Abstract
The purpose of this study was to probe the fate of a model antigen, a cysteine-free mutant of bacteriophage T4 lysozyme, to the level of fine structural detail, as a consequence of its interaction with an aluminum (Al)-containing adjuvant. Fluorescence spectroscopy and differential scanning calorimetry were used to compare the thermal stability of the protein in solution versus adsorbed onto an Al-containing adjuvant. Differences in accessible hydrophobic surface areas were investigated using an extrinsic fluorescence probe, 8-Anilino-1-naphthalenesulfonic acid (ANS). As has been observed with other model antigens, the apparent thermal stability of the protein decreased following adsorption onto the adjuvant. ANS spectra suggested that adsorption onto the adjuvant caused an increase in exposure of hydrophobic regions of the protein. Electrostatic interactions drove the adsorption, and disruption of these interactions with high ionic strength buffers facilitated the collection of two-dimensional 15N heteronuclear single quantum coherence nuclear magnetic resonance data of protein released from the adjuvant. Although the altered stability of the adsorbed protein suggested changes to the protein’s structure, the fine structure of the desorbed protein was nearly identical to the protein’s structure in the adjuvant-free formulation. Thus, the adjuvant-induced changes to the protein that were responsible for the reduced thermal stability were not observed upon desorption.
Keywords: aluminum-containing adjuvant, vaccine, NMR spectroscopy, protein structure, fluorescence spectroscopy, calorimetry (DSC), protein stability, adsorption
INTRODUCTION
Adjuvants are required for most subunit vaccines (i.e., vaccines with protein or virus-like particle antigens) to elicit a robust immune response. The aluminum (Al)-containing adjuvants account for the majority of the adjuvants found in U.S. Food and Drug Administration-approved vaccines. In these vaccines, the antigens are adsorbed onto the adjuvant. Several studies report that protein antigens have apparent reduced thermal stability when adsorbed onto Al-containing adjuvants.1–4 This phenomenon has been observed even when spectral data suggest that the proteins are native-like at relatively low temperatures (i.e., refrigerated to room temperature). These observations lead to speculations regarding what is occurring at the level of the fine structure that may account for the altered thermal stability. The purpose of the present study was to investigate stability and structural changes to a model antigen resulting from its interaction with an Al-containing adjuvant. In addition to structural stability information based on differential scanning calorimetry (DSC) and fluorescence data, this study was designed to provide, to the best of our knowledge, the first high-resolution solution-state nuclear magnetic resonance (NMR)-based description of a protein in the presence of the micron-size vaccine adjuvant particles.
Protein-based antigens that are used in licensed vaccines are too large to be suitable for conventional solution-state NMR; therefore, we selected a model antigen that was amenable to both solution-state NMR and biophysical studies. Wild-type (WT)* T4 lysozyme, a cysteine-free mutant of bacteriophage T4 lysozyme, was chosen as the model antigen in this study. The cysteines at positions 54 and 97 have been replaced by threonine and alanine, respectively; yet, the thermal stability of the protein has been reported to be identical to that of the WT protein.5 The use of a cysteine-free mutant eliminated the potential complication of aberrant disulfide bonds. T4 lysozyme has three tryptophan residues, making it amenable to intrinsic fluorescence spectroscopy, and an isoelectric point (pI) of approximately 10, suggesting that it should adsorb onto an aluminum phosphate adjuvant via electrostatic interactions. T4 lysozyme has served as the classic model for understanding how amino acid replacement impacts protein stability and structure, thus numerous X-ray crystal structures and full NMR assignments are available.5–8
To understand the biophysical impact of adjuvant adsorption, the consequences of the interaction between WT* T4 lysozyme and Adju-PhosR, a commercially available aluminum phosphate adjuvant, were explored using DSC, fluorescence spectroscopy (intrinsic and extrinsic probes), and two-dimensional 15N heteronuclear single quantum coherence (HSQC) NMR experiments. Calorimetry and intrinsic fluorescence studies were used to examine the effects of the interaction on the apparent thermal stability of model antigen. Fluorescence spectroscopy using 1-anilino8-naphthalene sulfonate (ANS) provided information about the effects of the binding interaction on the accessibility of the probe to hydrophobic regions. Finally, high-resolution NMR spectra of WT* T4 lysozyme in the presence and absence of adjuvant were obtained to investigate whether adsorption affected the fine structure of the protein. These complementary biophysical studies of the structure of the protein indicated that adsorption is accompanied by increased hydrophobic exposure of the protein, which likely contributes to the reduction in affinity. However, these changes were reversible in the desorbed protein. Protein that had been desorbed using high salt appeared indistinguishable from that of adjuvant-naive protein in the same buffer condition using the sensitive metric of 1H and 15N chemical shifts.
