Abstract
Leigh syndrome is a frequent, heterogeneous pediatric presentation of mitochondrial oxidative phosphorylation (OXPHOS) disease, manifesting with psychomotor retardation and necrotizing lesions in brain deep gray matter. OXPHOS occurs at the inner mitochondrial membrane through the integrated activity of five protein complexes, of which complex V (CV) functions in a dimeric form to directly generate adenosine triphosphate (ATP). Mutations in several different structural CV subunits cause Leigh syndrome; however, dimerization defects have not been associated with human disease. We report four Leigh syndrome subjects from three unrelated Ashkenazi Jewish families harboring a homozygous splice-site mutation (c.87 + 1G>C) in a novel CV subunit disease gene, USMG5. The Ashkenazi population allele frequency is 0.57%. This mutation produces two USMG5 transcripts, wild-type and lacking exon 3. Fibroblasts from two Leigh syndrome probands had reduced wild-type USMG5 mRNA expression and undetectable protein. The mutation did not alter monomeric CV expression, but reduced both CV dimer expression and ATP synthesis rate. Rescue with wild-type USMG5 cDNA in proband fibroblasts restored USMG5 protein, increased CV dimerization and enhanced ATP production rate. These data demonstrate that a recurrent USMG5 splice-site founder mutation in the Ashkenazi Jewish population causes autosomal recessive Leigh syndrome by reduction of CV dimerization and ATP synthesis.
Introduction
Leigh syndrome (OMIM 25600) is a progressive neurodevelopmental disorder characterized by deep brain gray matter necrotizing lesions (1,2). Genetic causes of Leigh syndrome are diverse: more than 90 nuclear and mitochondrial DNA (mtDNA) causative genes have been identified (3), most of which are required for mitochondrial oxidative phosphorylation (OXPHOS) (4). OXPHOS provides cellular energy by the activities of four electron transport complex enzymes (CI–IV) that generate a proton gradient within the inner mitochondrial membrane (IMM). Complex V (CV; ATP synthase) uses this gradient to produce ATP (5). CV has two domains: F1 projects into the mitochondrial matrix and Fo spans the IMM (6,7). These two domains contain at least 15 protein subunits: 2 mtDNA-encoded (ATP6, ATP8) and 13 nuclear DNA-encoded (8). CV exists as a monomer complex V (Vm) and a dimeric supercomplex (Vd). Vd helps shape mitochondrial cristae to optimize proton flow (9,10). Deficiency of Vm activity can cause Leigh syndrome (mutations in MT-ATP6) (11); however, defective Vd formation has not previously been linked to human disease.
USMG5 (DAPIT) is a 6.3 kDa protein identified in mammalian CV (12,13). Knock-out of USMG5 reduces ATP synthesis (14), and USMG5 role in CV dimerization has been hypothesized (12,15) and recently described (16).
Here, we report four affected individuals with Leigh syndrome from three unrelated Ashkenazi Jewish families whose disease results from a homozygous c.87+1G>C USMG5 mutation leading to reduced Vd formation and impaired ATP synthesis.
Results
USMG5 homozygous splice-site mutation was identified in three unrelated Ashkenazi Jewish kindreds with Leigh syndrome
In Probands 1–3, exome data analysis identified a homozygous single nucleotide substitution, c.87+1G>C, in USMG5 (upregulated during skeletal muscle growth 5; NM_032747.3) as the most likely pathogenic candidate. Segregation analysis confirmed that the homozygous status was limited to probands and asymptomatic parents of all probands were heterozygous. No occurrences of the homozygous mutation were found in the Exome Aggregation Consortium dataset (ExAC v0.3), gnomAD or sequencing data from more than 2000 exome samples in the CHOP Center for Applied Genomics database. The variant was present in the IBD dataset with a minor allele frequency (MAF) of 0.003697 in Ashkenazi-Jewish control cohort, in our Ashkenazi-Jewish control cohort with an MAF of 0.0057 and was reported in dbSNP134 (rs146599698). All kindreds shared 24 nearby homozygous rare single nucleotide variants composing a 720 kilobase (kb) identical region on all affected alleles. This highly indicates that the variant arose in a common founder and that the mutant alleles were identical by descent.
