Abstract
The elucidation of the genetic code remains among the most influential discoveries in biology. While innumerable studies have validated the general universality of the code and its value in predicting and analyzing protein coding sequences, established and emerging work has also suggested that full genome decryption may benefit from a greater consideration of a codon’s neighborhood within an mRNA than has been broadly applied. This review examines the evidence for context cues in translation, with a focus on several recent studies that reveal broad roles for mRNA context in programming translation start sites, the rate of translation elongation, and stop codon identity.
Introduction
The simple concept that sixty-four distinct nucleotide triplets can be unambiguously read by the ribosome as coding sequence starts, amino acid strings, and stop signals is powerful, although also incomplete. With knowledge of an organism’s gene sequences and a few simplifying assumptions, this code could in principle allow prediction of the full set of proteins produced in that organism. Indications that additional or alternate information may lurk in mRNA sequences beyond the simple codon conversion rules have been long recognized in at least some instances (for examples, see (Baranov et al., 2015; Gouy and Gautier, 1982)), although the importance and prevalence of most such additional cues has been unclear. Recent research, however, has revealed broad and surprising complexity to genome decoding, including evidence for efficient translation initiation at unexpected sites, non-additive effects of codon pairs on translation elongation, regulated and prevalent translation readthrough of stop codons, widespread modifications to mRNA bases, and context-dependent amino acid incorporation. Together, these findings suggest a higher-order set of rules for genome decoding by the ribosome than previously thought, with great relevance for our understanding of transcript- and condition- specific regulation of protein synthesis.
Following a refresher on the primary canonical rules for eukaryotic translation and established context contributions to translation, we will review here recent developments that have defined new types of mRNA context cues and provided evidence for the widespread influence of new and previously identified cues in programming translation. Finally, the emerging role of mRNA modification on translation will be discussed. While this review focuses primarily on eukaryotic translation, the translation mechanism is remarkably conserved and thus many of the principles discussed here are applicable to all forms of life. Examples of non-eukaryotic regulation will be specifically noted at some points, when deemed relevant and informative.
The canonical model for translation
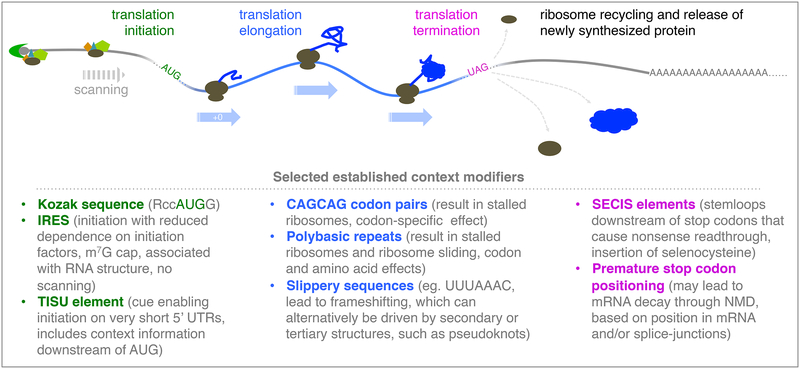
Translation of an mRNA by the eukaryotic ribosome begins with recognition of the 5’ m7G cap by a complex of proteins that recruit the small ribosomal subunit together with additional initiation factors, including an initiator tRNA conjugated to methionine and positioned in the ribosome’s peptidyl transfer (P-) site. This pre-initiation complex (PIC) scans in a 5’ to 3’ direction until it encounters an AUG start codon, which forms canonical base-pairs with the complementary initiator tRNA anticodon and triggers irreversible scanning arrest and release of a subset of initiation factors, enabling large ribosomal subunit recruitment and 80S ribosome assembly (reviewed in (Hinnebusch, 2011); Figure 1). The elongating ribosome traverses the mRNA, pausing by codon until recognition occurs by base-paring of the complementary aminoacyl-tRNA anticodon with the codon in the ribosome A-site, triggering peptide bond formation and transfer of the growing peptide chain to the A-site. Translocation moves the new tRNA-peptide conjugate, still base-paired to the mRNA, to the P-site and the initiator tRNA to the exit (E-) site, opening the A-site for the next mRNA codon to be recognized by its cognate aminoacyl-tRNA (reviewed in (Dever and Green, 2012); Figure 1). It is important to note that for much of this process, two tRNAs are closely juxtaposed within the ribosome. It has been proposed that their degree of ‘fit’ is likely to differ among specific tRNA pairs and that this may influence translation efficiency (Bossi and Ruth, 1980). The elongation process continues until one of three possible stop codons is reached, at which point a protein release factor will preferentially recognize this codon. Hydrolysis of the linkage between the elongated peptide chain and P-site tRNA releases the nascent protein, and the ribosome dissociates, aided by recycling factors (Dever and Green, 2012).
Figure 1:
Diverse context cues are known to regulate translation. The canonical model of translation is pictured above, with a selected set of established examples of context influence on translation below for initiation (green), elongation (blue), termination (pink). Some examples, such as Kozak sequences, are thought to be broadly influential in most eukaryotic organisms, while others, such as SECIS and IRES elements, are message-specific modulators and with usage that varies significantly among organisms.
Translation is largely unidirectional and extremely energy intensive as a result of multiple GTP hydrolysis steps. It is also quite precise, with errors in amino acid incorporation into the peptide chain predicted to occur at a rate between 0.01% to 0.1% (Loftfield and Vanderjagt, 1972; Ribas de Pouplana et al., 2014). Initiation at the most 5’ AUG is thought to occur in 90 to 95% of cases (Hinnebusch, 2011; Kozak, 1986) and termination efficiency at the first in-frame stop codon ranges from 90 to 99% (Arribere et al., 2016). The differences in these start and stop codon ‘efficiency’ ranges relative to those observed for amino acid incorporation during elongation was an early clue that context is a factor in programming translation initiation and termination codons. Subsequent studies defined some such cues, generally identified in an ad hoc manner or using reporter constructs, that were found to contribute to start and stop codon recognition beyond the initial simple rules put forth by the canonical translation model (Figure 1). A broad set of elegant studies thus defined global and specialized translation mechanisms and have also revealed gaps in our understanding of decoding rules and have suggested a greater role for context than is explained by standard translation rules (reviewed in (Baranov et al., 2015; Grosjean and Westhof, 2016)). Systematic approaches to define the degree to which codon context influences translation, however, have long been limited by available experimental and computational tools. This has changed with recent dramatic advances in sequencing technology that have resulted in thousands of largely complete genomes available for analysis and have enabled the development of genomic tools such as ribosome profiling, a method for systematic and in vivo translation measurement. Based on these and other advances, evidence has rapidly accumulated to suggest that mRNA context cues provide a layer of information that exceeds and occasionally overrides classical codon cues in translation initiation, elongation, and termination. Below, we discuss established historical and emergent evidence that context cues program translation initiation, elongation, and termination.
