Abstract
Background
Glioblastoma (GBM) is a difficult to treat brain cancer that nearly uniformly recurs, and recurrent tumors are largely therapy resistant. Our prior work has demonstrated an important role for the tumor necrosis factor–like weak inducer of apoptosis (TWEAK) receptor fibroblast growth factor–inducible 14 (Fn14) in GBM pathobiology. In this study, we investigated Fn14 expression in recurrent GBM and in the setting of temozolomide (TMZ) resistance.
Methods
Fn14 mRNA expression levels in nonneoplastic brain, primary (newly diagnosed) GBM, and recurrent GBM (post-chemotherapy and radiation) specimens were obtained from The Cancer Genome Atlas data portal. Immunohistochemistry was performed using nonneoplastic brain, patient-matched primary and recurrent GBM, and gliosarcoma (GSM) specimens to examine Fn14 protein levels. Western blot analysis was used to compare Fn14 expression in parental TMZ-sensitive or matched TMZ-resistant patient-derived xenografts (PDXs) established from primary or recurrent tumor samples. The migratory capacity of control and Fn14-depleted TMZ-resistant GBM cells was assessed using the transwell migration assay.
Results
We found that Fn14 is more highly expressed in recurrent GBM tumors than their matched primary GBM counterparts. Fn14 expression is also significantly elevated in GSM tumors. GBM PDX cells with acquired TMZ resistance have higher Fn14 levels and greater migratory capacity than their corresponding parental TMZ-sensitive cells, and the migratory difference is due, at least in part, to Fn14 expression in the TMZ-resistant cells.
Conclusions
This study demonstrates that the Fn14 gene is highly expressed in recurrent GBM, GSM, and TMZ-resistant GBM PDX tumors. These findings suggest that Fn14 may be a valuable therapeutic target or drug delivery portal for treatment of recurrent GBM and GSM patients.
Keywords: Fn14, glioblastoma, gliosarcoma, migration, temozolomide
Importance of the study
The current treatments for GBM patients do not fully eradicate the invasive cancer cells that remain following tumor resection, resulting in most patients developing recurrent disease that is almost universally fatal. Previous studies have established that the TWEAK receptor Fn14 is overexpressed in primary GBM tumors and in GBM cells invading the normal brain parenchyma. In this study, we report that Fn14 is actually expressed at higher levels in recurrent GBM tumors compared with patient-matched, untreated primary tumors, and is also overexpressed at particularly high levels in GSM, an aggressive variant of GBM. Additionally, we show that Fn14 expression is higher in GBM PDX cells with acquired resistance to TMZ (GBM-TMZ) compared with their parental TMZ-sensitive counterparts and that Fn14 contributes to GBM-TMZ cell migration. Collectively, these findings that Fn14 may have suggest an important role in recurrent and highly aggressive GBMs, and may be a valuable receptor for targeted therapeutics.
Glioblastoma (GBM) is the most common and deadly adult primary brain cancer. There are approximately 15000 new cases of GBM per year in the USA.1,2 The current standard of care for patients with GBM consists of maximal safe surgical resection followed by high-dose radiation and concomitant oral chemotherapy using the DNA alkylating agent temozolomide (TMZ).3,4 Despite this combination treatment strategy, tumor recurrence is nearly universal and the median overall survival of GBM patients is ~18 months after diagnosis.3,4 GBM is one of the most difficult to treat human cancers because extensive tumor cell invasion into surrounding functioning brain tissue prevents complete surgical removal of tumor cells and limits cytotoxic treatments, which risk injuring nearby neural components.5–9
Recurrent GBM tumors tend to be more aggressive and treatment resistant than their primary (ie, newly diagnosed) counterparts,9,10 and generally only 20%–30% of patients with these tumors are considered eligible for repeat surgery.10 Although there have been numerous clinical trials testing chemotherapeutics and molecularly targeted agents in the recurrent disease setting,11 only 2 pharmaceutical treatments have been approved for recurrent GBM: Gliadel, a surgically implanted, biodegradable chemotherapy (carmustine) wafer,12 and bevacizumab, a monoclonal antibody targeting the pro-angiogenic factor vascular endothelial growth factor-A.13 Neither of these treatments was specifically designed to treat unique features of recurrent GBM or has had a major impact on patient survival.10 A better understanding of recurrent GBM biology would offer new opportunities for tailoring treatments following standard-of-care chemoradiation therapy. Toward this goal, recent genomic and transcriptomic studies have shown that this therapy can promote the evolution of hypermutated, therapy-resistant tumor cell populations, frequently subclonal and stemlike in nature, that drive tumor recurrence.9,14–19 In particular, TMZ treatment introduces genetic alterations and biological changes into GBM cells, with a high frequency of mutations affecting DNA mismatch repair,18,20 tumor suppressor,17,18,21,22 and phosphatidylinositol-3 kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) signaling pathway19,22 genes.
We have reported that the tumor necrosis factor receptor (TNFR) family member fibroblast growth factor–inducible 14 (Fn14)23–25 is expressed at low levels in nonneoplastic brain but upregulated in malignant brain cancers,26–28 especially GBM tumors exhibiting the highly aggressive and invasive mesenchymal molecular subtype.28 Additionally, Fn14 is significantly overexpressed in the invasive cells that are responsible for tumor recurrence.27–29 In this study, we report for the first time that (i) Fn14 is highly expressed in recurrent GBM tumors and gliosarcomas (GSMs), (ii) Fn14 expression in GBM patient-derived xenograft (PDX) tumor tissue increases with TMZ exposure in vivo, and (iii) GBM PDX cells with acquired TMZ resistance have higher migratory capacity than their corresponding parental TMZ-sensitive cells, and this difference is due, at least in part, to their higher levels of Fn14 gene expression.