Clinical description of four pediatric Leigh syndrome subjects
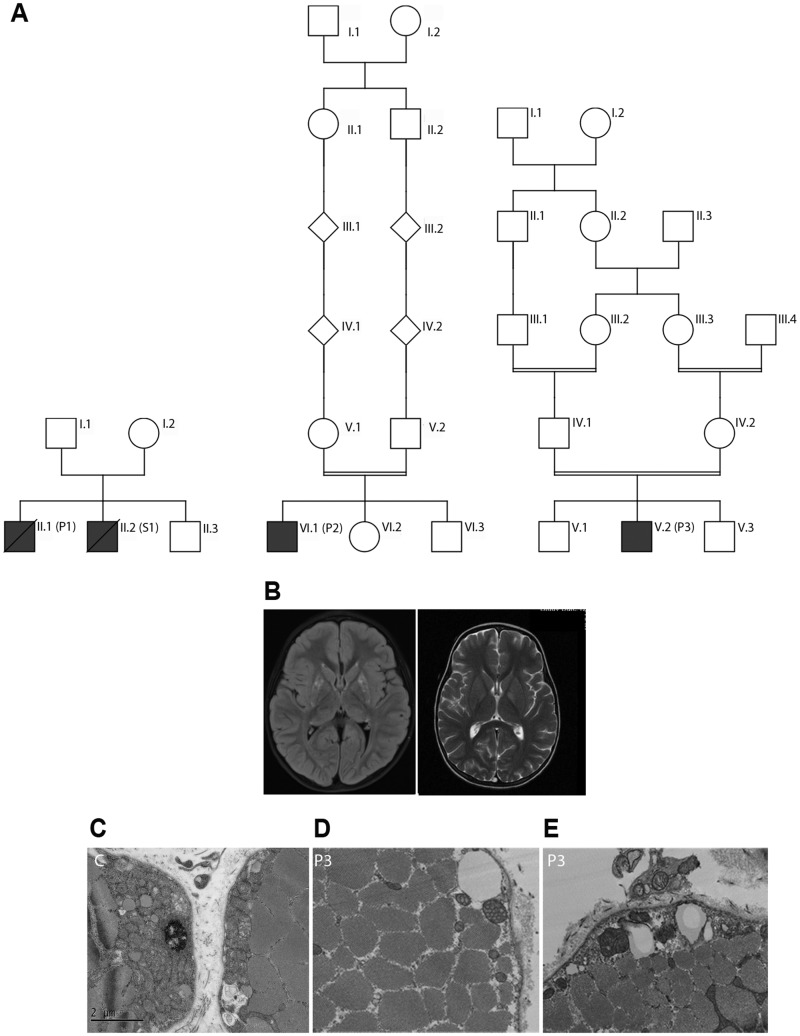
The 4 affected subjects were from 3 unrelated Ashkenazi Jewish kindreds, where each was diagnosed with Leigh syndrome between 18 months and 5 years of age (Fig. 1A;Table 1). Each presented by 18 months of age with gross motor developmental delay and subsequently manifested developmental regression in the setting of fever between 2.5 and 5 years of age. All subjects experienced recurrent, episodic gross motor regressions during metabolic stress, particularly febrile illnesses and general anesthesia. Full clinical details are provided in Supplementary Material, S1. MRI scans in Probands 2 and 3 and autopsy in Proband 1 were consistent with Leigh syndrome (Fig. 1B). Muscle biopsy of vastus lateralis in Proband 3 showed no alterations in COX and succinate dehydrogenase (SDH) histochemical staining or ragged-red fibers. Respiratory chain enzyme activities in muscle on Probands 1 and 3 and in fibroblasts in Proband 2, performed in clinical diagnostic laboratories (Baylor—Probands 1 and 2; CUMC—Proband 3), were normal (>Supplementary Material, Fig. S2); however, altered mitochondrial cristae shape with increased mitochondrial volume was seen by electron microscopy (Fig. 1D and E).
Figure 1.
(A) Pedigrees of the three kindereds (P1, Proband 1; P2, Proband 2; P3, Proband 3; S1, Subject 1). (B) T2-weighted brain MRI scans showing bilateral hyper-intensity of basal ganglia in P2 (T2-TIRM; obtained prior to developmental regression onset) and P3 (obtained after the onset of developmental regression). (C) Transmission electron micrograph showing normal mitochondria. (D and E) Transmission electron micrographs of P3 skeletal muscle showing altered mitochondria with abnormal arrangement of cristae (magnification D: ×25 000; C and E: ×20 000).
Table 1.
Clinical characteristics of Leigh syndrome subjects
| Individual | P1 | S1 | P2 | P3 |
|---|---|---|---|---|
| Kindred | 1 | 1 | 2 | 3 |
| USMG5 homozygous mutation | c.87 + 1G>C | Not available | c.87 + 1G>C | c.87 + 1G>C |
| Recognized consanguinity | – | – | + | + |
| Sex | Male | Male | Male | Male |
| Age at presentation | 18 months | Early infancy | 6 months | 14 months |
| Deceased (age at death or last evaluation) | +(9 years old) | +(6 years old) | −(6 years old) | −(6 years old) |
| Developmental delay | + | + | + | + |
| Developmental regression (age) | +(23 months; 9 years) | +(30 months; multiple others) | +(4 years) | +(30 months; multiple others) |
| Setting of developmental regression | Febrile illness | Febrile illness, general anesthesia | Febrile illness | Febrile illness, general anesthesia |
| Ataxia | + | + | + | −(non-ambulatory) |
| Movement disorder | generalized bradykinesia, arms choreiform movements | − | − | Armschoreiform movements |
| Hypertrophic cardiomyopathy | − | − | − | +(transient) |
| Ophthalmoplegia | − | + | − | + |
| Brainstem lesionsa | + | + | + | − |
| Basal ganglia lesionsa | + | − | − | + |
| Electron transport chain complex I−IV enzyme activities analysis | Reduced citrate synthase, increased activities of CI–IV (muscle tissue) | ND | Reduced citrate synthase, otherwise normal (fibroblast) | Normal (muscle tissue) |
| Other | NA | Accessory spleen, testicular atrophy, fatty liver | NA | G-tube dependent, tracheostomy/ventilator dependent |
P1 and S1 are siblings in kindred 1, where DNA was only available for mutation confirmation in P1.