Initiation: context can influence start site selection
Pioneering studies by Marilyn Kozak in the 1980s defined the epononymous eukaryotic sequence cues that bias an AUG start codon towards higher initiation efficiencies ((Kozak, 1986), reviewed in (Hinnebusch, 2011)). These cues include a purine in the position three nucleotides before the first start codon nucleotide and a G in the position immediately after the start codon AUG. AUGs that were bypassed by the scanning PIC were generally found to lack these context cues, allowing ‘leaky scanning’ and initiation at the next AUG encountered by the PIC. Translation initiation in prokaryotes differs from that in eukaryotes, with no scanning stage and an even stronger role for context. In E.coli, AGGAGG (commonly termed the ‘Shine-Delgarno sequence’) located 4 to 7 nucleotides upstream of the initiator AUG positions the ribosome to initiate translation by base-pairing directly to the 16S rRNA (Marintchev and Wagner, 2004).
Subsequent studies have identified additional types of viral and eukaryotic translation initiation that depend on mRNA context cues, including at Internal Ribosome Entry Sites (IRESes), generally associated with RNA structure that can functionally substitute for translation initiation factors or the 5’ mRNA cap. Translation Initiator of Short 5’UTR (TISU) elements allow initiation to occur on 5’UTR (leader) regions that are fewer than 15 nucleotides long, and thus too short to allow a canonical scanning mechanism of initiation (reviewed in (Komar et al., 2012)). The broad importance of alternate context driven initiation mechanisms is highlighted by the mRNA for the conserved gene, histone H4, which has a very short 5’UTR and is translated in a manner that depends on RNA secondary structures within the ORF rather than the classic scanning mechanism (Martin et al., 2011).
Emergent evidence of pervasive non-canonical translation initiation
Ribosome profiling involves capturing and sequencing every ~30 nucleotide mRNA region (‘ribosome footprint’) that is protected from nuclease digestion by in vivo translating ribosomes in a cell population. The first ribosome profiling study (Ingolia et al., 2009), performed in budding yeast cells, largely supported predictions based on the prevailing view of translation mechanism, but also revealed efficient translation initiation at dozens of non-AUG start codons, suggesting more complex translation initiation context cues than anticipated. These alternate initiation sites were near-cognate start codons, meaning that they differed from AUG at just one nucleotide position and were positioned in 5’ leader regions, resulting in translation of generally short upstream open reading frames (uORFs; Figure 2A). Near-cognate initiation had been observed previously, but was thought to be inefficient and rare under wild-type conditions (Clements et al., 1988; Kozak, 1989; Peabody, 1989). Subsequent ribosome profiling studies also identified much more widespread near-cognate initiation than expected in a variety of organisms, including in mice (Ingolia et al., 2011) and meiotic budding yeast cells, the latter of which displayed at least 8000 instances of regulated near-cognate translation initiation in 5’ leader regions (Brar et al., 2012). Oddly, in these meiotic cells, ~3000 transcripts—or roughly half of the mRNA species present in the cell—exhibited efficient translation initiation at near-cognate codons in 5’ leaders whereas half did not, suggesting that condition-specific regulation might dramatically modulate fidelity of translation initiation for distinct pools of transcripts. While little is known about the mechanism underlying these many newly identified cases of near-cognate translation initiation, a recent mass spectrometry study found that the resultant stable amino acid chains from uORF translation in mammalian cells are initiated by methionine, indicating that such initiation events share at least some key features of the canonical scanning mechanism (Menschaert et al., 2013).
Figure 2:
Translation initiation at non-AUG codons is more common than traditionally thought and leads to unexpected protein products. A) A general model for near-cognate initiated uORF translation is shown with outstanding questions noted, including the stability of the resultant peptide and the context cues that program premature translation initiation in these cases. B) Translation initiation at in-frame near-cognate codons prior to the canonical AUG start codon can produce an alternate protein product. C) and D) A working model for RAN translation in multiple frames, based on the case for CGG repeats in the 5’ leader of FMR1. C) illustrates translation in the GGC frame, resulting in poly-glycine peptides. D) illustrates translation in the GCG frame, resulting in poly-alanine peptides. Translation initiation at non-AUG codons in both of these cases can occur simultaneously on different transcript molecules in the same cell population. RAN translation of FMR1 is cap-dependent and proposed to involve downstream RNA structures within the tri-nucleotide repeat stretch that affect start site selection stringency.
AUG-initiated uORF translation was similarly seen to be widespread in several other ribosome profiling studies, including in yeast, plants, human, mouse, and zebrafish and such cases have been more heavily investigated for context cues than near-cognate initiated uORFs to date (Brar et al., 2012; Chew et al., 2013, 2016; Fields et al., 2015; Ingolia et al., 2011, 2014; Johnstone et al., 2016; Liu et al., 2013). Analyses of vertebrate cases have found evidence for positional conservation for translated uORFs, indicating functional importance for these unexpected translation events (Chew et al., 2016; Fields et al., 2015; Johnstone et al., 2016). Studies of neighboring sequences that allow translation initiation at non-AUG start codons or normally bypassed AUG codons have not yet yielded a clear set of context signals, however. Some analyses, for example, have suggested that translated uORFs generally conform to Kozak sequence rules (Menschaert et al., 2013), while others have not observed this trend (Chew et al., 2016), suggesting the influence of as-yet unidentified context cues. Similar cues may influence the translation initiation efficiency of traditional ORFs, as the uORF initiation codons that were most likely to be translated in zebrafish had upstream context that was most like that seen upstream of known coding sequence start codons (Johnstone et al., 2016).
A subset of newly identified AUG and near-cognate translation initiation sites in 5’ leader regions are in frame with the canonical ORF and lack intervening stop codons. Their translation results in a protein with an N-terminal extension of the canonical ORF-encoded protein product, rather than the short, possibly unstable peptide products expected to result from uORF translation (Figure 2B;(Brar et al., 2012; Fields et al., 2015; Ingolia et al., 2011; Ivanov et al., 2011; Menschaert et al., 2013)). The context cues leading to these cases of alternative initiation remain undefined and are likely to be similar to those seen for uORFs, but their biological significance is clearly different, as these translation initiation events result in extensions to proteins that may affect their properties, including localization (Baranov et al., 2015; Touriol et al., 2003).