MATERIALS AND METHODS
Materials
Commercial aluminum phosphate adjuvant, AdjuPhos®, was purchased from Accurate Chemical (Westbury, New York). Deuterium oxide (D2O, 99.9%), 4-morpholinepropanesulfonic acid (MOPS, >99.5%), MOPS sodium salt (>99.5%), and ANS were purchased from Sigma–Aldrich (St. Louis, Missouri). Sodium chloride was purchased from Fisher Scientific (Pittsburgh, Pennsylvania). Reagents were used without further purification. All buffers were filtered through0.2or0.45 μm nylon membrane filters (What-man, Piscataway, New Jersey). Plasmid for WT* T4 lysozyme was a generous gift from Professor Brian Matthews (University of Oregon). Amicon Ultra-15, 5000 molecular weight cutoff (MWCO), centrifugal filters (Millipore, Burlingame, Massachusetts) were used to concentrate the protein, when necessary. Slide-A-Lyzer® dialysis cassettes, 7000 MWCO (Pierce, Rockford, Illinois), were used for buffer exchanges.
Protein Preparation
WT* T4 lysozyme (unlabeled and 15N labeled) was expressed in BL21 Escherichia coli cells and purified using ion-exchange chromatography as previously described.9 Isotopically labeled protein was obtained using standard techniques by expression in minimal media, with (15NH4)2SO4 as the sole nitrogen source. Following purification, the protein was concentrated to over 10mg/mL with an Amicon Ultra-15 5000 MWCO centrifugal filter (Millipore) at 4000g (4°C). The protein was then exhaustively dialyzed into 10mM MOPS (pH 6.6) using a Slide-A-Lyzer® dialysis cassette 7000 MWCO (Pierce). Precipitates were pelleted and discarded. Concentrated protein was stored at 4°C until needed.
Protein Adsorption/Desorption
First, a 1mg Al/mL working stock of Adju-Phos® in 10% D2O was prepared by diluting the adjuvant from the supplier. Next, a series of WT* T4 lysozyme solutions (from 0.2 to 1.4mg/mL) in 10mM MOPS (pH 6.6) + 10% D2O was prepared. Triplicate samples of 0.1mL adjuvant working stock combined with 0.1mL of protein solution were prepared and allowed to settle at room temperature for 15min for adsorption. Afterwards, the samples were centrifuged at 2000g at 4°C for 2 min to pellet the adjuvant and adsorbed protein. The supernatant was then analyzed using the ultraviolet (UV) absorbance at 280nm [A280 = 1.36mL/ (mg cm)] to determine the amount of protein that remained associated with the adjuvant pellet.
Desorption of adjuvantwith sodium chloride (NaCl) was investigated by first preparing samples containing protein and adjuvant at a final concentration of 0.6mg protein/mg Al. After 20min, the samples were centrifuged. The supernatant was replaced with solutions containing up to 1M NaCl + 10% D2O in distilled deionized water. The pellets were resuspended. The samples were then centrifuged again, and the supernatants were assayed using UV absorbance to determine protein content.
Differential Scanning Calorimetry
For solution-state DSC, the samples were prepared by diluting the concentrated protein to 0.5 mg/mL in 10mM MOPS (pH 6.6) + 10% D2O (D2O, which was included to allow comparison with the NMR results, was added after pH adjustment). For DSC of the adsorbed protein, the samples were prepared by first diluting the concentrated WT* T4 Lys to 1.6mg/mL in 10mM MOPS (pH 6.6) + 10% D2O. Next, aluminum phosphate adjuvant, with a stock concentration of 4.5mg Al/mL, was diluted to 2mg Al/mL in 10mM MOPS (pH 6.6) + 10% D2O. The WT* T4 Lys and adjuvant were then combined in a 1:1 volume ratio for a final concentration of 0.8mg/mL T4 and 1mg Al/mL. The protein was allowed to adsorb onto the adjuvant for 20min at room temperature prior to further manipulation. Samples were then centrifuged for 2 min at 2374g, and the supernatant containing any unbound protein was removed and replaced with fresh buffer.
A MicroCal VP DSC (MicroCal LLC, Northhampton, Massachusetts) was used for all calorimetric studies. The pre-equilibration time was set to 15 minutes then samples were subjected to thermal scans from 10°C to 90°C at a rate of 90°C/h. A protein-free buffer solution was used as the reference for the solution state samples. For the samples that contained adjuvant, a protein-free suspension of Adju-Phos® in buffer was used as the reference. The DSC data for WT* T4 Lys in solution and adsorbed were analyzed using Origin-7 software (OriginLab, Northampton, Massachusetts) with built-in macros for DSC analysis. All DSC data fit best using a single-peak, two-state model. Data were collected in triplicate.