See Supplemental Material, S3 for full details.
ND, not done; NA, not applicable.
USMG5 mRNA and protein expression was reduced in USMG5 proband fibroblasts
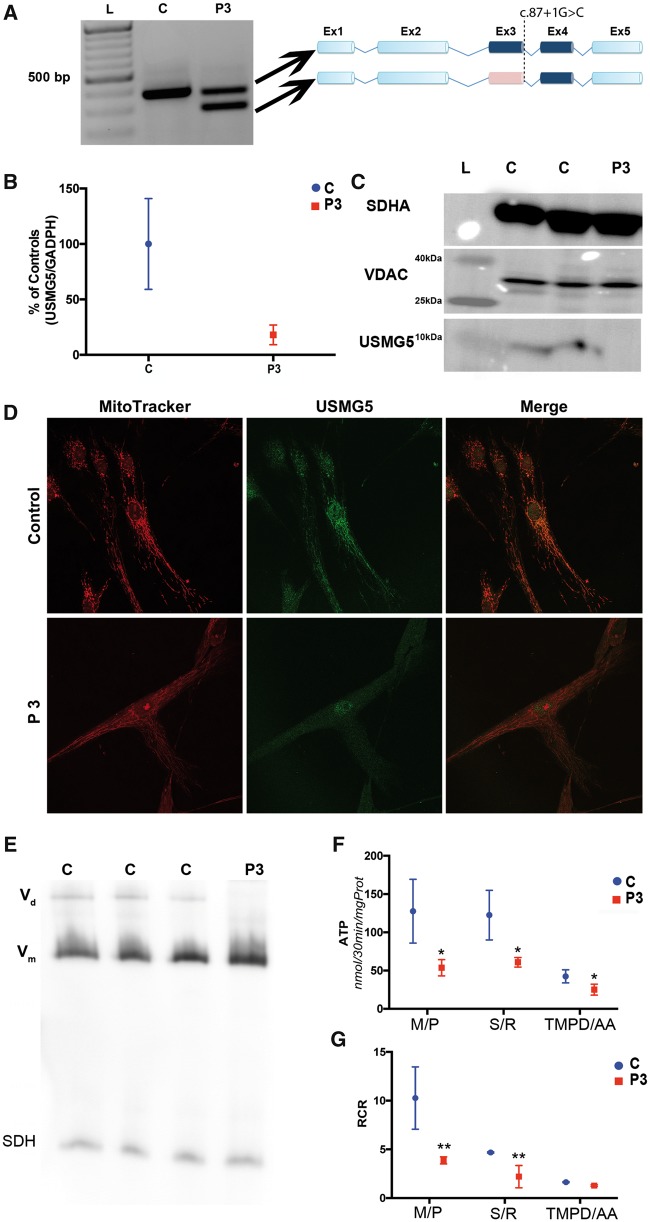
The Human Splicing Finder 3 tool (17) predicted that the c.87+1G>C mutation in USMG5 would cause skipping of exon 3, which was confirmed by reverse transcriptase-polymerase chain reaction (RT-PCR) analysis in Probands 2 and 3. Probands fibroblasts had two cDNA species: a 400 basepair (bp) band consistent with wild-type transcript, and a shorter 300 bp band that lacked exon 3 (Fig. 2A). Probands 2 (data not shown) and 3 showed markedly reduced USMG5 mRNA levels (18 ± 8.9% relative to controls) (Fig. 2B) by quantitative (qRT-PCR). USMG5 protein was normal in control cells but undetectable by western blot in Proband 2 (not shown) and three fibroblasts (Fig. 2C). Moreover, confocal fluorescence microscopy localized USMG5 to mitochondria in control fibroblasts, but failed to detect the protein in proband cells (Fig. 2D).
Figure 2.