RAN translation: initiation driven entirely by context?
In parallel with the discovery of widespread translation initiation in 5’ leader regions, a disparate type of context-dependent translation initiation was found to be prevalent in repeat expansion diseases, a group of neurodegenerative or neuromuscular conditions that are associated with expanded triplet or sextuplet repeats. The prevailing models for toxicity in these diseases posit that translation of the expanded regions, which frequently repeat glutamine codons, result in proteins that are aggregation prone or that the repeat RNAs cause toxicity by binding to and limiting free pools of other proteins. (Cleary and Ranum, 2014; Green et al., 2016). Zu et al. (Zu et al., 2011) first identified a type of aberrant translation initiation from within CAGCUG repeats of ATXN8 (associated with spinocerebellar ataxia). The resultant poly-alanine protein products can be observed in cell types that typically degenerate as part of SCA8 progression in mice and in human autopsy tissue from SCA8 patients. Surprisingly, the translation of these polymers was found to be independent of an AUG or near-cognate codon and was thus termed ‘repeat-associated non-ATG’ (RAN) translation. RAN translation was subsequently observed in other repeat expansion genes, including most strikingly, the amyotrophic lateral sclerosis- and frontotemporal dementia- associated C9ORF72, in which both sense and antisense transcripts were found to be translated in all three frames without any initiating AUG codon, resulting in three distinct peptide polymers from the GGGGCC or CCCGG repeats (Cleary and Ranum, 2014). The degree to which RAN translation products contribute to pathology remains unclear, but the prevalence of RAN translation in these repeat contexts has raised interesting questions about the non-canonical mechanism of translation initiation underlying the phenomenon.
IRES-based initiation has been proposed as a potential RAN translation mechanism, but recent work has shown that RAN translation from FMR1 (associated with fragile X associated tremor and ataxia syndrome; Figure 2C, 2D) requires an m7G mRNA cap and ribosome scanning components, suggesting a modification only in start site selection cues rather than an entirely alternative initiation mechanism (Cleary and Ranum, 2014; Green et al., 2016; Kearse et al., 2016). In the case of FMR1, as with other cases of RAN translation, efficient repeat-based initiation is highly dependent on long repeats, which result in stable mRNA secondary structures. It has been proposed that such secondary structures downstream of RAN translation initiation sites may stall the ribosome pre-initiation complex, resulting in a higher probability of a non-canonical initiation event at the stall site. Interestingly, it has also been suggested that aspects of this mechanism may be more general and could explain the translation initiation at near-cognate codons in 5’ leaders discussed above (Kearse et al., 2016). This model is intriguing, given that a number of IRES-based mechanisms have been proposed for observed cases of non-canonical translation initiation but rigorous in vivo proof of IRES activity outside of viral systems has been elusive in many cases (reviewed in (Komar et al., 2012; Kozak, 2002)). A model in which secondary structure kinetically modifies the classical scanning mechanism rather than allowing internal ribosome entry may prove to be applicable to some such inconclusive cases and is consistent with trends seen at AUG start codons in suboptimal context (Hinnebusch, 2011).
The long-standing enigma of context-influenced translation elongation
Despite established roles for specific context cues in initiation and termination (discussed below), the general importance of context in translation elongation has been difficult to rigorously determine outside of important but infrequently observed mechanisms such as frameshifting (reviewed in (Baranov et al., 2015)). This lack of clarity is best exemplified by the deliberate codon optimization approaches that are a common step in designing constructs for efficient heterologous expression. Early strategies for codon optimization were based on the idea that codons that were infrequently used in a given organism, relative to synonymous options, should be avoided to achieve maximum translation elongation speed and protein production. The rationale was that rare codons are associated with low abundance cognate tRNAs and that translation elongation should be slowed at these sites while the ribosome waits for these aminoacyl-tRNAs to diffuse to the A-site. It was observed, however, that often fully optimized open reading frames (ORFs) were not translated as efficiently as expected, leading to an array of hypotheses, including those implicating neighboring codon effects and competition for tRNAs (Elena et al., 2014; Gustafsson et al., 2004; Hatfield and Roth, 2007). Advances in codon optimization algorithms for higher protein expression have, by necessity, largely come from empirical observations and have tended to lack concrete mechanistic explanations. Based on these algorithms, it has however become clear that most of the commonly seen differences in protein expression cannot be accounted for by our existing understanding of codon effects on translation elongation. It has been proposed that this may be due to a lack of importance of codon context within ORFs relative to the effects of initiation rate, but also may reflect the existence of context cues that we do not yet understand (Pop et al., 2014; Shah et al., 2013). Whether elongation rate sets protein production rates under normal cellular conditions, greater understanding of context effects in translation would clearly have significant utility in advancing diverse synthetic biology and biochemical approaches.
New evidence that ribosome-tRNA interaction influence translation elongation
Codon context influence on translation elongation has been extremely challenging to study due to the highly constrained nature of the nucleotide sequence of mRNA, which has evolved to accommodate secondary structures, splicing cues, binding sites for proteins, the encoded protein sequence, and also reflects mutational biases (Letzring et al., 2010; Moura et al., 2007). For these reasons, although it has long been known that individual and codon pair biases exist in all organisms, assessing the broad role of codon context in translation has been challenging. Moreover, no consensus has emerged on the relative importance of various factors in programming translation elongation rate, including tRNA availability, tRNA-ribosome interactions, and interactions between the ribosome and amino acids conjugated to a tRNA or traversing the ribosome exit tunnel.
Ribosome profiling has provided some insight into this issue, while also revealing surprising complexity to the impact of specific codons on translation. Each ribosome footprint can be mapped to the codon representing the A-site position of the translating ribosome that protected it, theoretically allowing interpretation of the amount of time taken to decode each codon (Ingolia et al., 2012). After known artifacts based on drug treatment during sample harvesting and biases in footprint sizes are taken into account (discussed in (Brar and Weissman, 2015; Hussmann et al., 2015; Lareau et al., 2014)), several groups have observed reproducible codon-specific variability in ribosome dwell time. One such analysis concluded that ribosome occupancy levels at a given position was influenced by tRNA:mRNA interactions and the amino acid being encoded (Lareau et al., 2014). Specifically, as observed in nematodes (Stadler and Fire, 2011), some wobble anticodon:codon interactions were associated with markedly higher ribosome footprint occupancy levels, interpreted to reflect slower decoding than canonical Watson-Crick anticodon: codon interactions. It was also found that codons for small, polar amino acids showed greater ribosome occupancy than those for large hydrophobic amino acids, suggesting that interactions between the ribosome and the amino acid in the A-site contribute to translation elongation rate (Lareau et al., 2014). In vivo mRNA structure has been weakly implicated in the elongation rate of an mRNA, Pro-Pro and polybasic repeats have been shown to modulate translation elongation, and analyses of multiple published ribosome profiling data sets to show that there is likely to be an additional contribution of tRNA abundance on translation elongation rate, as broadly predicted (Artieri and Fraser, 2014a; Bazzini et al., 2016; Brandman et al., 2012; Hussmann et al., 2015; Ingolia et al., 2011; Koutmou et al., 2015; Lareau et al., 2014; Pop et al., 2014; Weinberg et al., 2016).