Intrinsic Fluorescence Spectroscopy
For intrinsic fluorescence experiments, a protein concentration of 0.3 mg/mL was used. As with the DSC study, the buffer was 10mM MOPS (pH 6.6) + 10% D2O. For the samples that contained adjuvant, the preparation was similar to that described above but the final concentrations of protein and adjuvant were 0.3mg/mL and 0.5mg Al/mL, respectively. All samples were pipetted into triangular cuvettes. The samples for the protein in solution were assayed immediately. Adsorbed protein was allowed to settle overnight at 4°C to form a dense cake at the bottom of the cuvette.
A QuantaMaster 4™ (Photon Technology International, Birmingham, New Jersey) fluorometer with a Peltier-controlled cuvette holder (Quantum Northwest, Liberty Lake, Washington) was used for all fluorescence studies. Thermal unfolding data were obtained using a user-written macro to collect emission spectra every 2.5°C from 10°C to 80°C, with a 5min equilibration time at each temperature prior to the start of the spectral scan. An excitation wavelength of 295nm and an emission range of 305–400nm were used. For the solution state data, the slits were set to excitation wavelength of 4nm and emission wavelength of 3.5nm. The excitation and emission slits were set to 3nm for the adsorbed protein studies. Buffer scans at 10°C were collected in all cuvettes for spectral correction.
To analyze the data, text formats of raw data were exported to Excel (Microsoft). The data for the samples were corrected using the buffer spectral data. The corrected data were then imported into Origin-7 software (OriginLab), and the wavelengths of the peaks were determined using derivative plot analysis. The final graphs were produced by plotting the peak positions as a function of temperature using Prism 5 (GraphPad Software, La Jolla, CA). The data points were fit to nonlinear functions to determine the apparent melting temperature(Tm). Fivesetsofsamples were analyzed.
ANS Binding Studies
A stock solution of 0.015mM ANS was prepared using the final dialysis buffer (10mM MOPS, pH 6.6) to ensure that buffer in all samples was matched as closely as possible. Samples, both with and without protein, were prepared by mixing equal parts ANS stock with buffer or 2× concentrated protein, adjuvant, or protein and adjuvant stocks. Fluorescence spectra were collected using an excitation wavelength of 350nm and an emission range of 405–600nm. Slit settings for excitation and emission monochrometers were set at 4 nm. Dialysis buffer to which no ANS was added was used to correct all spectra. The spectra for ANS in the presence of protein, in solution or adsorbed, were corrected further by subtracting the signal of ANS in either buffer or adjuvant, respectively. Triplicate samples were analyzed.
Nuclear Magnetic Resonance
Data were collected on 500MHz Varian Inova NMR spectrometer (Varian Inc., Palo Alto, California) equipped with a standard HCN probe using Varian pulse sequences with minor modification. Samples were generally ~16 μM. Data were processed using NMRPipe (http://spin.niddk.nih.gov/nmrpipe) and visualized with NMRDraw, a component of NMRPipe. A reference 15N HSQC spectrum was collected for WT* T4 Lys in 30mM Tris, 100mM KCl, 10% D2O, 0.02% NaN3, pH 5.6. All observable nonoverlapping cross-peaks could be assigned using values for the protein obtained from Professor Frederick Dahlquist (University of California, Santa Barbara, California). Because of the minimal perturbation as a function of buffer, these assignments could be readily transferred to the protein in MOPS buffer (5mM MOPS, 10% D2O, 0.02% NaN3, pH 6.5). To assess the effects of adjuvant on the protein structure, NMR spectra were collected at 25°C in the MOPS buffer containing 0–250mM NaCl. The change in chemical shift (Δδ) was calculated using the following equation:
where ΔHN indicates the difference in chemical shift values in the proton dimension for an amide hydrogen and Δ15N indicates the difference in chemical shift values in the nitrogen dimension for the amide nitrogen, which is weighted by 0.154.10
RESULTS AND DISCUSSION
The goal of the study was to investigate how adsorption onto the adjuvant affected the structure and stability of the adsorbed antigen. It was important therefore to establish the experimental conditions that resulted in the adsorption of the antigen onto an Al-containing adjuvant. As predicted based on the pI of the protein, WT* T4 lysozyme readily adsorbed onto the aluminum phosphate adjuvant. At the ratio of 0.6mg WT* T4 lysozyme per milligram of Al, there was no measurable protein in the supernatant in the NaCl-free MOPS buffer (UV absorbance spectroscopy—datanotshown).Thisratioforcompleteadsorptionwasusedtoguidethesubsequentbiophysical studies.