(A) Two percent agarose gel demonstrating the presence of an abnormal USMG5 transcript in Proband 3 (P3) compared with control (C) fibroblasts. L=molecular weight ladder. Schematic representations of the wild-type and abnormal USMG5 transcripts are shown on the right. In pink: exon 3 deleted in the Proband 3 aberrant transcript. (B) USMG5 mRNA levels analyzed by qRT-PCR demonstrating the reduction of transcript levels in proband fibroblasts. Data are normalized to GAPDH and expressed as percentage of controls (mean±SD). (C) Tricine–SDS gel demonstrating the lack of USMG5 protein in proband fibroblasts (P3) compared with controls (C). SDHA and VDAC were used as loading controls for mitochondrial protein (full western blot available in Supplementary Material, Fig. S5). (D) Confocal fluorescence microscopy showing the presence of USMG5 (green) in controls (upper panels) with positive localization in the mitochondrial compartment (red). Proband 3 cells did not show USMG5 signal in mitochondria. Scale bar 50 µm. (E) Representative high-resolution clear native gel showing the absence of CV dimer (Vd) and normal levels of CV monomer (Vm) in proband (P3) fibroblasts. Vd is present in control lanes (C). SDH was used as loading control. (F) ATP levels assessed by HPLC showed reduction of ATP production in Proband 3 (P3) cells compared with controls (C) with all substrates tested. M/P, malate/pyruvate; S/R, succinate/rotenone; TMPD/AA, tetramethyl-p-phenylenediamine/ascorbic acid. Results are expressed as mean±SD of three biological replicate experiments. *P < 0.05. (G) RCR, calculated as the ratio of oxygen consumption in State 3ADP of respiration divided by oxygen consumption in State 4o, confirmed the reduction of substrate oxidation capacity in Proband 3 (P3) compared with controls (C) fibroblasts. Results are expressed as mean±SD of three biological replicate experiments. *P < 0.05; **P < 0.01.
CV dimerization was reduced by USMG5 mutation
Fibroblasts from Proband 3 and controls were analyzed to determine respiratory chain complex assembly and super-assembly. Vm was identified at the expected molecular weight (≃750 kDa) (Supplementary Material, Fig. S3) at the same intensity between proband and controls; however, the band corresponding to Vd was detected only in control fibroblasts (≃1000 kDa) but not in the proband’s cells (Fig. 2E).
USMG5 mutated fibroblasts have reduced ATP synthesis
Spectrophotometric assays of mitochondrial respiratory enzymes and citrate synthase were normal in Proband 3 fibroblasts (Supplementary Material, Fig. S4). Cell respiration showed no differences between proband fibroblasts and controls in coupling efficiency (85.8 ± 3.2% controls versus 87.5 ± 2.6% proband). ATP synthesis was consistently decreased in permeabilized proband fibroblasts with all substrates tested (127.6 ± 41.70 nmol ATP/30 min/mg-prot in controls and 53.67 ± 10.62 nmol ATP/30 min/mg-prot in proband cells (P = 0.035) using CI substrates; 122.5 ± 32.4 nmol ATP/30 min/mg-prot in controls and 60.87 ± 6.26 nmol ATP/30 min/mg-prot in proband fibroblasts (P = 0.03) with Complex II (CII) substrates; and 42.49 ± 8.64 in controls versus 25.10 ± 7.1 nmol ATP/30 min/mg-prot (P = 0.023) in proband fibroblasts using CIV substrates] (Fig. 2F). Reduced substrate oxidation was also confirmed by analysis of respiration in permeabilized fibroblasts (Fig. 2G). The alteration of ATP production regardless of substrate utilized, together with the evidence of normal respiratory chain enzyme complexes indicate impairment of ATP production at the level of CV (18).
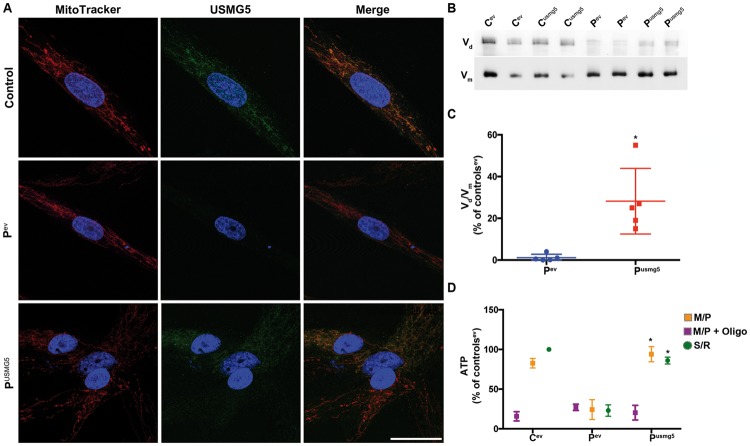
Transfection with wild-type USMG5 rescues CV dimerization and ATP production
Transient transfection produced USMG5 protein in Proband 3 fibroblasts [USMG5/VDAC 11 ± 4% of controls transfected with empty vector (EV), controlev] by western blot (data not shown). By confocal fluorescence microscopy, wild-type USMG5 was detected in the mitochondrial compartment in transfected proband fibroblasts (Fig. 3A). Native gel analysis of mitochondrial complexes revealed a faint complex Vd in proband fibroblasts transfected with EV (Pev) (dimer/monomer ratio 1.2 ± 1.6% of controls transfected with EV, controlsev), and clear increase in the dimerization in proband cells transfected with wild-type USMG5: Vm/Vd ratio of 28.2 ± 15.7% of controlsev (P = 0.017) (Fig. 3B and C). Subject fibroblasts expressing wild-type USMG5 construct (Pusmg5) demonstrated recovery of ATP production with substrates for Complex I (CI) (Pev 24.2 ± 12.5% versus Pusmg5 94 ± 9.5% of controlsev, P = 0.035) and CII (Ptev 23 ± 7.1% versus Pusmg5 86 ± 4.3% of controlsev, P = 0.035) (Fig. 3D).
