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, Harvey AR, Hodgetts SI, Fox AH. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA. 2017;23(6):872–881. doi: 10.1261/rna.059477.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolova T, Kiweler N, Krämer OH. Interstrand Crosslink Repair as a Target for HDAC Inhibition. Trends Pharmacol Sci. 2017;38(9):822–836. doi: 10.1016/j.tips.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Mateo J, Boysen G, Barbieri CE, et al. DNA Repair in Prostate Cancer: Biology and Clinical Implications. Eur Urol. 2017;71(3):417–425. doi: 10.1016/j.eururo.2016.08.037. [DOI] [PubMed] [Google Scholar]
- 31.Foy V, Schenk MW, Baker K, et al. Targeting DNA damage in SCLC. Lung Cancer. 2017;114:12–22. doi: 10.1016/j.lungcan.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Li Z, Shu FJ, Xiong H, Phillips AC, Dynan WS. Double-strand break repair deficiency in NONO knockout murine embryonic fibroblasts and compensation by spontaneous upregulation of the PSPC1 paralog. Nucleic Acids Res. 2014;42(15):9771–9780. doi: 10.1093/nar/gku650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9(8):1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]