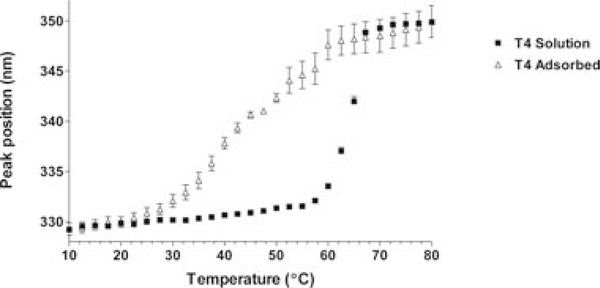
Thermal denaturation monitored by both DSC and fluorescence spectroscopy data indicated that the thermal stability of WT* T4 lysozyme was significantly reduced when it is adsorbed onto the adjuvant (Fig.1).Under the studyconditions, WT*T4lysozyme in solution had an apparent Tm of approximately 65°C, according to either technique (DSC, 65°C ± 0.5°C; fluorescence, 64°C ± 0.5°C). This agrees well with a previously reported Tm value of 65°C reported for WT* T4 lysozyme at pH 6.5 derived from circular dichroism studies.5,11 In contrast, when WT* T4 lysozyme was adsorbed onto the adjuvant, the thermal stability decreased significantly. DSC suggested that the thermal stability was reduced by approximately 20–45°C ± 0.10°C. An equivalent apparent Tm for the adsorbed protein could not be measured as precisely by fluorescence spectroscopy, due to the heterogeneity of the bound state in the samples. The data in the transition region for the adsorbed protein were more variable than those for the protein in the solution state. Nevertheless, the analyses of the data suggested that an initial transition indicative of protein unfolding occurred at a significantly lower temperature for the case of the adsorbed protein with a midpoint of the transition around 43°C.
Figure 1.
Thermal unfolding based on intrinsic fluorescence data. Squares—T4 lysozyme in solution. Triangles—T4 lysozyme adsorbed onto adjuvant. The apparent Tms are approximately 64 ± 0.5°C and 43°C for the solution state and adsorbed protein, respectively. For comparison, the apparent Tms of these formulations according to DSC data (thermograms not shown) were 65 ± 0.5°C (solution state) and 45 ± 0.10°C (adsorbed).
Although the thermal stability studies indicated that the Tm of WT* T4 lysozyme had shifted to a lower temperature as result of adsorption, the initial intrinsic fluorescent emission peak position for the adsorbed protein was similar to that of the protein in solution, ~ 330nm (Fig.1 data points corresponding to 10°C). This lack of a shift in peak maximum has also been observed for other adsorbed antigens.2,3 WT* T4 lysozyme consists of two domains joined by a long α-helix.9 The N-terminal domain, spanning residues 1–60, contains two α-helices and all of the β-sheet content of the protein. The C-terminal domain comprises residues 80–164 and is a barrel of seven α-helices. The three tryptophan residues in the protein are all located within the C-terminal domain. Because intrinsic fluorescence spectra only provided details of the local environment of the tryptophan residues, structural perturbations to any part of the protein in the bound state that did not result in changes to the environment of these three residues would not have resulted in a shift of the intrinsic fluorescence spectra. Additional techniques, therefore, were necessary to probe more fully the ways in which adsorption may have affected the structure of T4 lysozyme in the absence of thermal stress.
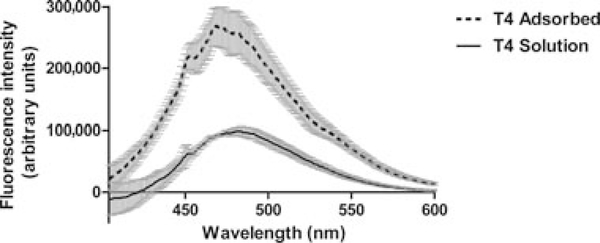
ANS binding studies were performed to probe for increased exposure of hydrophobic surfaces on the protein following adsorption onto the adjuvant. The fluorescence of ANS in the presence of T4 lysozyme in solution is low, indicative of little hydrophobic surface exposure, which is consistent with the behavior of this protein in solution (Fig. 2).12,13 The addition of ANS to a sample containing adsorbed T4 lysozyme resulted in a significant increase in the intensity of the ANS spectral signal. The intensities of the ANS fluorescence emission spectra for the buffer or adjuvant in buffer were significantly lower (data not shown) than that of the ANS in the presence of the protein, and the spectra in Figure 2 were corrected using the corresponding protein-free ANS background spectrum. Thus, the spectral changes of the ANS signal in the presence of the adsorbed protein relative to that of the protein in solution did not appear to be because of simply increased light scattering or interaction of the ANS with the adjuvant. The increased intensity and spectral blue shift of the ANS signal were most likely due to interactions between ANS and newly accessible hydrophobic regions of the protein when it was adsorbed onto the adjuvant. WT* T4 lysozyme is a water-soluble protein that is stabilized by the burial of hydrophobic residues.14–16 The decrease in the thermal stability of the adsorbed protein is likely attributable, at least in part, to the increase in exposed hydrophobic surface of the adsorbed protein.