Figure 3.
(A) Confocal fluorescence microscopy demonstrating restoration of USMG5 protein (green) expression in Proband 3 fibroblasts (lower panels). The expressed protein localized to mitochondria (red); nuclei are in blue. Proband fibroblasts transfected with plasmid backbone (EV) lack detectable USMG5 protein. Scale bar 30 µm. (B) Representative clear native gel showing the increased CV dimer (Vd) in proband fibroblasts transfected with USMG5 plasmid (Pusmg5) compared with proband cells expressing the EV (Pev). For comparison, controls transfected with EV (Cev) and with USMG5 (Cusmg5) were analyzed. Vm indicates the band corresponding to the CV monomer (full membrane shown in Supplementary Material, Fig. S6). (C) Quantification of CV dimerization was calculated as the ratio of CV dimer (Vd) to CV monomer (Vm) and expressed as the percentage of controls transfected with EV (Cev) (mean±SD of four biological replicate experiments). *P < 0.05. (D) ATP levels assessed by HPLC showed the recovery of ATP synthesis in proband fibroblasts transfected with USMG5 (Pusmg5) compared with proband fibroblasts transfected with the EV (Pev). Data are expressed as percentage of controls transfected with EV (Cev) (mean±SD of four biological replicate experiments). M/P, Malate/pyruvate; M/P+oligo, malate/pyruvate+oligomycin; S/R, succinate/rotenone; *P < 0.05.
Discussion
Here, we report a homozygous splice-site mutation, c.87+1G>C, in the USMG5 CV subunit, not previously linked to human disease, as a novel and recurrent cause of pediatric Leigh syndrome. Although, mutations in the mtDNA-encoded CV subunit MT-ATP6 were previously associated with Leigh syndrome and known to impair complex Vm activity (11), Vd defects have not previously been associated with human disease.
We identified the mutation in three unrelated Ashkenazi Jewish kindreds. This common mutation leads to shortened USMG5 transcripts lacking exon 3, mRNA decay and severe reduction of USMG5 protein. USMG5 thus represents the first nuclear-encoded CV subunit gene linked to the Leigh syndrome. It is plausible that the relative lack of known autosomal causes of CV-deficient Leigh syndrome stems from the paucity of diagnostic assays for CV function. Indeed, while OXPHOS complexes I–IV activities are routinely performed in diagnostic laboratories, biochemical evaluation of CV function is performed only on a research basis. From a clinical perspective, USMG5-deficient Leigh syndrome shares multiple phenotypic features common to MT-ATP6-deficient Leigh syndrome, including absence of lactic acidosis, occasional hypertrophic cardiomyopathy and relatively prolonged survival. The reason for survival into mid-childhood is not clear, but may relate to relative ability to tolerate defects in CV compared with other components of the mitochondrial respiratory chain (similar to the relatively prolonged life expectancy of patients with Leigh Syndrome owing to mutations in MT-ATP6) (19), ecogenomic interactions where normal function is maintained until one encounters life stressors (such as anesthesia or infection), and/or the presence of residual wild-type USMG5 transcript, as we observed in proband fibroblasts. However, all four affected individuals in our series did experience recurrent, severe neurodevelopmental regression with fever and general anesthesia sensitivity was particularly prominent, leading to the childhood death of one subject in kindred 1 and permanent ventilator dependence in Proband 3. Therefore, minimizing anesthesia exposure, as has been done in Proband 2, appears to be clinically important to optimize outcomes and survival.
CV activity utilizes the proton gradient across the IMM to generate chemical energy in form of ATP. ATP production is mainly based on the activity of Vm (20); however, CV exists also in a higher complex assembly, such as dimers. Vd is postulated to be involved in shaping IMM cristae structure. The IMM is a specialized structure essential for energy production and mitochondrial function (21). Several proteins are involved in IMM formation, and among them, Vd ribbons are important in determining cristae edge curvature (22–24) that has been hypothesized to facilitate protons flow across CV (25).
Here, we established that a novel mutation in USMG5 does not affect Vm activity but rather reduces Vd steady-state quantity. Based upon the evidence of impaired ATP synthesis in the context of normal levels of CV monomer, we hypothesize that altering Vd directly impairs mitochondrial energy production leading to clinical disease. Moreover, the alterations in mitochondrial cristae geometry observed in Proband 3 skeletal muscle are consistent with this postulated role for Vd. It remains uncertain whether the observed ATP synthesis defect is owing to a decrease in cristae surface or to defective biophysical proton/ATP coupling. Overall, the identification of a human neurologic disease primarily caused by a Vd defect expands the pathophysiological basis of mitochondrial diseases and widens the clinical and diagnostic spectrum of this complex group of disorders. Furthermore, USMG5 c.87+1G>C represents an Ashkenazi Jewish founder mutation that should be included in diagnostic testing and carrier screening panels for pediatric Leigh syndrome.