Materials and Methods
Nonneoplastic Brain, Primary GBM, and Recurrent GBM mRNA Expression Analysis
Fn14 (TNFRSF12A) mRNA expression data corresponding to 10 nonneoplastic brain, 535 primary GBM, and 13 recurrent GBM specimens were downloaded from the data portal of The Cancer Genome Atlas (TCGA) on February 18, 2018 (level 3 data). Patients included in this analysis were limited to the provisional TCGA microarray dataset with gene expression availability for the Fn14 gene. Primary GBM and recurrent GBM samples were selected by filtering the “Sample Type” column in the patient clinical information table. Gene expression data are presented as normalized z-scores, which reflect the number of standard deviations away from the mean of expression in the reference population. In this context, the reference population refers to either all tumors that are diploid for the gene in question, or matched nonneoplastic brain tissue. A univariate analysis of survival was performed using the Kaplan–Meier method and compared by the log-rank test. The upper and lower quartiles of Fn14 expression for the primary GBM specimens were used for comparison. “Fn14 low” expressors were defined as patients with Fn14 expression levels less than the lower quartile of all patients, while “Fn14 high” expressors were defined as patients with Fn14 expression levels greater than the upper quartile.
Immunohistochemistry of Tissue Specimens
We obtained formalin-fixed, paraffin-embedded, nonneoplastic brain specimens, patient-matched primary and recurrent GBM specimens (8 patients), and GSM specimens (3 patients) from the University of Maryland School of Medicine Department of Pathology and prepared tissue sections (5 µm). Approval by the University of Maryland School of Medicine institutional research board was obtained prior to collecting the archived tissue. Patient characteristics and treatment regimens are summarized in Supplementary Table S1. The specimens were immunostained using the UltraVision Quanto horseradish peroxidase detection system (Thermo Fisher Scientific). After routine deparaffinization with a series of xylene and alcohols, antigen retrieval was performed using 90% formic acid. Slides were then rinsed with distilled H2O and wash buffer. Endogenous peroxidase activity was blocked with H2O2 solution (TA-125-HP, Thermo Fisher Scientific) for 10 min prior to incubation with a rabbit anti-TWEAKR (Fn14) monoclonal antibody (Abcam) at 1:200 for 60 min at room temperature. The primary antibody signal was developed with Quanto detection reagents and 3,3ʹ-diaminobenzidine chromogen as per the manufacturer’s instructions. Virtual slides were produced by scanning the immunohistochemical (IHC) glass slides using the Aperio CS2 digital pathology scanner (Leica Biosystems). Digital quantitative analysis of Fn14 immunoreactivity in neoplastic cells was performed by a neuropathologist (J.E.H.) in a blinded manner with Aperio ImageScope software v12.2.2.5015 (Leica Biosystems) using a customized positive pixel count algorithm. Stain intensity values are provided as an “Fn14 Score” using a linear scale.
Western Blot Analysis of GBM PDX Tumor Tissue and Cells
TMZ-sensitive and TMZ-resistant PDX subcutaneous tumor samples as well as GBM12 cells were provided by Dr Jann Sarkaria (Mayo Clinic Arizona). The parental TMZ-sensitive lines were derived from primary or recurrent tumor resections. Short tandem repeat analysis was used to compare the original patient tumor sample, when available, with the derivative parental PDX model to ensure the provenance of the PDX line. The O6-methylguanine-DNA methyltransferase promoter methylation status and TMZ sensitivity of the parental PDX lines have been described,30,31 and a complete description of the parental lines can be found at the Mayo Clinic Brain PDX National Resource Portal.32 The TMZ-resistant PDX lines were established by subjecting parental TMZ-sensitive GBM subcutaneous xenografts to in vivo selection with escalating TMZ doses (20 mg/kg/d for 3 days, followed by 66 mg/kg/d for 3 days after initial tumor regrowth).33 The resulting TMZ-resistant tumor lines were completely resistant to a 120 mg/kg/d dose over a 5 day treatment period. Mouse survival studies confirmed that this TMZ selection method generated highly TMZ-resistant tumor lines.33 All mouse experiments were conducted in accordance with protocols approved by the Mayo Clinic Institutional Animal Care and Use Committee and followed NIH guidelines for animal welfare.
Tumor tissue and GBM12 cells were lysed in 1× radioimmunoprecipitation assay buffer containing 1 mM phenylmethylsulfonyl fluoride. Lysates were then sonicated and total protein concentration was determined using the bicinchoninic acid assay kit (Pierce), using bovine serum albumin as a standard. Equal protein amounts were loaded per well, and sodium dodecyl sulfate–polyacrylamide gel electrophoresis was performed. Protein was transferred to a nitrocellulose membrane (Invitrogen) using the Bio-Rad Trans-Blot Turbo Transfer System, followed by blocking for 1 h with Odyssey Blocking Buffer (LI-COR Biosciences). Primary antibodies detecting Fn14 (Abcam) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Cell Signaling Technology) were diluted in Odyssey Blocking Buffer containing 0.2% Tween 20 and incubated overnight with gentle shaking at 4°C. An IRDye 800CW secondary antibody was used to detect primary antibodies and was visualized using the Odyssey CLx Near-Infrared Western Blot Detection System (LI-COR Biosciences). Densitometric analysis of the western blot data was performed using Image Studio Lite v5.2 or ImageJ software, and all Fn14 expression values were normalized to GAPDH values.
Transwell Migration Assay
GBM12 and GBM12 TMZ-resistant cells (GBM12 TMZ) cells were seeded in 60 mm diameter culture dishes and incubated overnight at 37 °C. GBM12 TMZ cells were transiently transfected with nontargeting (control) or Fn14 small interfering (si)RNA (10 nM) using Lipofectamine RNAiMax (Thermo Fischer Scientific) following the manufacturer’s protocol. At 48 h posttransfection, the cells were serum starved for 16 h at 37°C. Cells were then harvested and plated in Costar collagen-coated transwell chambers (8 μm pore size) and allowed to migrate in the presence of 10% fetal bovine serum. After incubation for 4 h at 37°C, nonmigrated cells were scraped off the upper side of the membrane and cells that migrated to the other side of the membrane were fixed with 4% paraformaldehyde (Affymetrix) and stained with 4′,6′-diamidino-2-phenylindole (Invitrogen). Nuclei of migrated cells were counted in 5 high power fields with a 10x objective. Data represent the average of triplicate transwells.