Gamble et. al performed a large-scale, systematic screen aimed at assessing the degree to which codon context may modulate eukaryotic translation elongation rates beyond effects seen at individual codons (Gamble et al., 2016). The authors screened yeast cell populations housing libraries containing random sets of triplet codons within an ORF encoding superfolder GFP. Controlling for reading frame and excluding variants predicted to give rise to inhibitory RNA structures, it was found that 17 codon pairs, rather than single codons, were strongly associated with reduced GFP expression. This set included the known inhibitory codon pair CGA-CGA (Letzring et al., 2010), and was enriched for codons decoded by wobble interactions. Ribosome profiling data were consistent with the predicted translation elongation slowing over these codon pair sets and GFP expression could be rescued by overexpression of the complementary tRNA to the 3’ (A-site) codon in the pair or, to a greater degree, by mutation of the tRNA such that wobble interactions were no longer required for codon recognition. In codon pair cases in which the 5’ (P-site) codon required wobble decoding, expression of a mutant tRNA version that no longer required wobble tRNA:codon recognition also suppressed the GFP expression defect. Moreover, among the 12 inhibitory codon pairs that were not repeats of the same codon, in all cases the translation inhibition depended on the ordering of the pair, suggesting that the specific positioning of their complementary tRNAs base-paired to these mRNA codon pairs within the ribosome mediates the inhibitory effect (Figure 3;(Gamble et al., 2016)). A model by which tRNA interactions within the ribosome can affect translation is consistent with findings from E.coli, in which authors observed that P-site tRNA:codon mismatches can trigger a quality control pathway that reduces the fidelity of subsequent A-site tRNA:codon recognition, ultimately triggering premature translation termination (Zaher and Green, 2009). Moreover, analyses of the thermodynamics of aminoacyl-tRNA binding within the ribosome decoding site suggests that all natural tRNAs show similar binding energies despite strongly differing codon-anticodon pairing stabilities, suggesting significant interactions among—and coevolution of— the ribosome structure, tRNA sequence, and codon usage within mRNAs (reviewed in (Grosjean and Westhof, 2016)).
Figure 3:
Codon pair orientation can influence translation elongation rate. Interactions between P- and A-site tRNAs within the ribosome can result in slowed translation elongation for codon pairs in an orientation-dependent manner. This effect is rescued by replacement of wobble codon:anticodon pairing with exclusively Watson-Crick base-pairing for either the A-site (3’) or P-site (5’) tRNA:mRNA interaction.
These findings highlight the need to consider adjacent codon sequences in the context of a translating ribosome (also in (Chevance et al., 2014); reviewed in (Grosjean and Westhof, 2016)). It is interesting to note that, while there are strong codon pair biases known to exist all organisms, the inhibitory pairs identified in this study are not generally the rarest codon pairs present in the yeast ORFeome (Moura et al., 2005). This is likely to partially reflect evolutionary constraints including mutational bias, but it may also support a model in which translation speed modulation may be selected for due to a regulatory function, perhaps in enabling proper co-translational protein folding (Gingold and Pilpel, 2011). The recent observation that the codon composition of the abundant set of mRNAs, in concert with corresponding tRNAs, can be dynamic, shifting in response to cellular stress or proliferation status (Gingold et al., 2012, 2014) adds even greater complexity to analyses of the functional significance of codon pair composition in translation speed.
In vivo single molecule imaging of translation detects elongation heterogeneity
A major limitation of the in vivo methods discussed thus far to dissect codon context effects in translation elongation is that they require averaging, sometimes among mRNAs for different genes and always among different mRNA molecules for a given gene. These approaches can be valuable in identifying trends, but they are unable to identify complexity based on heterogeneity. Single molecule biophysical approaches have been valuable in dissecting bacterial translation dynamics and can detect molecular heterogeneity, but typically allow study of only a small number of messages and in an ex vivo system (reviewed in (Bustamante et al., 2014; Tsai et al., 2016)). Recently, several groups converged in development of a suite of tools for live imaging of in vivo translation of single transcripts that allows direct monitoring of variability in translation elongation rates (Morisaki et al., 2016; Wang et al., 2016; Wu et al., 2016; Yan et al., 2016). These methods use simultaneous fluorescent tagging of individual mRNAs and the nascent peptide produced from their translation, enabling quantitative analyses of diverse aspects of translation. Application of the very similar methodologies to different mRNAs in different biological systems revealed average translation elongation rates that ranged from 3 to 10 codons per second, roughly similar to each other and also the rate of 5.6 codons per second determined using run-off analyses by ribosome profiling (Ingolia et al., 2011). The differences in the measurements may reflect condition- or mRNA- specific variability in translation elongation. In fact, one of the studies reported elongation speed variability based on codon composition, with a faster elongation rate measured for a codon optimized reporter construct relative to a non-optimized transcript (Yan et al., 2016). The same study also identified apparently bimodal heterogeneity in elongation rates, even among different molecules of the same mRNA. These findings suggest surprising variability in translation dynamics among individual mRNA molecules, and also highlight the potential of these approaches for future mechanistic dissection of context effects on translation elongation.