Figure 2.
Averaged emission spectra from ANS-binding experiments. Solid line—ANS in WT* T4 solution. Dashed line—ANS with adsorbed WT* T4 lysozyme. The spectra have been corrected by subtracting the appropriate blank spectrum: ANS in buffer, with or without adjuvant.
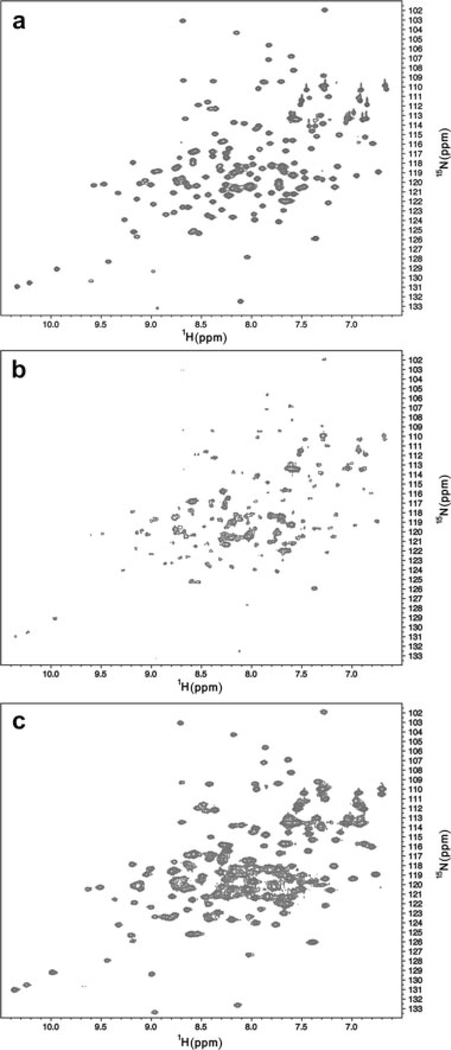
In order to probe the structural changes that accompany adsorption of protein onto adjuvant at higher resolution, we obtained 15N HSQC spectra on free and adjuvant-bound T4 lysozyme. Free T4 lysozyme provided outstanding quality spectra as anticipated, and could be readily assigned using existing data (Fig. 3a).8 Upon titration of adjuvant, which has an average diameter of 2 μm, we observed loss of the free protein signal (Fig. 3b). This observation suggests that adjuvant-bound protein has a correlation time associated with a fully bound state and that resonances are broadened beyond the limit of observation. Furthermore, there is no evidence in the spectra that regions of the protein become disordered with respect to the adjuvant particle, suggesting that the global correlation time dominates the behavior of the protein resonances.
Figure 3.
15N HSQC spectra of WT* T4 lysozyme in solution. (a) The spectrum of the protein in MOPS buffer containing 50mM NaCl in the absence of adjuvant. (b) The spectrum obtained in the same MOPS buffer containing 50mM NaCl in the presence of adjuvant. The reduction in signal intensity is due to the binding of protein to adjuvant. (c) The spectrum obtained in the presence of the same MOPS buffer in 200mM NaCl and adjuvant. Recovery of the signal is due to the desorption of protein from adjuvant. The spectrum suggests minimal perturbation to the protein as a result of binding.
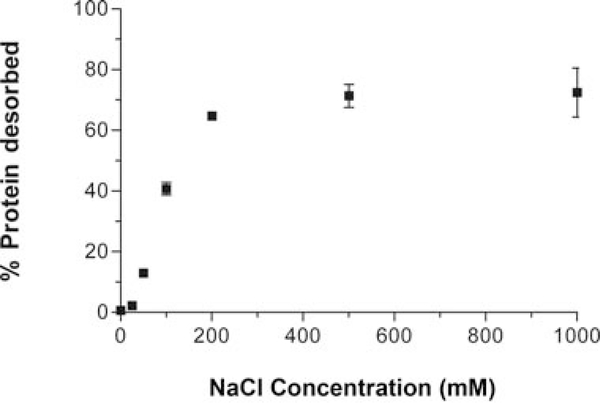
Not all antigens remain adsorbed following vaccine administration, they are desorbed from the adjuvant because of the physiological environment.17–20 Moreover, recent evidence suggests that tight binding of the antigen to the adjuvant in the vial may not be desirable with respect to eliciting the highest antibody titers following vaccine administration.21–24 Thus, the ability of the adjuvant to irreversibly alter the tertiary structure of the desorbed antigen may be important in the quality of the vaccine. Because we have shown that adsorption perturbs (and likely partially unfolds) the antigen, an outstanding question is whether these conformational alterations are maintained in the desorbed state. We determined that increasing ionic strength could be used to desorb the protein. When the NaCl-free MOPS buffer used to prepare the adsorbed protein samples was removed and replaced with increasing amounts of NaCl from 50 to 200 mM, UV analysis of the supernatant revealed that protein content increased as the concentration of NaCl increased (Fig. 4). Protein desorbed from adjuvant using NaCl was also monitored by NMR (Figs. 3b and 3c). When the protein, in excess to allow equilibrium exchange between the free and bound states, was adsorbed onto the adjuvant in the presence of MOPS buffers containing increasing concentrations of NaCl (50–250 mM), the resultant NMR spectra showed an increasing recovery of the signal intensity due to the presence of more observable protein with higher levels of desorption.