Statistics
For TCGA data mining results, statistical analysis was performed using an analysis of variance (ANOVA) with a post-hoc Tukey’s honest significance test (expression analysis) or the log-rank test (survival data). For the Fn14 IHC staining and the western blot signal quantitation results, statistical analysis was performed using 2-way ANOVA. For the migration assay, significance was measured by Student’s t-test. P-values <0.05 were considered significant. All tests were performed using JMP Pro 12 (SAS Institute), GraphPad Prism, or R 3.2.2 software.
Results
Fn14 mRNA Is Overexpressed in Both Primary and Recurrent GBM Tumors Compared with Nonneoplastic Brain and High Fn14 mRNA Expression Levels in Primary GBM Tumors Correlate with Poor Patient Outcome
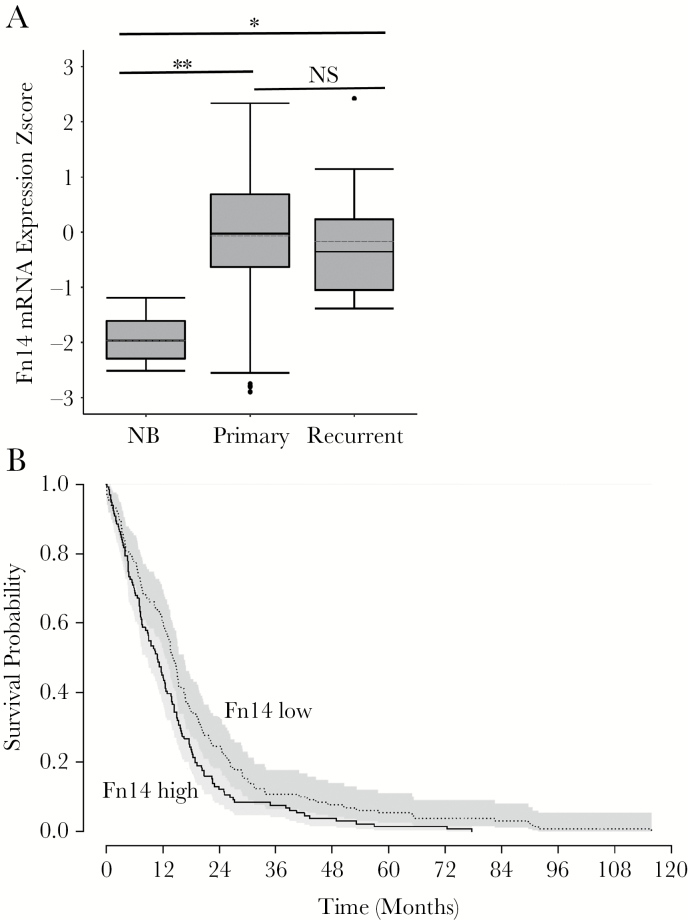
We first determined the relative level of Fn14 mRNA in nonneoplastic brain, primary GBM, and recurrent GBM tissue specimens (not patient-matched) by mining the GBM data portal of TCGA. Fn14 expression was significantly higher in both primary and recurrent GBM specimens in comparison to nonneoplastic brain tissue (Fig. 1A). Fn14 mRNA levels were not significantly different between the primary and recurrent GBM tumors. We next performed a patient survival analysis using the primary GBM data and an algorithm based on the log-rank test. We compared the upper and lower quartiles of Fn14 expression and found that high Fn14 expression correlates with poor survival (P < 0.01) (Fig. 1B).
Fig. 1.
Analysis of Fn14 mRNA expression levels in nonneoplastic brain, primary GBM tumors, and recurrent GBM tumors. (A) Fn14 (queried as TNFRSF12A) mRNA expression data in 10 nonneoplastic brain (NB), 535 primary GBM, and 13 recurrent GBM specimens were downloaded from the data portal of TCGA and converted to z-scores. Patients used in this analysis were limited to the provisional TCGA dataset with gene expression availability for the Fn14 gene. Patient/sample sets for each expression cluster were plotted as box-and-whisker plots. The whiskers of the plot map the maximum and minimum z-score for each expression cluster. The bar and dotted bar in each box represent the median and mean value, respectively, for the expression z-score of each group. The top and bottom of each box represent the 25th and 75th percentiles, respectively, of the expression z-score values for each group. Fn14 expression was significantly different in NB vs primary GBM (**P < 0.0001) as well as in NB vs recurrent GBM (*P < 0.005) as determined using an ANOVA with a post-hoc Turkey’s test. However, Fn14 expression was not significantly different (NS, P = 0.939) in primary GBM vs recurrent GBM. (B) Kaplan–Meier survival curves were generated comparing primary GBM patients with low or high Fn14 mRNA expression. Comparisons were performed using the log-rank test with upper and lower quartiles of Fn14 expression (P < 0.01).