Termination: context can define the meanings of stop codons
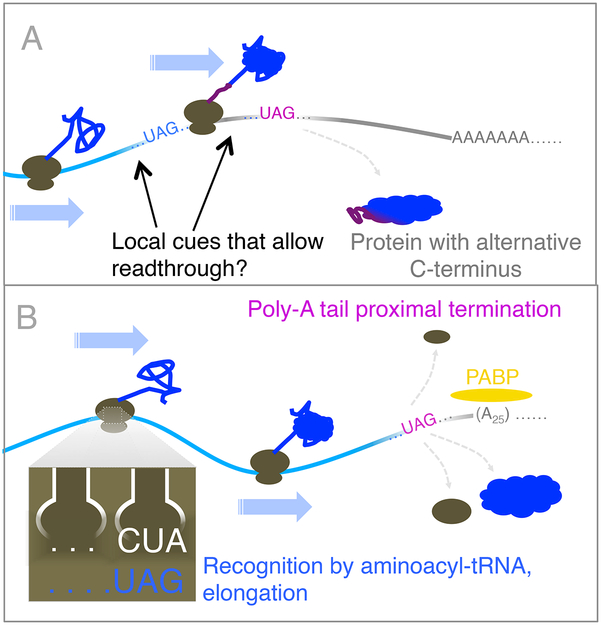
The first hint that context affects termination came from a study of nonsense suppression (Bossi and Ruth, 1980), in which a mutant tRNA with an anticodon sequence complementary to the UAG stop codon competed with the release factor to suppress termination at that stop codon. It was observed that there was variability over an order of magnitude in suppression efficiency that depended on the nucleotide immediately 3’ of the stop codon. Since then, numerous context-dependent modifiers of termination have been identified. For example, the position of a stop codon relative to other features of an mRNA can commit the mRNA to destruction by nonsense-mediated decay (NMD; reviewed in (Hug et al., 2016)); the highly specific, selenocysteine insertion sequence (SECIS)-dependent insertion of the amino acid selenocysteine at a stop codon (reviewed in (Baranov et al., 2015; Gonzalez-Flores et al., 2013)) allows production of full-length, functional proteins from coding regions that use them; and readthrough, in which termination at the stop codon yields wild-type protein and translation through the stop codon yields an extended protein isoform that my have altered or even opposite function (Figure 4A; (Baranov et al., 2015; Eswarappa and Fox, 2015; Steneberg et al., 1998)). While the mechanisms and significance of NMD and selenocysteine insertion are largely established, with few exceptions the causes and consequences of readthrough remain enigmatic. Nonetheless, biologically significant examples of readthrough in non-pathological conditions have been observed, and cases have even been identified in which readthrough is essential to life (Baranov et al., 2015; Bekaert et al., 2010; True et al., 2004), indicating that low termination efficiency, rather than a stochastic error, may be an evolutionarily selected property.
Figure 4:
Stop codon readthrough may be programmed or suppressed by mRNA context cues. A) In most organisms, stop codon readthrough is thought to be rare, but can occur in a message-specific manner, dependent on local mRNA context, and can result in a C-terminal extension to a protein encoded by a canonical ORF. B) In the marine ciliate, C. magnum, recognition of canonical stop codons as sense codons is the default state within an ORF, and recognition of such codons as termination cues is determined by their proximity to the 3’ mRNA end, potentially determined through steric interactions of the poly-A binding protein with the translation apparatus.
Emergent evidence for context as a major determinant of termination
The pervasive influence of context on translation termination has been highlighted by recent phylogenetic and ribosome profiling analyses. Elegant genomic analysis of 12 Drosophila species predicted ~280 cases of conserved stop codon readthrough, based on signatures of protein-coding conservation (Jungreis et al., 2011). Ribosome profiling studies provided experimental evidence for many of these predicted cases, but also found translational readthrough in flies to be generally much more common than previously thought, and often regulated. The majority of these novel readthrough cases did not show signs of conservation across species, although they did show signs of selection within D. melanogaster, suggesting lineage specificity. In several cases, it was shown that the C-terminal extensions resulting from these readthrough events (Figure 4A) shifted the cellular localization of the encoded protein, implying biological significance. The efficiency of readthrough relative to termination for the new cases identified in flies was seen to vary over at least three orders of magnitude, though the context cues enabling these readthrough events remain largely mysterious (Dunn et al., 2013). Subsequent analyses continue to identify additional cases of readthrough in fungi (Artieri and Fraser, 2014b), insects (Jungreis et al., 2016), and mammals (Loughran et al., 2014), suggesting roles for translational readthrough in protein regulation, localization, and evolution, and consistent with earlier reports that elevated readthrough can play a functional role in organismal fitness (Baranov et al., 2015; Bekaert et al., 2010; True et al., 2004).
Using computational screening, the Nowacki lab recently identified a marine ciliate, C. magnum, that contains no dedicated stop codons. Ciliates have previously been seen to have stop codon reassignments—in fact, studies dating back decades noted that ciliate genes could not be efficiently expressed in other organisms due to this phenomenon (Caron and Meyer, 1985)—but Nowacki and colleagues present the first organism in which all stop codons appear to have been (conditionally) reassigned to sense codons. Analyses of substitution rates for essential genes, along with ribosome profiling data, provided strong evidence that in C. magnum, all three stop codons are used to encode amino acids. tRNA sequencing identified natural ‘nonsense suppressor’ aminoacyl-tRNAs for two of the three stop codons, providing mechanistic clues to how stop-codon reassignment occurred in this organism (Swart et al., 2016). It was observed that the three standard stop codons are depleted from the last 50 nucleotides of coding sequences except at the terminal position, where ribosome profiling data confirmed that protein synthesis efficiently terminates. Remarkably, the positioning of these codons at the 3’ end of transcripts was a sufficient context cue to define them as either sense or termination codons, resulting in readthrough rates that are similar to those seen in organisms with dedicated stop codons. The authors noted that the 3’UTRs of C. magnum are unusually short and therefore propose proximity to polyA tail binding protein, PABP, as a trigger for stop codon recognition (Figure 4B;(Swart et al., 2016)). This is an intriguing potential mechanism, as PABP interactions with NMD machinery have also been widely proposed to explain the ability of this pathway to distinguish between premature and normal stop codons (Hug et al., 2016).
Although simultaneous, fully context-dependent regulation of all three stop codons may not exist outside of ciliate lineages, the case demonstrates intriguing parallels to the standard translation initiation mechanism. In translation initiation, the largely unidirectional nature of scanning complex progression determines that proximity to the 5’ mRNA end is a primary factor specifying the utilized start codon (Kozak, 2002). Also similarly, while near-cognate initiated uORFs are translated on some messages and some AUG codons are bypassed during scanning, both are depleted from directly prior to the canonical start codon (Brar et al., 2012; Chew et al., 2016; Johnstone et al., 2016; Kozak, 1987; Zur and Tuller, 2013).
mRNA modifications are widespread and impact translation at multiple levels
Modifications to the four standard RNA nucleotides have long been recognized to influence RNA structure and function, but until recently these effects were thought to be mostly limited to noncoding RNAs (Machnicka et al., 2013). A recent flurry of genome-wide studies, however, have identified widespread methylation of mRNA bases (m6A, m5C, m1A) beyond the well characterized m7G cap, and isomerization of uridine to pseudouridine (Ψ) within mRNA populations in bacterial, yeast, and mammalian cells (Carlile et al., 2014; Deng et al., 2015; Dominissini et al., 2012, 2012, 2016; Edelheit et al., 2013; Ke et al., 2015; Khoddami and Cairns, 2013; Li et al., 2015, 2016; Liu and Pan, 2015; Lovejoy et al., 2014; Meyer et al., 2012; Schwartz et al., 2013, 2014; Squires et al., 2012). Most of these newly-identified modifications are of unknown function, but there is strong evidence that they may influence multiple aspects of mRNA structure, stability, and function (reviewed in (Gilbert et al., 2016)) and their study may shed light on aspects of translation that thus far cannot be explained by sequence analyses alone.