Materials and Methods
Human subject enrollment and fibroblast cell line studies
Subjects provided written consent and all studies were performed in accordance with Children’s Hospital of Philadelphia (CHOP) IRB-approved protocol (#08-6177, M.J.F. PI). Kindred 3 also provided written consent for Columbia University Medical Center (CUMC) IRB-approved protocol (#AAAJ8651, W.K.C. PI). Skin biopsy was performed following informed consent to establish fibroblast cell lines.
Whole exome sequencing and bioinformatics analytic methods
FASTQ clinical trio-based whole exome sequence data from Proband 2 was re-analyzed on a research basis, excluding synonymous variants, variants with MAF >1% and biallelic variants previously identified in controls in the exome variant dataset in the CHOP Center for Applied Genomics. BAM files from research-based exome sequencing were obtained from fibroblasts from Proband 1 and blood from his unaffected parents; biological samples were not available from his affected deceased brother (Table 1). Exome data from these kindreds were analyzed in a larger dataset of neurologic disease within Genesis (21), enabling identification of a shared USMG5 variant in both kindreds, confirmed by Sanger sequencing (Supplementary Materials, S2 and S3). Clinical exome sequencing in Proband 3 (GeneDx) identified the same homozygous USMG5 variant.
Haplotype and frequency analysis
We analyzed the genotype of single nucleotide variants flanking the USMG5 variant to determine whether the mutation was part of the same haplotype in all kindreds. Frequency of the variant in the Ashkenazi Jewish population was confirmed in an independent cohort using allelic discrimination, as previously described (26).
Cell culture and transient transfection
Fibroblasts from Probands 2 and 3 were cultured following standard techniques. Cells below passage 10 were used for all experiments. Transient expression of wild-type USMG5 was performed by nucleofection (Nucleofector®, Lonza, Germany), according to published protocols (27). USMG5 expressing vector with pCMV6 promoter was purchased from Origene® (NM_0322747); Proband 3 and controls fibroblasts were also transfected with a complete pCMV6 plasmid without the USMG5 open reading frame (EV). Functional studies were performed at 48h post-transfection.
RT-PCR for USMG5 transcript analysis and transcription levels
RNA was extracted from the probands and control fibroblasts using PureLink™ RNA MiniKit (Ambion) according to the manufacturer’s instructions. Total RNA was reverse-transcribed to cDNA with SuperScript® VILO™ cDNA Synthesis Kit (Invitrogen) after digestion with DNase I (Roche). cDNA amplification was performed using specific primers (Supplementary Material, S3) and visualized on a 2%, w/v, agarose gel. For transcript sequencing, cDNA bands were excised from the gel using QIAquick Gel Extraction Kit according to the manufacturer’s instructions, cloned by universal TA cloning (28) and Sanger sequenced. qRT-PCR was performed using TaqMan® Assays (NM_032747.3 for USMG5) (Applied Biosystems, Invitrogen). Expression of target genes was calculated by the Δ-Ct method and normalized to GAPDH expression.
Protein separation and western blotting
For USMG5 levels, proteins were separated using SDS 4–20% Tris–glycine gels (Novex, Invitrogen), transferred onto a polyvinylidene difluoride (PVDF) membrane, and probed with specific antibodies (Supplementary Material, S4). To assess CV assembly, mitochondrial membranes were digested using digitonin (2 g/g) and loaded onto a 3–10% native gel (Novex, Invitrogen) using high-resolution clear native-3 buffer (29). For size determination, NativeMark™ Unstained Protein Standard was used (Invitrogen, LC0725). Gels were blotted onto PVDF membranes, and probed with specific antibodies (Supplementary Material, S4). Protein bands were visualized by chemiluminescence (GE Healthcare). Image intensity was measured by ImageJ. Results are expressed as percentage of average controls intensity. CV dimerization was calculated as the ratio between the intensity of the higher molecular weight band, relative to CV dimer (Vd), normalized for the intensity of monomer band (Vm) and expressed as percentage of controls.
Fluorescence microscopy in cells and tissues
For mitochondrial staining, patient and control fibroblasts were incubated in a medium containing 20 nM MitoTracker® Red CMXRos (ThermoFisher Scientific, M7512), and probed with a purified rabbit polyclonal antibody against the N-terminal of USMG5 (kindly provided by I. Wittig; 1:10). After nuclear staining with Hoescht, slides were imaged using an SP5 Leica confocal microscope (Leica Microsystems, Wetzlar, Germany).
Respiratory chain enzyme activities assay
Biochemical activities of cytochrome c oxidase (COX, Complex IV), NADH–cytochrome c reductase (CI + CIII), succinate–cytochrome c reductase (CII + CIII), NADH–CoQ reductase (CI), SDH (CII) and citrate synthase, as an index of mitochondrial mass, were assayed spectrophotometrically, as previously described (30).