Fn14 Protein Is Highly Expressed in Recurrent GBM and Gliosarcoma Tumors
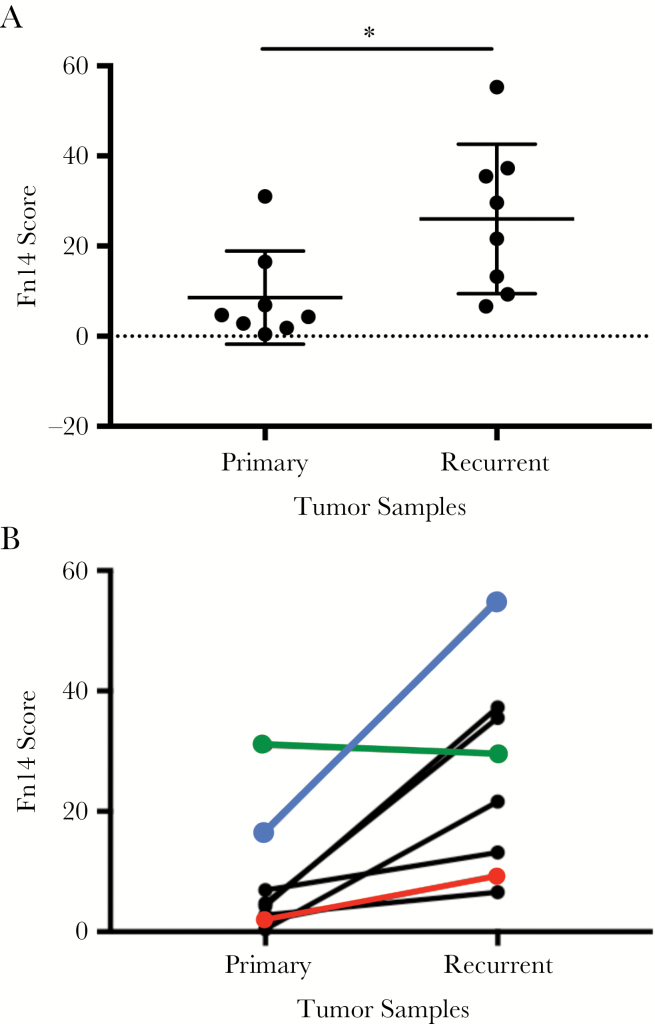
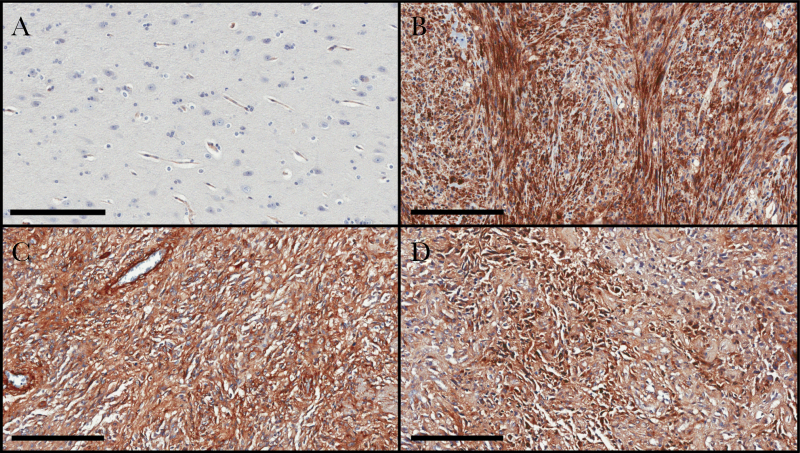
We obtained archived patient-matched primary and recurrent GBM specimens (8 patients) and performed IHC analysis using an Fn14 monoclonal antibody. Patient characteristics and treatment information can be found in Supplementary Table S1. Fn14 expression was detected in both tumor cells and vascular cells. Quantitative analysis of Fn14 immunoreactivity in tumor cells was performed using the Aperio ImageScope slide viewing software. We found that Fn14 expression was significantly higher in the recurrent tumors in comparison to the newly diagnosed primary tumors (Fig. 2A). Additionally, 7 of the 8 patients in this study exhibited higher levels of Fn14 expression at relapse (Fig. 2B). Representative IHC images from 3 of the patients are shown in Fig. 3. Of note, for patient #6, the recurrent tumor had histopathological features that are typical of GSM, a rare variant of GBM that can develop de novo (primary GSM) or following standard-of-care treatment (secondary GSM).34,35 Since Fn14 overexpression in GSM had not yet been reported in the literature, we performed additional IHC analyses comparing Fn14 levels in nonneoplastic brain tissue and 3 additional GSM specimens (1 secondary GSM and 2 primary GSMs). In the nonneoplastic brain specimen, we detected Fn14 expression in vascular cells, consistent with our previous studies.27,28 In contrast, Fn14 was highly expressed in all 3 GSM patient specimens, with expression found in both the glial and mesenchymal tumor areas (Fig. 4).
Fig. 2.
Quantitative analysis of Fn14 protein expression in patient-matched primary and recurrent GBM tumors. (A) Patient-matched primary and recurrent specimens were immunostained with an Fn14 antibody and stain intensity was quantitated using the Aperio imaging system. Each specimen is indicated by a black circle. Horizontal bars indicate the median (long bar) and interquartile (short bar) range. *P = 0.01 using 2-way ANOVA. (B) Fn14 staining intensity for the 8 sets of resected tumors is indicated. Colored lines are used for the 3 patients whose immunostaining results are included in Fig. 3 (patient #8, red line; patient #3, green line; patient #6, blue line).
Fig. 3.
Representative examples of Fn14 protein expression levels in patient-matched primary and recurrent GBM tumors. The Fn14 immunostaining results of the 3 patient specimens indicated by the colored lines in Fig. 2B are shown. The Fn14-positive cells are brown (scale bar = 200 µm).
Fig. 4.
Analysis of Fn14 protein expression levels in nonneoplastic brain and GSM tumors. (A) One nonneoplastic brain tissue specimen from an epilepsy patient, (B) 1 secondary GSM patient specimen (ie, the patient was initially diagnosed with GBM, but 5 years later the tumor recurred as GBM with focal features of GSM), and (C–D) 2 primary GSM patient specimens were immunostained with an Fn14 antibody. The Fn14-positive cells are brown (scale bar = 200 µm).
Fn14 Expression in GBM PDX Tumor Tissue Increases with TMZ Exposure In Vivo
Prior evidence demonstrates that adjuvant TMZ therapy produces subclonal, drug-resistant GBM cells that constitute the recurrent tumor mass.9,14–19 We have shown that Fn14 signaling can promote TMZ resistance.29,36,37 In fact, Fn14 knockdown in a GBM PDX model prolonged the survival of animals in combination with TMZ treatment.36 Thus, glioma cells with high Fn14 levels, and as a consequence active Fn14 signaling,27,38,39 may have a survival advantage in the setting of TMZ treatment. To investigate whether TMZ exposure generated TMZ-resistant tumors with elevated Fn14 gene expression, we compared Fn14 levels in 9 pairs of matched TMZ-sensitive or TMZ-resistant GBM PDX subcutaneous tumor tissue samples by western blot analysis. We found that Fn14 expression was at least 1.5-fold higher in 6/9 (67%) tumors exhibiting acquired TMZ resistance compared with their matched parental counterpart
(Fig. 5A–C). When all 9 PDX tumor pairs were considered, there was a statistically significant difference in Fn14 expression between the TMZ-sensitive and -resistant tumors (Fig. 5D).