Several such mRNA modifications have been shown to directly influence translation initiation, elongation, and termination. In the case of m6A, the most abundant of the known mRNA modifications, the position of base modification affects its impact on translation. Under most conditions examined thus far, m6A is seen to be enriched near 3’ ends of mRNA ORFs and 3’ UTRs, where it has been shown to affect mRNA stability, translation, and polyA site choice (Ke et al., 2015; Wang et al., 2014, 2015; Zhang et al., 2016). Under heat shock and other cellular stress conditions, there is removal of 3’ m6A modifications and a relative increase in 5’ leader m6A modifications for a subset of messages, including that for the chaperone Hsp70 (Figure 5A, 5B;(Dominissini et al., 2012; Meyer et al., 2015; Zhou et al., 2015)). These new 5’ m6A bases are capable of directly recruiting the translation initiation factor eIF3 and enabling translation initiation independent of the canonical eIF4E cap-binding complex. It is interesting to note that, in contrast to the canonical cap-dependent translation initiation model, which requires eIF4E-cap interaction, recent work has shown that eIF3 can form an alternate cap-binding complex (Lee et al., 2016). This result suggests that pervasive perception of ‘cap-dependent’ as generally synonymous with eIF4E-dependent may require revisiting, and that choice between these two types of cap-dependent initiation may represent a broad new mode of selective translation initiation control. The eIF4E-independent translation initiation seen for HSP70 is highly specific to m6A modifications and also highly dependent on the context of the modification. A non-structured 5’ mRNA end is required, implying a scanning rather than IRES-based mechanism of translation initiation, and the effect was seen to be strongest when the methylated A was flanked by a 5’G and 3’C base. m6A modifications outside of the 5’ leader did not retain this activity (Meyer et al., 2015). Given the reversibility of this modification, these results suggest a mechanism by which mRNA initiation cues can be dynamically and selectively modulated.
Figure 5:
mRNA modifications can reprogram translation initiation and termination cues. A) HSP70 translation under steady-state conditions, with no m6A 5’ modification is low, and initiated by the canonical m7G mRNA cap and eIF4E-based mechanism. B) Cap-independent translation of HSP70 is efficient after heat shock, and depends on a 5’ m6A modification, through eIF3 recognition. C) and D) Stop codon readthrough and serine/threonine incorporation can be seen to be enhanced by pseudouridylation of the stop codon.
Pseudouridylation, unlike m6A modification, is not reversible and thus seems likely to mediate less dynamic roles in translation. The clearest function established thus far for Ψ modifications in mRNA is in nonsense suppression (Karijolich and Yu, 2011). Site-specific targeting of Ψ to a premature stop codon in yeast reporter system led to amino acid incorporation at all three stop codons (Figure 5C, 5D). Mass spectrometry analysis determined that incorporation of either serine or threonine, or phenylalanine or tyrosine, occurred at the pseudouridylated codon, depending on the stop codon sequence that was modified. Interestingly, the amino acids incorporated show similarity in structure, but the tRNAs that supply these amino acids do not show similar anticodon sequences, leading the authors to propose either the presence of an unidentified modification of the tRNAs or a change in the local structure of the A-site that enables recognition driven partially by the conjugated amino acid rather than tRNA anticodon sequence (Karijolich and Yu, 2011). A subsequent crystal structure of the bacterial ribosome with a ΨAG codon stably bound to a tRNA(Ser) revealed the unusual purine-purine basepairing that can enable such cases of nonsense suppression (Fernández et al., 2013). The degree to which nonsense suppression is seen at endogenous sites of Ψ modification remains unclear, as stop codons were not enriched for Ψ modification in large-scale datasets (Carlile et al., 2014; Lovejoy et al., 2014; Schwartz et al., 2014).
m5C mRNA modifications can be seen in mammals and bacteria, and a recent study showed that m5C at the second nucleotide of the CCC proline-encoding codon resulted in leucine or isoleucine misincorporation at a rate of 4% in bacteria (Hoernes et al., 2016). The effects of this type of recoding on protein structure and translation elongation, and the degree of in vivo amino acid misincorporation at endogenously modified sites, remain intriguing open questions. It is interesting to consider that RNA modifications, including m5C, pseudouridylation, and m6A, are now known to be abundant on all three major cellular components that intimately interact during translation—mRNA, tRNAs, and the rRNA of the ribosome. Given the apparent universality and abundance of such modifications, it seems likely that their study will prove to be critical to unraveling the complex set of structural and sequence cues that program translation.
Prospects
A recent surge of analytical and experimental tool development has catalyzed new directions in translation research that continue to reveal important and often conserved aspects of this process that have been inaccessible or difficult to systematically assess by previously available approaches. Experimental tools for genome-wide monitoring of gene expression have allowed us to identify features of translation—including broad and prevalent effects due to mRNA context—that fall outside of our expectations without having to know the mechanisms or prevalence of such behaviors. As these approaches have allowed us to begin filling gaps in our understanding of the role of mRNA sequence and structure context in translation, they also pave the way for tackling a new and exciting set of mechanistic questions on how changes in the ribosome, mRNAs, aminoacyl, tRNAs, and other factors impact each other and the polypeptide product in the extremely close quarters of a translating ribosome.