Figure 4.
Effect of increasing amounts of salt on adsorbed WT* T4 lysozyme. The graph shows the percentage of the previously adsorbed protein that is desorbed as when sodium chloride is added to the sample—determined by to UV–visible spectroscopy analysis of the supernatant.
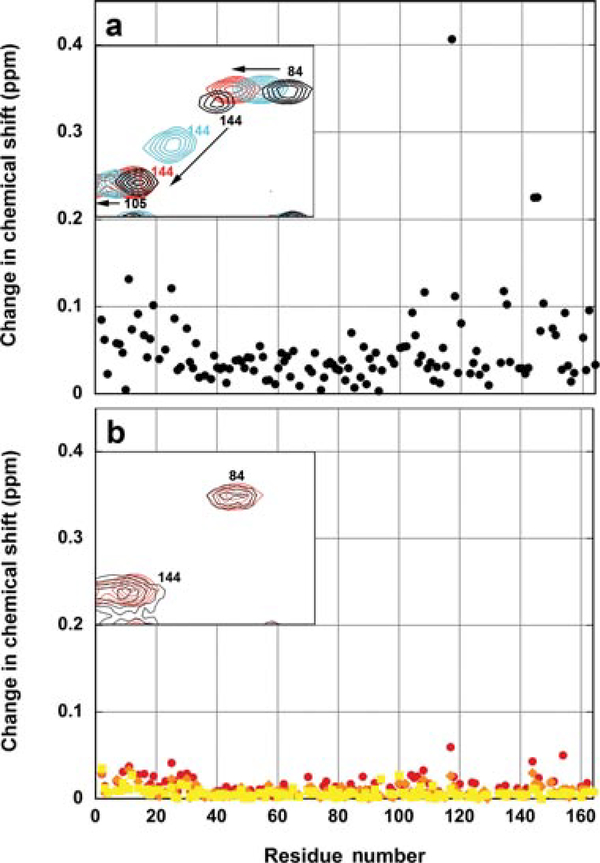
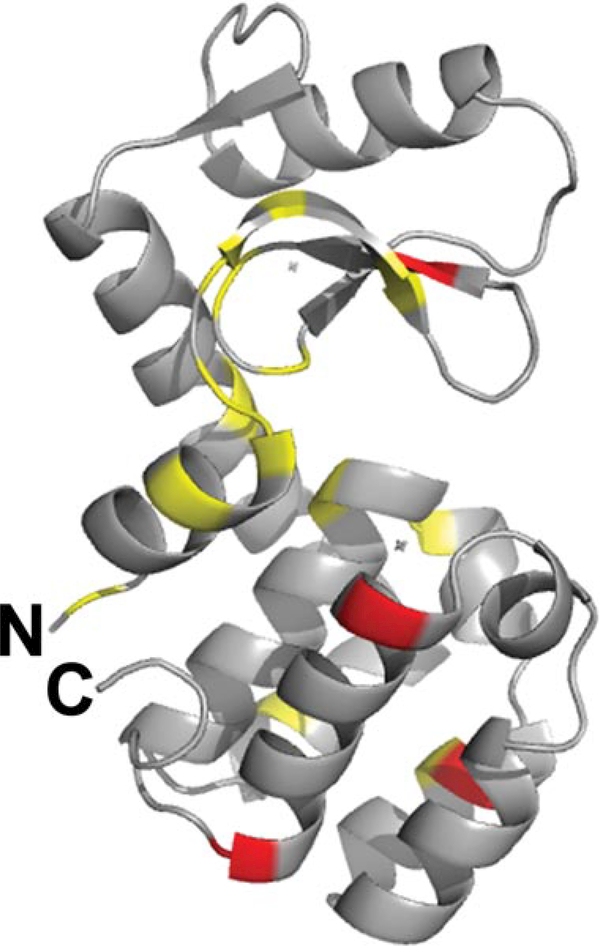
The ability to obtain NMR data directly on the desorbed model antigen in the presence of adjuvant by adding NaCl was exploited to probe the detailed structure of the desorbed protein. Although the chemical shifts obtained on the adjuvant-bound sample in increasing concentration of ionic strength were significant, these chemical shift changes nearly precisely mirrored the inherent ionic strength-dependent chemical shifts changes observed for WT* T4 lysozyme in the absence of adjuvant (Fig. 5a). The largest changes were observed for residues in the C-terminal domain of the protein. Thus, to observe only adjuvant-dependent chemical shifts requires comparison of the free and bound samples at identical ionic strengths. These changes, quantified in Figure 5b at three concentrations of NaCl (100, 150, and 200 mM), were much smaller than the simple salt effect. The residues that experience the largest differences in chemical shifts due to adjuvant were also those that generally had the greatest salt-dependent shifts in the absence of adjuvant. In Figure 6, the residues that experienced the largest adjuvant-dependent difference in shifts (in red are changes between 0.04 and 0.06ppm and in yellow are changes between 0.025 and 0.04 ppm) in the presence of 100mM NaCl are mapped onto the structure of T4 lysozyme (PDB 1L63). The affected residues are dispersed throughout the protein. Overall, the relative magnitude of the chemical shift change for each residue as a function of adjuvant at a given salt concentration is small in comparison with the change caused by salt alone (Figs. 5a and 5b). As the N HSQC spectrum is exquisitely sensitive to local chemical environment, the overall lack of chemical shift difference between the free and desorbed protein indicates that conformational changes that occur in the protein upon adsorption are mostly reversible after the protein desorbs from the adjuvant. The shifts are therefore likely attributable to differences in the electrostatics of the environment around those residues rather than structural changes in the molecule. Although the fluorescence and DSC data suggest that WT* T4 lysozyme experiences structural perturbation upon binding to the adjuvant, the NMR data reveal that the desorbed protein is native like.
Figure 5.