Fig. 5.
Analysis of Fn14 protein expression levels in parental and TMZ-resistant GBM PDX tumor tissue. Parental TMZ-sensitive (P) and TMZ-resistant (T) PDX subcutaneous tumors derived from (A) primary tumor resections or (B) recurrent tumor resections were isolated and lysed. Fn14 and GAPDH levels were then examined by western blot analysis. (C) Densitometry was used to quantitate the western blot results. Fn14 levels were normalized to GAPDH levels and presented as fold expression relative to parental cells. (D) Densitometry results are plotted as the Fn14/GAPDH ratio with each PDX tumor indicated by a black circle. Horizontal bars indicate the median (long bar) and interquartile (short bar) range. *P = 0.02 using 2-way ANOVA.
GBM12 TMZ-Resistant Cells Have Increased Migratory Capacity Compared with Their Parental Counterpart and This Difference Is Abrogated by Fn14 Depletion
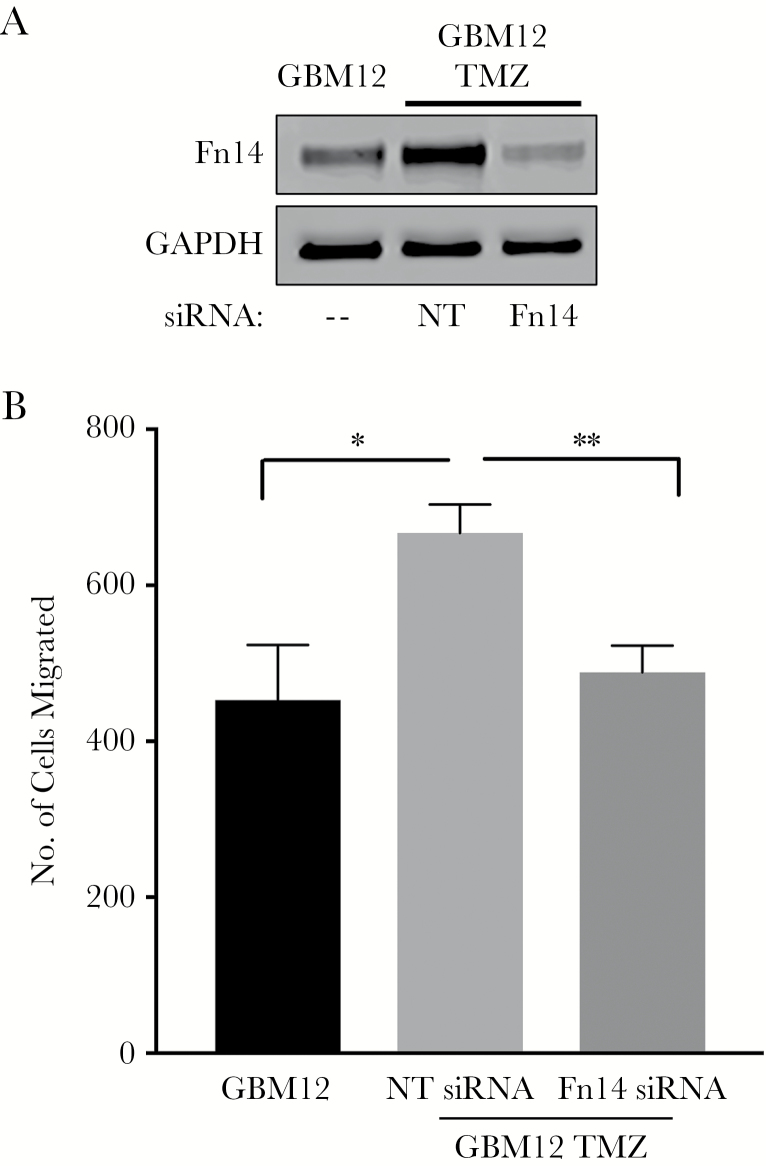
Several studies have indicated that the tumor necrosis factor–like weak inducer of apoptosis (TWEAK)–Fn14 signaling axis can regulate glioma cell migration and invasion.28 For example, forced Fn14 overexpression in glioma cells can promote migration27,40 and invasion,27,41 while Fn14 siRNA-mediated downregulation of endogenously expressed Fn14 in glioma cells reduces invasive capacity.27 Accordingly, we used the GBM12 TMZ-sensitive or TMZ-resistant cell lines,33 which express relatively low or high Fn14 levels, respectively (Fig. 5A), as a model to investigate Fn14’s role in the migratory capacity of TMZ-resistant cells. First, we transiently transfected the GBM12 TMZ-resistant cell line with control non-silencing or Fn14 mRNA-specific siRNA duplexes and examined Fn14 levels by western blot analysis (Fig. 6A). Fn14 expression was higher in the GBM12 TMZ-resistant line compared with the parental GBM12 line (~2.2-fold increase), consistent with the data in Fig. 5A. Additionally, Fn14 siRNA transfection of the GBM12 TMZ-resistant cells reduced the Fn14 expression level by ~80%. Second, we compared the migratory capacity of the parental GBM12 cells, the control siRNA-transfected GBM12 TMZ-resistant cells, and the Fn14 siRNA-transfected GBM12 TMZ-resistant cells using a transwell migration assay. We found that the migratory capacity of the control GBM12 TMZ-resistant cells was greater than that of the parental GBM12 cells and that this difference was abrogated by siRNA-mediated Fn14 depletion (Fig. 6B).
Fig. 6.