High resolution respirometry
Oxygen consumption rates (OCRs) were measured in intact fibroblasts using an XF24 Extracellular Flux Analyzer (Agilent Technologies) per standard protocols (31) for Proband 3 and Oxygraph 2k (Oroboros Instruments) for Proband 2. Coupling efficiency and cell respiratory control ratio (RCR) were calculated as previously reported (32). To study OCR after addition of specific oxidative substrates fibroblasts was permeabilized using 2% digitonin, and respiration was assessed during basal state and after addition of each specific substrate (33). All assays were performed in ADP excess.
ATP synthesis
Fresh fibroblasts pellets were permeabilized with 2% digitonin and incubated for 30 min with specific respiratory chain substrates and excess ADP. Specifically, substrates used to assess ATP production driven by CI were malate/pyruvate, by CII were succinate/rotenone and by CIV were TMPD/ascorbic acid. ATP levels were assessed by high-performance liquid chromatography, per previously described technique (18).
Statistical analysis
All experiments were conducted in at least biologic triplicates. Western blot bands quantifications were compared using Mann–Whitney test. P < 0.05 was considered as statistically significant. Data are expressed as mean ±standard deviation (SD).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank all families involved in this study for their participation. We are thankful to I. Wittig for generously providing USMG5 antibodies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Conflict of Interest statement: None declared.
Funding
E.B. and V.E. are supported by the North American Mitochondrial Disease Consortium (NAMDC). NAMDC is part of the Rare Diseases Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), National Center for Advanced Translational Sciences (NCATS), National Institutes of Health (NIH). NAMDC is jointly funded through an NIH U54 grant mechanism by NCATS (U54 NS078059), the National Institute of Neurological Disorders and Stroke (NINDS), the Eunice Kennedy Shriver National Institute of Child Health and Development (NICHD) and the Office of Dietary Supplements (ODS). Research reported in this publication was supported by the NIH (R03-DK082446 to M.J.F.; T32-GM008638 and K08-DK113250 to R.G.; P01-HD032062 to M.H.), The Angelina Foundation Fund from the Division of Child Development and Metabolic Disease at the Children’s Hospital of Philadelphia (M.J.F. and R.G.); the Foerderer Award for Excellence from the Children’s Hospital of Philadelphia Research Institute (X.G. and M.J.F.); the Center for Mitochondrial and Epigenomic Medicine (CMEM) at The Children’s Hospital of Philadelphia; the Genesis Foundation; Institutional Development Funds to the Center for Applied Genomics (CAG) at the Children’s Hospital of Philadelphia (CHOP); the Marriott Mitochondrial Disorders Clinical Research Network (M.H.); the Nicholas Nunno Fund (M.H.); the Mileti Fund (M.H.), the Simons Foundation and JPM foundation (W.K.C.).
References
- 1. Rahman S., Thorburn D. (2015) Nuclear gene-encoded Leigh Syndrome Overview. In Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A.. (eds), GeneReviews(R). Seattle, WA. [PubMed] [Google Scholar]
- 2. Leigh D. (1951) Subacute necrotizing encephalomyelopathy in an infant. J. Neurol. Neurosurg. Psychiatry, 14, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McCormick E., Place E., Falk M.J. (2013) Molecular genetic testing for mitochondrial disease: from one generation to the next. Neurotherapeutics, 10, 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lake N.J., Compton A.G., Rahman S., Thorburn D.R. (2016) Leigh syndrome: one disorder, more than 75 monogenic causes. Ann. Neurol., 79, 190–203. [DOI] [PubMed] [Google Scholar]
- 5. DiMauro S., Schon E.A., Carelli V., Hirano M. (2013) The clinical maze of mitochondrial neurology. Nat. Rev. Neurol., 9, 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jonckheere A.I., Smeitink J.A., Rodenburg R.J. (2012) Mitochondrial ATP synthase: architecture, function and pathology. J. Inherit. Metab. Dis., 35, 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vinothkumar K.R., Montgomery M.G., Liu S., Walker J.E. (2016) Structure of the mitochondrial ATP synthase from Pichia angusta determined by electron cryo-microscopy Proc. Natl. Acad. Sci. U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuhlbrandt W., Davies K.M. (2016) Rotary ATPases: a new twist to an ancient machine. Trends Biochem. Sci., 41, 106–116. [DOI] [PubMed] [Google Scholar]
- 9. Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D.M., Brethes D., di Rago J.P., Velours J. (2002) The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J., 21, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hahn A., Parey K., Bublitz M., Mills D.J., Zickermann V., Vonck J., Kuhlbrandt W., Meier T. (2016) Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol. Cell, 63, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thorburn D.R., Rahman J., Rahman S. (2003) Mitochondrial DNA-associated Leigh Syndrome and NARP. In Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Mefford H.C., Stephens K., Amemiya A., Ledbetter N. (eds), GeneReviews(R). Seattle, WA. [Google Scholar]