Intimately linked with the increased understanding of the complexity of the mechanism of translation suggested by recent studies, has been emerging evidence that broad and long-standing categorizations of coding versus noncoding regions may be flawed, with a large and diverse set of data suggesting surprising prevalence to regulated translation in unexpected regions. These include 5’ and 3’ mRNA regulatory regions, multiple reading frames within long tri-nucleotide repeats, and short regions within RNAs previously thought to be noncoding (Aspden et al., 2014; Bazzini et al., 2014; Bekaert et al., 2010; Brar and Weissman, 2015; Brar et al., 2012; Chew et al., 2013, 2016; Dunn et al., 2013; Fields et al., 2015; Green et al., 2016; Ingolia et al., 2011, 2014; Ji et al., 2015; Jungreis et al., 2011; Ma et al., 2014; Menschaert et al., 2013; Stern-Ginossar et al., 2012; Young et al., 2015; Zu et al., 2011). This rich body of recent research has allowed us to begin to form more complete set rules for predicting the full set of genomic regions that are translated and the levels of the translation of these regions.
Key outstanding questions include the specific set of context cues that specify uORF translation, alternative start codon selection, mRNA modifications, and translational readthrough, as well as the functional importance of these phenomena and their degree of condition-specific regulation. We are just beginning to be able to identify and verify sequences and additional cues that affect translation elongation, an area of study that is likely to be important for advancing our understanding of translation fidelity and protein folding. Future breakthroughs are likely to result from integrating the findings of genome-scale studies with single molecule analyses of in vivo translation, the continued development and application of computational tools to new and existing gene expression and genome sequence datasets, functional screening, as well as advances in our ability to solve in vivo mRNA structures. Together these approaches will bring us yet closer to full decryption of the complex, layered information encoded in mRNA sequences.
Acknowledgements:
I thank Joshua Dunn and Calvin Jan for critical reading of this manuscript and helpful discussions. I am grateful for the NIH funding (DP2-GM-119138 and P50-GM102706) that supports research in my lab, along with investigator awards from the Sloan and Pew Foundations.
Sources cited
- Arribere JA, Cenik ES, Jain N, Hess GT, Lee CH, Bassik MC, and Fire AZ (2016). Translation readthrough mitigation. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri CG, and Fraser HB (2014a). Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 24, 2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri CG, and Fraser HB (2014b). Evolution at two levels of gene expression in yeast. Genome Res. 24, 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MAS, Brocard M, and Couso J-P (2014). Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3, e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Atkins JF, and Yordanova MM (2015). Augmented genetic decoding: global, local and temporal alterations of decoding processes and codon meaning. Nat. Rev. Genet 16, 517–529. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al. (2014). Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 33, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, and Giraldez AJ (2016). Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. e201694699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M, Firth AE, Zhang Y, Gladyshev VN, Atkins JF, and Baranov PV (2010). Recode-2: new design, new search tools, and many more genes. Nucleic Acids Res. 38, D69–D74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L, and Ruth JR (1980). The influence of codon context on genetic code translation. Nature 286, 123–127. [DOI] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li G-W, Zhou S, King D, Shen PS, Weibezahn J, et al. (2012). A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, and Weissman JS (2015). Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol 16, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, and Weissman JS (2012). High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CJ, Kaiser CM, Maillard RA, Goldman DH, and Wilson CAM (2014). Mechanisms of cellular proteostasis: insights from single-molecule approaches. Annu. Rev. Biophys 43, 119–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, and Gilbert WV (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron F, and Meyer E (1985). Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature 314, 185–188. [DOI] [PubMed] [Google Scholar]
- Chevance FFV, Le Guyon S, and Hughes KT (2014). The effects of codon context on in vivo translation speed. PLoS Genet. 10, e1004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew G-L, Pauli A, Rinn JL, Regev A, Schier AF, and Valen E (2013). Ribosome profiling reveals resemblance between long non-coding RNAs and 5’ leaders of coding RNAs. Dev. Camb. Engl 140, 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew G-L, Pauli A, and Schier AF (2016). Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat. Commun 7, 11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, and Ranum LPW (2014). Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders. Curr. Opin. Genet. Dev 26, 6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements JM, Laz TM, and Sherman F (1988). Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol. Cell. Biol 8, 4533–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Chen K, Luo G-Z, Weng X, Ji Q, Zhou T, and He C (2015). Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 43, 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, and Green R (2012). The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol 4, a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. (2016). The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, and Weissman JS (2013). Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2, e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, and Sorek R (2013). Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena C, Ravasi P, Castelli ME, Peirú S, and Menzella HG (2014). Expression of codon optimized genes in microbial systems: current industrial applications and perspectives. Front. Microbiol 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarappa SM, and Fox PL (2015). Antiangiogenic VEGF-Ax: A New Participant in Tumor Angiogenesis. Cancer Res. 75, 2765–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández IS, Ng CL, Kelley AC, Wu G, Yu Y-T, and Ramakrishnan V (2013). Unusual base pairing during the decoding of a stop codon by the ribosome. Nature 500, 107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Rodriguez EH, Jovanovic M, Stern-Ginossar N, Haas BJ, Mertins P, Raychowdhury R, Hacohen N, Carr SA, Ingolia NT, et al. (2015). A Regression-Based Analysis of Ribosome-Profiling Data Reveals a Conserved Complexity to Mammalian Translation. Mol. Cell 60, 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble CE, Brule CE, Dean KM, Fields S, and Grayhack EJ (2016). Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV, Bell TA, and Schaening C (2016). Messenger RNA modifications: Form, distribution, and function. Science 352, 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, and Pilpel Y (2011). Determinants of translation efficiency and accuracy. Mol. Syst. Biol 7, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Dahan O, and Pilpel Y (2012). Dynamic changes in translational efficiency are deduced from codon usage of the transcriptome. Nucleic Acids Res. 40, 10053–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sørensen KD, et al. (2014). A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Flores JN, Shetty SP, Dubey A, and Copeland PR (2013). The molecular biology of selenocysteine. Biomol. Concepts 4, 349–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, and Gautier C (1982). Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10, 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Linsalata AE, and Todd PK (2016). RAN translation-What makes it run? Brain Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, and Westhof E (2016). An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, and Minshull J (2004). Codon bias and heterologous protein expression. Trends Biotechnol. 22, 346–353. [DOI] [PubMed] [Google Scholar]