Analysis of GBM12 and GBM12 TMZ-resistant cell migration capacity and the effect of Fn14 depletion on GBM12 TMZ-resistant cell migration. (A) Parental GBM12 cells and matched GBM12 TMZ-resistant cells (GBM12 TMZ) transfected with either nontargeting (NT) siRNA or Fn14 siRNA were harvested and lysed. Fn14 and GAPDH levels were examined by western blot analysis. (B) GBM12 TMZ cells were transfected with either NT siRNA or Fn14 siRNA and 48 h later these cells and the parental GBM12 cells were cultured in serum-free medium for 16 h and then plated on transwell migration chambers. Medium containing 10% serum was placed in the lower wells to act as a chemoattractant. After 4 h of incubation, the number of cells that migrated through the membrane was determined. Values shown are the mean ± SD of triplicate chambers. Significance was measured by Student’s t-test. *P < 0.05, **P < 0.01. The difference between the GBM12 and GBM12 TMZ plus Fn14 siRNA values is not significant.
Discussion
The TWEAK receptor Fn14 has been implicated in GBM pathobiology and identified as a potential therapeutic target for this difficult to treat disease.28 Prior studies have established that (i) Fn14 is significantly overexpressed in primary GBM cells residing in the tumor core and invasive rim26–29 and (ii) high levels of Fn14 gene expression in glioma cells can activate intracellular signaling pathways and promote cell migration, invasion, and chemotherapy resistance.26–29,36,37,40–43 Intracellular molecular signaling mediators of Fn14 activity include the RhoGTPases Rac1, Cdc42, and RhoG, and the guanine exchange factors Ect2, Trio, and SGEF.27,36,37,41 Although many GBM patients treated with adjuvant TMZ and radiation develop recurrent tumors,9,10 there have been no prior reports examining Fn14 gene expression in recurrent GBMs or investigating whether TMZ exposure could affect Fn14 levels in GBM cells. Accordingly, we initiated the current study to address these important considerations.
We first examined Fn14 mRNA expression levels in nonneoplastic brain, primary GBM, and recurrent GBM specimens by interrogating the GBM dataset of TCGA. We found that Fn14 mRNA was overexpressed in primary GBM compared with nonneoplastic brain, consistent with prior reports.26–28 A similar degree of Fn14 mRNA overexpression was also found in the recurrent GBM specimens. Although this finding is not consistent with our Fn14 protein analysis (see below), the primary and recurrent Fn14 mRNA data in the database of TCGA are from different patients, whereas the protein data were obtained from patient-matched primary and recurrent GBM samples. Additionally, it would be difficult to detect a statistically significant difference between the primary and recurrent GBMs in the mRNA analysis because of the high patient-to-patient variability in the primary GBM Fn14 mRNA expression level and the large difference in sample size (535 primary GBM samples vs 13 recurrent GBM samples). We also used the database of TCGA to investigate whether there was a correlation between Fn14 mRNA expression levels in primary GBM and patient survival. We found that high Fn14 expression correlates with poor patient outcome, consistent with our 2006 report using the REMBRANDT database and a much smaller sample size.27
We also examined Fn14 protein expression levels in a set of 8 patient-matched primary and recurrent GBM tissue samples by IHC and detected Fn14 expression in all 16 samples, with 7/8 patients (87.5%) exhibiting the highest levels of Fn14 expression at relapse. Although our sample size was relatively low, this finding suggests that Fn14 may be a particularly promising therapeutic target for recurrent GBM compared with other cell surface receptor targets such as epidermal growth factor receptor variant III (EGFRvIII) and interleukin-13Rα2, which are frequently expressed at much lower levels in recurrent GBM tumors compared with untreated primary tumors.18,44–46 For example, Montano et al examined EGFRvIII mRNA expression in 14 patient-matched primary and recurrent GBMs.44 Ten patients had EGFRvIII mRNA-positive primary tumors but only 3 of the corresponding recurrent tumors were EGFRvIII mRNA positive.44
There are at least 3 potential explanations for our observation that Fn14 expression is higher in recurrent GBM tumor tissue compared with primary GBM tissue. One possibility is that Fn14 expression is upregulated in recurrent GBM via the preferential activation of certain signaling pathways in the recurrent setting. One candidate pathway would be PI3K/Akt/mTOR, which is activated in recurrent GBM tumors19,22 and is a known regulator of Fn14 gene expression.47 A second possibility is that adjuvant chemoradiotherapy increases Fn14 gene expression in the tumor cells, and those cells with the highest levels of Fn14 expression may have a survival advantage. This explanation is supported by studies indicating that (i) whole body γ-irradiation can increase Fn14 mRNA levels in intestinal cells,48 (ii) TWEAK stimulation of Fn14-positive glioma cells can promote TMZ resistance in vitro,29,36 and (iii) Fn14-depleted glioma cells exhibit enhanced TMZ sensitivity in vivo.36 The third possible explanation is that the primary GBM treatment regimen has no direct effect on Fn14 gene expression, but since GBM tumors are heterogeneous in nature, those tumor cells that naturally express the highest level of Fn14 have a survival advantage. As an initial approach to address whether Fn14 expression may evolve during the standard-of-care treatment of newly diagnosed GBMs, we compared Fn14 levels in 9 pairs of matched TMZ-sensitive or TMZ-resistant GBM PDX tumor tissue samples and found that Fn14 expression was higher in 6/9 of the tumors with acquired TMZ resistance. This result supports the general concept shared by the last 2 proposed explanations described above that there is a link between high Fn14 levels and chemotherapy resistance.