- 12. Chen R., Runswick M.J., Carroll J., Fearnley I.M., Walker J.E. (2007) Association of two proteolipids of unknown function with ATP synthase from bovine heart mitochondria. FEBS Lett., 581, 3145–3148. [DOI] [PubMed] [Google Scholar]
- 13. Lee J., Ding S., Walpole T.B., Holding A.N., Montgomery M.G., Fearnley I.M., Walker J.E. (2015) Organization of subunits in the membrane domain of the bovine F-ATPase revealed by covalent cross-linking. J. Biol. Chem., 290, 13308–13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ohsakaya S., Fujikawa M., Hisabori T., Yoshida M. (2011) Knockdown of DAPIT (diabetes-associated protein in insulin-sensitive tissue) results in loss of ATP synthase in mitochondria. J. Biol. Chem., 286, 20292–20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meyer B., Wittig I., Trifilieff E., Karas M., Schagger H. (2007) Identification of two proteins associated with mammalian ATP synthase. Mol. Cell. Proteomics, 6, 1690–1699. [DOI] [PubMed] [Google Scholar]
- 16. He J., Ford H.C., Carroll J., Douglas C., Gonzales E., Ding S., Fearnley I.M., Walker J.E. (2018) Assembly of the membrane domain of ATP synthase in human mitochondria Proc. Natl. Acad. Sci. U S A., 115, 2988–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desmet F.O., Hamroun D., Lalande M., Collod-Beroud G., Claustres M., Beroud C. (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res., 37, e67.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manfredi G., Spinazzola A., Checcarelli N., Naini A. (2001) Assay of mitochondrial ATP synthesis in animal cells. Methods Cell. Biol., 65, 133–145. [DOI] [PubMed] [Google Scholar]
- 19. Morava E., Rodenburg R.J., Hol F., de Vries M., Janssen A., van den Heuvel L., Nijtmans L., Smeitink J. (2006) Clinical and biochemical characteristics in patients with a high mutant load of the mitochondrial T8993G/C mutations. Am. J. Med. Genet. A, 140, 863–868. [DOI] [PubMed] [Google Scholar]
- 20. Seelert H., Dencher N.A. (2011) ATP synthase superassemblies in animals and plants: two or more are better. Biochim. Biophys. Acta, 1807, 1185–1197. [DOI] [PubMed] [Google Scholar]
- 21. Cogliati S., Enriquez J.A., Scorrano L. (2016) Mitochondrial Cristae: where beauty meets functionality. Trends Biochem. Sci., 41, 261–273. [DOI] [PubMed] [Google Scholar]
- 22. Strauss M., Hofhaus G., Schroder R.R., Kuhlbrandt W. (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J., 27, 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dudkina N.V., Heinemeyer J., Keegstra W., Boekema E.J., Braun H.P. (2005) Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett., 579, 5769–5772. [DOI] [PubMed] [Google Scholar]
- 24. Davies K.M., Strauss M., Daum B., Kief J.H., Osiewacz H.D., Rycovska A., Zickermann V., Kuhlbrandt W. (2011) Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. U S A, 108, 14121–14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rieger B., Junge W., Busch K.B. (2014) Lateral pH gradient between OXPHOS complex IV and F(0)F(1) ATP-synthase in folded mitochondrial membranes. Nat. Commun., 5, 3103.. [DOI] [PubMed] [Google Scholar]
- 26. Fedick A.M., Jalas C., Swaroop A., Smouha E.E., Webb B.D. (2016) Identification of a novel pathogenic OTOF variant causative of nonsyndromic hearing loss with high frequency in the Ashkenazi Jewish population. Appl. Clin. Genet., 9, 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koster J., Waterham H.R. (2017) Transfection of primary human skin fibroblasts for peroxisomal studies. Methods. Mol. Biol., 1595, 63–67. [DOI] [PubMed] [Google Scholar]
- 28. Zhou M.Y., Gomez-Sanchez C.E. (2000) Universal TA cloning. Curr. Issues Mol. Biol., 2, 1–7. [PubMed] [Google Scholar]
- 29. Wittig I., Karas M., Schagger H. (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell Proteomics, 6, 1215–1225. [DOI] [PubMed] [Google Scholar]
- 30. Paull D., Emmanuele V., Weiss K.A., Treff N., Stewart L., Hua H., Zimmer M., Kahler D.J., Goland R.S., Noggle S.A.. et al. (2012) Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature, 493, 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Invernizzi F., D'Amato I., Jensen P.B., Ravaglia S., Zeviani M., Tiranti V. (2012) Microscale oxygraphy reveals OXPHOS impairment in MRC mutant cells. Mitochondrion, 12, 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brand M.D., Nicholls D.G. (2011) Assessing mitochondrial dysfunction in cells. Biochem. J, 435, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salabei J.K., Gibb A.A., Hill B.G. (2014) Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc., 9, 421–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.