- Hatfield GW, and Roth DA (2007). Optimizing scaleup yield for protein production: Computationally Optimized DNA Assembly (CODA) and Translation Engineering. Biotechnol. Annu. Rev 13, 27–42. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (2011). Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. MMBR 75, 434–467, first page of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoernes TP, Clementi N, Faserl K, Glasner H, Breuker K, Lindner H, Hüttenhofer A, and Erlacher MD (2016). Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 44, 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug N, Longman D, and Cáceres JF (2016). Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 44, 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, and Press WH (2015). Understanding Biases in Ribosome Profiling Experiments Reveals Signatures of Translation Dynamics in Yeast. PLoS Genet. 11, e1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, and Weissman JS (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJS, Jackson SE, Wills MR, and Weissman JS (2014). Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8, 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Firth AE, Michel AM, Atkins JF, and Baranov PV (2011). Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 39, 4220–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, and Struhl K (2015). Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4, e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, and Giraldez AJ (2016). Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 35, 706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I, Lin MF, Spokony R, Chan CS, Negre N, Victorsen A, White KP, and Kellis M (2011). Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 21, 2096–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungreis I, Chan CS, Waterhouse RM, Fields G, Lin MF, and Kellis M (2016). Evolutionary dynamics of abundant stop codon readthrough in Anopheles and Drosophila. [DOI] [PMC free article] [PubMed]
- Karijolich J, and Yu Y-T (2011). Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 29, 2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, and Todd PK (2016). CGG Repeat-Associated Non-AUG Translation Utilizes a Cap-Dependent Scanning Mechanism of Initiation to Produce Toxic Proteins. Mol. Cell 62, 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, and Cairns BR (2013). Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol 31, 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Mazumder B, and Merrick WC (2012). A new framework for understanding IRES-mediated translation. Gene 502, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, and Green R (2015). Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1986). Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Kozak M (1987). An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1989). Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol 9, 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (2002). Pushing the limits of the scanning mechanism for initiation of translation. Gene 299, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, and Brown PO (2014). Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 3, e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Kranzusch PJ, Doudna JA, and Cate JHD (2016). eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Dean KM, and Grayhack EJ (2010). Control of translation efficiency in yeast by codon-anticodon interactions. RNA N. Y. N 16, 2516–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, and Yi C (2015). Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol 11, 592–597. [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, and Yi C (2016). Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol 12, 311–316. [DOI] [PubMed] [Google Scholar]
- Liu N, and Pan T (2015). Probing RNA Modification Status at Single-Nucleotide Resolution in Total RNA. Methods Enzymol. 560, 149–159. [DOI] [PubMed] [Google Scholar]
- Liu M-J, Wu S-H, Wu J-F, Lin W-D, Wu Y-C, Tsai T-Y, Tsai H-L, and Wu S-H (2013). Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25, 3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftfield RB, and Vanderjagt D (1972). The frequency of errors in protein biosynthesis. Biochem. J 128, 1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Chou M-Y, Ivanov IP, Jungreis I, Kellis M, Kiran AM, Baranov PV, and Atkins JF (2014). Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 42, 8928–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP, and Brown PO (2014). Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PloS One 9, e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ward CC, Jungreis I, Slavoff SA, Schwaid AG, Neveu J, Budnik BA, Kellis M, and Saghatelian A (2014). Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. J. Proteome Res. 13, 1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. (2013). MODOMICS: a database of RNA modification pathways−−2013 update. Nucleic Acids Res. 41, D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, and Wagner G (2004). Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys 37, 197–284. [DOI] [PubMed] [Google Scholar]
- Martin F, Barends S, Jaeger S, Schaeffer L, Prongidi-Fix L, and Eriani G (2011). Cap-assisted internal initiation of translation of histone H4. Mol. Cell 41, 197–209. [DOI] [PubMed] [Google Scholar]
- Menschaert G, Van Criekinge W, Notelaers T, Koch A, Crappé J, Gevaert K, and Van Damme P (2013). Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics MCP 12, 1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, and Jaffrey SR (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S-B, and Jaffrey SR (2015). 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T, Lyon K, DeLuca KF, DeLuca JG, English BP, Zhang Z, Lavis LD, Grimm JB, Viswanathan S, Looger LL, et al. (2016). Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429. [DOI] [PubMed] [Google Scholar]
- Moura G, Pinheiro M, Silva R, Miranda I, Afreixo V, Dias G, Freitas A, Oliveira JL, and Santos MAS (2005). Comparative context analysis of codon pairs on an ORFeome scale. Genome Biol. 6, R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura G, Pinheiro M, Arrais J, Gomes AC, Carreto L, Freitas A, Oliveira JL, and Santos MAS (2007). Large scale comparative codon-pair context analysis unveils general rules that fine-tune evolution of mRNA primary structure. PloS One 2, e847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS (1989). Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem 264, 5031–5035. [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, and Koller D (2014). Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol 10, 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas de Pouplana L, Santos MAS, Zhu J-H, Farabaugh PJ, and Javid B (2014). Protein mistranslation: friend or foe? Trends Biochem. Sci 39, 355–362. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. (2013). High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. (2014). Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, and Plotkin JB (2013). Rate-limiting steps in yeast protein translation. Cell 153, 1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, and Preiss T (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M, and Fire A (2011). Wobble base-pairing slows in vivo translation elongation in metazoans. RNA N. Y. N 17, 2063–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steneberg P, Englund C, Kronhamn J, Weaver TA, and Samakovlis C (1998). Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the drosophila trachea. Genes Dev. 12, 956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VTK, Hein MY, Huang S-X, Ma M, Shen B, Qian S-B, Hengel H, et al. (2012). Decoding human cytomegalovirus. Science 338, 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Serra V, Petroni G, and Nowacki M (2016). Genetic Codes with No Dedicated Stop Codon: Context-Dependent Translation Termination. Cell 166, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats A-C, and Vagner S (2003). Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell Auspices Eur. Cell Biol. Organ 95, 169–178. [DOI] [PubMed] [Google Scholar]
- True HL, Berlin I, and Lindquist SL (2004). Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431, 184–187. [DOI] [PubMed] [Google Scholar]
- Tsai A, Puglisi JD, and Uemura S (2016). Probing the Translation Dynamics of Ribosomes Using Zero-Mode Waveguides. Prog. Mol. Biol. Transl. Sci 139, 1–43. [DOI] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, and Zhuang X (2016). Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells. Cell 165, 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, and He C (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, and Bartel DP (2016). Improved Ribosome-Footprint and mRNA Measurements Provide Insights into Dynamics and Regulation of Yeast Translation. Cell Rep. 14, 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, and Singer RH (2016). Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, and Tanenbaum ME (2016). Dynamics of Translation of Single mRNA Molecules In Vivo. Cell 165, 976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, and Green R (2015). Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3’UTRs In Vivo. Cell 162, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, and Green R (2009). Quality control by the ribosome following peptide bond formation. Nature 457, 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, and Semenza GL (2016). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A 113, E2047–E2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, and Qian S-B (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MAC, et al. (2011). Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U. S. A 108, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur H, and Tuller T (2013). New universal rules of eukaryotic translation initiation fidelity. PLoS Comput. Biol 9, e1003136. [DOI] [PMC free article] [PubMed] [Google Scholar]