We then used the GBM12 TMZ-sensitive and TMZ-resistant PDX cells,33 which express relatively low and high Fn14 levels, respectively, to determine if the level of Fn14 expression correlates with cell migration capacity. We found that the GBM12 TMZ-resistant cells had increased migratory capacity compared with their parental counterpart, and that Fn14 depletion reduces GBM12 TMZ-resistant cell migration, suggesting that Fn14 may be responsible for the increased migration of the TMZ-resistant cells. These findings are consistent with previous studies indicating that the Fn14 expression level regulates glioma cell migration and invasion.27,28,40,41
Finally, in the context of this work it became apparent that the recurrent GBM specimen exhibiting the highest level of Fn14 expression in the IHC analysis (patient #6) had histological features of GSM. GSM is a rare GBM variant with a poor prognosis.49 These tumors are characterized by a biphasic tissue pattern with areas that display either glial or mesenchymal differentiation.34,35 GSMs that develop de novo are referred to as primary GSMs, while those that arise following standard-of-care primary GBM treatment, such as the recurrent tumor specimen mentioned above, are referred to as secondary GSMs. We evaluated Fn14 expression in an additional secondary GSM specimen as well as 2 primary GSM specimens and again found high Fn14 levels in comparison to nonneoplastic brain tissue. Although the expression of some proteins, such as transcription factors involved in the epithelial-mesenchymal transition,50 is restricted to the glial or mesenchymal areas of GSM tumors, we found a similar degree of Fn14 expression throughout the neoplastic tissue.
GBM recurrence is a near universal outcome for patients undergoing standard-of-care treatment, and current therapies for recurrent tumors are largely ineffective.9,10 New therapeutic targets and drug delivery strategies are sorely needed. The TNFR superfamily member Fn14 has been described as a potential GBM drug target, and in particular a good target for the invasive cell population left behind after surgery,28 but its expression in recurrent GBM tumors or in other, less well understood GBM variants such as GSM has not been reported. This study demonstrates that Fn14 gene expression increases in recurrent GBM compared with untreated newly diagnosed GBM and that Fn14 expression is higher in TMZ-resistant GBM cells compared with matched TMZ-sensitive cells. Several TWEAK- or Fn14-targeted therapeutic agents are currently in development for potential use in cancer patients.23,25,28 These current findings, coupled with prior work, suggest that Fn14 may be a valuable therapeutic target or drug delivery portal for recurrent GBM and GSM treatment.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported in part by National Institutes of Health Grant K08 NS09043 (G.F.W.), American Cancer Society Research Scholar Grant 128970-RSG-16-012-01-CDD (G.F.W.), Elsa U. Pardee Foundation Cancer Research Grant EP24503 (A.J.K. and J.A.W.), and the Ben and Catherine Ivy Foundation (N.L.T.).
Acknowledgments
We thank Dr Jann Sarkaria for providing the GBM PDX flank tumors and cell lines.
Conflict of interest statement. The authors declare no conflict of interest.
Authorship statement. D.S.H., G.F.W., N.L.T., J.A.W. conceived the project and designed the experiments. B.G.H., A.R., S.P. performed experiments and collected the data. J.E.H., T.L. reviewed the pathology slides and quantified the IHC data. D.S.H., S.P., A.J.K., G.F.W., N.L.T., J.A.W. analyzed the data. D.S.H., A.J.K., G.F.W., N.L.T., J.A.W. wrote the manuscript.
References
- 1. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. [DOI] [PubMed] [Google Scholar]
- 2. Ostrom QT, Gittleman H, Liao P et al. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol. 2017;19(Suppl 5):v1–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stupp R, Mason WP, van den Bent MJ et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 4. Stupp R, Hegi ME, Mason WP et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 5. Xie Q, Mittal S, Berens ME. Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol. 2014;16(12):1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64(4):458–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vehlow A, Cordes N. Invasion as target for therapy of glioblastoma multiforme. Biochim Biophys Acta. 2013;1836(2):236–244. [DOI] [PubMed] [Google Scholar]
- 8. Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15(7):455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campos B, Olsen LR, Urup T, Poulsen HS. A comprehensive profile of recurrent glioblastoma. Oncogene. 2016;35(45):5819–5825. [DOI] [PubMed] [Google Scholar]
- 10. Weller M, Cloughesy T, Perry JR, Wick W. Standards of care for treatment of recurrent glioblastoma—are we there yet?Neuro Oncol. 2013;15(1):4–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lau D, Magill ST, Aghi MK. Molecularly targeted therapies for recurrent glioblastoma: current and future targets. Neurosurg Focus. 2014;37(6):E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mangraviti A, Tyler B, Brem H. Interstitial chemotherapy for malignant glioma: Future prospects in the era of multimodal therapy. Surg Neurol Int. 2015;6(Suppl 1):S78–S84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mar N, Desjardins A, Vredenburgh JJ. CCR 20th anniversary commentary: bevacizumab in the treatment of glioblastoma—the progress and the limitations. Clin Cancer Res. 2015;21(19):4248–4250. [DOI] [PubMed] [Google Scholar]
- 14. Osuka S, Van Meir EG. Overcoming therapeutic resistance in glioblastoma: the way forward. J Clin Invest. 2017;127(2):415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Auffinger B, Tobias AL, Han Y et al. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ 2014;21(7):1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Orzan F, De Bacco F, Crisafulli G et al. Genetic evolution of glioblastoma stem-like cells from primary to recurrent tumor. Stem Cells. 2017;35(11):2218–2228. [DOI] [PubMed] [Google Scholar]
- 17. Kim H, Zheng S, Amini SS et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25(3):316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J, Cazzato E, Ladewig E et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Byron SA, Tran NL, Halperin RF et al. Prospective feasibility trial for genomics-informed treatment in recurrent and progressive glioblastoma. Clin Cancer Res. 2018;24(2):295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Thuijl HF, Mazor T, Johnson BE et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129(4):597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nickel GC, Barnholtz-Sloan J, Gould MP et al. Characterizing mutational heterogeneity in a glioblastoma patient with double recurrence. PLoS One. 2012;7(4):e35262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson BE, Mazor T, Hong C et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winkles JA. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat Rev Drug Discov. 2008;7(5):411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burkly LC. TWEAK/Fn14 axis: the current paradigm of tissue injury-inducible function in the midst of complexities. Semin Immunol. 2014;26(3):229–236. [DOI] [PubMed] [Google Scholar]
- 25. Cheng E, Armstrong CL, Galisteo R, Winkles JA. TWEAK/Fn14 axis-targeted therapeutics: moving basic science discoveries to the clinic. Front Immunol. 2013;4:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tran NL, McDonough WS, Donohue PJ et al. The human Fn14 receptor gene is up-regulated in migrating glioma cells in vitro and overexpressed in advanced glial tumors. Am J Pathol. 2003;162(4):1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tran NL, McDonough WS, Savitch BA et al. Increased fibroblast growth factor-inducible 14 expression levels promote glioma cell invasion via Rac1 and nuclear factor-kappaB and correlate with poor patient outcome. Cancer Res. 2006;66(19):9535–9542. [DOI] [PubMed] [Google Scholar]
- 28. Perez JG, Tran NL, Rosenblum MG et al. The TWEAK receptor Fn14 is a potential cell surface portal for targeted delivery of glioblastoma therapeutics. Oncogene. 2016;35(17):2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fortin SP, Ennis MJ, Savitch BA et al. Tumor necrosis factor-like weak inducer of apoptosis stimulation of glioma cell survival is dependent on Akt2 function. Mol Cancer Res. 2009;7(11):1871–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kitange GJ, Carlson BL, Mladek AC et al. Evaluation of MGMT promoter methylation status and correlation with temozolomide response in orthotopic glioblastoma xenograft model. J Neurooncol. 2009;92(1):23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta SK, Kizilbash SH, Carlson BL et al. Delineation of MGMT hypermethylation as a biomarker for veliparib-mediated temozolomide-sensitizing therapy of glioblastoma. J Natl Cancer Inst 2015;108(5):djv369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mayo Clinic Brain PDX National Resource Portal http://www.mayo.edu/research/labs/translational-neuro-oncology/mayo-clinic-brain-tumor-patient-derived-xenograft-national-resource. Accessed May 1, 2018.