(a) The effect of salt on the chemical shift values for residues in WT* T4 lysozyme. The graph shows the magnitude of the change in chemical shift due to salt (0 vs. 200mM) as a function of position in the protein in the absence of adjuvant. The inset shows a small region of the spectra containing the peaks for three residues (84, 105, and 144) at three salt concentrations 0mM NaCl (black), 100mM NaCl (cyan), and 200mM NaCl (red) in MOPS buffer. (b) Chemical shift changes of desorbed WT* T4 lysozyme in the presence of adjuvant relative to the adjuvant-free solution state sample in the presence of identical concentrations of NaCl. Red—100mM NaCl. Orange—150mM NaCl. Yellow—200mM NaCl. The inset shows a small region of the 15N HSQC spectrum of the protein as in panel (a) in the presence of 200mM NaCl with adjuvant (black) and without adjuvant (red).
Figure 6.
The adjuvant-dependent differences in chemical shifts at 100mM NaCl mapped onto the structure of the protein. Residues with the largest chemical shift changes (0.04–0.06 ppm) are shown in red and residues with moderate changes (0.025–0.04 ppm) are shown in yellow. The figure was generated using Pymol (The PyMOL Molecular Graphics System, version 1.2r3pre; Schrödinger, LLC (Portland, Oregon) using the PDB file 1L63.
CONCLUSIONS
This manuscript describes the consequences, to thermal stability and structure, to the level of fine detail, of a model antigen that has been adsorbed onto an Al-containing adjuvant. The intrinsic fluorescence spectrum of the adjuvant-adsorbed protein indicated native-likestructureat10°C≤T≤25°C;however,the increased ANS binding and decreased apparent Tm of the adsorbed protein in comparison with the protein in the adjuvant-free formulation indicated alterations to the protein structure. The massive particle size of the adjuvant precluded elucidation of the detailed structure of the protein in the adsorbed state. High-resolution NMR spectra did reveal, however, that this protein readily refolded following desorption and the desorbed state exhibited native-like fine structure. Recent evidence suggests that vaccine formulations in which the antigens are not tightly adsorbed onto the adjuvant are more native like and elicit higher antibody responses than those formulations exhibiting a tight interaction between the protein and adjuvant. Thus, our findings are important in the context of a rational approach to optimize such vaccine formulations in general in that in that they provide for the first time evidence that the fine structure can be recovered for the desorbed protein, even if the spectral data for the adsorbed state suggest perturbations. If an antigen readily refolds following desorption in the in vivo environment, the destabilization and structural changes that are observed for the adsorbed antigen in the vial may have little consequence with respect to the ability of the vaccine to elicit antibodies against conformational epitopes.