- 33. Kitange GJ, Mladek AC, Carlson BL et al. Inhibition of histone deacetylation potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clin Cancer Res. 2012;18(15):4070–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Damodaran O, van Heerden J, Nowak AK et al. Clinical management and survival outcomes of gliosarcomas in the era of multimodality therapy. J Clin Neurosci. 2014;21(3):478–481. [DOI] [PubMed] [Google Scholar]
- 35. Cachia D, Kamiya-Matsuoka C, Mandel JJ et al. Primary and secondary gliosarcomas: clinical, molecular and survival characteristics. J Neurooncol. 2015;125(2):401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roos A, Dhruv HD, Mathews IT et al. Identification of aurintricarboxylic acid as a selective inhibitor of the TWEAK-Fn14 signaling pathway in glioblastoma cells. Oncotarget. 2017;8(7):12234–12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ensign SP, Roos A, Mathews IT et al. SGEF is regulated via TWEAK/Fn14/NF-κB signaling and promotes survival by modulation of the DNA repair response to temozolomide. Mol Cancer Res. 2016;14(3):302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tran NL, McDonough WS, Savitch BA, Sawyer TF, Winkles JA, Berens ME. The tumor necrosis factor-like weak inducer of apoptosis (TWEAK)-fibroblast growth factor-inducible 14 (Fn14) signaling system regulates glioma cell survival via NF-kappaB pathway activation and BCL-XL/BCL-W expression. J Biol Chem. 2005;280(5): 3483–3492. [DOI] [PubMed] [Google Scholar]
- 39. Brown SA, Cheng E, Williams MS, Winkles JA. TWEAK-independent Fn14 self-association and NF-κB activation is mediated by the C-terminal region of the Fn14 cytoplasmic domain. PLoS One. 2013;8(6): e65248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Winkles JA, Tran NL, Berens ME. TWEAK and Fn14: new molecular targets for cancer therapy?Cancer Lett. 2006;235(1):11–17. [DOI] [PubMed] [Google Scholar]
- 41. Fortin SP, Ennis MJ, Schumacher CA et al. Cdc42 and the guanine nucleotide exchange factors Ect2 and trio mediate Fn14-induced migration and invasion of glioblastoma cells. Mol Cancer Res. 2012;10(7):958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cherry EM, Lee DW, Jung JU, Sitcheran R. Tumor necrosis factor-like weak inducer of apoptosis (TWEAK) promotes glioma cell invasion through induction of NF-κB-inducing kinase (NIK) and noncanonical NF-κB signaling. Mol Cancer. 2015;14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dhruv H, Loftus JC, Narang P et al. Structural basis and targeting of the interaction between fibroblast growth factor-inducible 14 and tumor necrosis factor-like weak inducer of apoptosis. J Biol Chem. 2013;288(45):32261–32276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Montano N, Cenci T, Martini M et al. Expression of EGFRvIII in glioblastoma: prognostic significance revisited. Neoplasia. 2011;13(12):1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van den Bent MJ, Gao Y, Kerkhof M et al. Changes in the EGFR amplification and EGFRvIII expression between paired primary and recurrent glioblastomas. Neuro Oncol. 2015;17(7):935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bozinov O, Kalk JM, Krayenbühl N, Woernle CM, Sure U, Bertalanffy H. Decreasing expression of the interleukin-13 receptor IL-13Ralpha2 in treated recurrent malignant gliomas. Neurol Med Chir (Tokyo). 2010;50(8):617–621. [DOI] [PubMed] [Google Scholar]
- 47. Asrani K, Keri RA, Galisteo R et al. The HER2- and heregulin β1 (HRG)-inducible TNFR superfamily member Fn14 promotes HRG-driven breast cancer cell migration, invasion, and MMP9 expression. Mol Cancer Res. 2013;11(4):393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kawashima R, Kawamura YI, Oshio T et al. Interleukin-13 damages intestinal mucosa via TWEAK and Fn14 in mice—a pathway associated with ulcerative colitis. Gastroenterology. 2011;141(6):2119–2129.e8. [DOI] [PubMed] [Google Scholar]
- 49. Smith DR, Wu CC, Saadatmand HJ et al. Clinical and molecular characteristics of gliosarcoma and modern prognostic significance relative to conventional glioblastoma. J Neurooncol. 2018;137(2): 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nagaishi M, Paulus W, Brokinkel B et al. Transcriptional factors for epithelial-mesenchymal transition are associated with mesenchymal differentiation in gliosarcoma. Brain Pathol. 2012;22(5):670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.