It is our opinion that this study represents an important step forward in improving our understanding of the impact of the interaction between to an Al-containing adjuvant and a model protein adjuvant to the thermal stability and fine structure of the protein. Nevertheless, unanswered questions remain. One important point of consideration for the present study is that fine structural detail was only obtained for WT* T4 lysozyme that was capable of being desorbed from the adjuvant. Monitoring the recovery of signal from the adjuvant adsorbed state as a function of ionic strength (data not shown) indicated that the time-scale of the exchange between adjuvant-bound and desorbed-protein was slow compared to the NMR time-scale. Therefore, this strategy could not be used to indirectly obtain information about the bound state. No information is available for protein that did not desorb. It is possible therefore that there are protein molecules that do not desorb that have a large degree of structural perturbation which could not be observed due to the limitations of the HSQC experiment. Therefore, the details about the structure of the adsorbed protein remain elusive. Second, this study did not evaluate the fine structure of desorbed protein from a vaccine formulation that had been aged prior to desorption. In an actual vaccine, the antigen and adjuvant have been formulated together in a single vial for months or years before administration to the patient. Thus, presumably, aging the WT* T4 lysozyme model vaccine would have yielded data that more closely resembled real-world formulations. It has previously been reported that the ability of an adsorbed protein to readily desorb from the adjuvant can decrease over time.4,25 The ease of protein desorption from a surface, including adjuvants, appears to correlate with the conformational stability of the protein, suggesting that protein that is more difficult to desorb is more likely to be non-native.4,25,26 Desorption of the protein from the adjuvant was essential for obtaining information about the fine structure. Thus, any observations from an aging study using the present study design would likely yield fine-structure data biases towards the stable, native-like conformational states within the ensemble of conformations that might exist on the adjuvant surface. Finally, the model antigen used in this study is a relatively small, well-behaved protein that has frequently been used as a model in thermodynamic studies of proteins. Antigens in vaccines and vaccine candidates that might utilize an Al-containing adjuvant range from small peptides to large virus-like particles. Whether the observations made using WT* T4 lysozyme are representative of those that would be made for a vaccine relevant to human health will likely depend on the inherent conformational stability of the antigen as well as the nature (i.e., strength and mechanism) of the adsorption interaction. Only future studies utilizing such antigens will address these questions.
ACKNOWLEDGMENTS
The authors acknowledge funding for this study from the University of Colorado Butcher Biotechnology Seed Grant (L.J.B. and D.S.W.). The authors also thank Professor Brian Matthews for providing plasmid containing WT* T4 lysozyme and Professor Rick Dahlquist for providing chemical shift values.
REFERENCES
- 1.Ausar SF, Chan J, Hoque W, James O, Jayasundara K, Harper K. Application of extrinsic fluorescence spectroscopy for the high throughput formulation screening of aluminumadjuvanted vaccines. J Pharm Sci 100(2):431–440. [DOI] [PubMed] [Google Scholar]
- 2.Jones LS, Peek LJ, Power J, Markham A, Yazzie B, Middaugh CR. 2005. Effects of adsorption to aluminum salt adjuvants on the structure and stability of model protein antigens. J Biol Chem 280(14):13406–13414. [DOI] [PubMed] [Google Scholar]
- 3.Peek LJ, Martin TT, Elk Nation C, Pegram SA, Middaugh CR.2007. Effects of stabilizers on the destabilization of proteins upon adsorption to aluminum salt adjuvants. J Pharm Sci 96(3):547–557. [DOI] [PubMed] [Google Scholar]
- 4.Vessely C, Estey T, Randolph TW, Henderson I, Cooper J,Nayar R, Braun LJ, Carpenter JF. 2009. Stability of a trivalent recombinant protein vaccine formulation against botulinum neurotoxin during storage in aqueous solution. J Pharm Sci 98(9):2970–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumura M, Matthews BW. 1989. Control of enzyme activity by an engineered disulfide bond. Science 243(4892):792–794. [DOI] [PubMed] [Google Scholar]
- 6.Bell JA, Wilson KP, Zhang XJ, Faber HR, Nicholson H, Matthews BW. 1991. Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths. Proteins 10(1):10–21. [DOI] [PubMed] [Google Scholar]
- 7.Fischer MW, Majumdar A, Dahlquist FW, Zuiderweg ER.1995. 15N,13C, and1H NMR assignments and secondary structure for T4-lysozyme. J Magn Reson B 108(2):143–154. [DOI] [PubMed] [Google Scholar]
- 8.McIntosh LP, Wand AJ, Lowry DF, Redfield AG, Dahlquist FW. 1990. Assignment of the backbone1H and15N NMR resonances of bacteriophage T4 lysozyme. Biochemistry 29(27):6341–6362. [DOI] [PubMed] [Google Scholar]
- 9.Alber T, Matthews BW. 1987. Structure and thermal stability of phage T4 lysozyme. Methods Enzymol 154:511–533. [DOI] [PubMed] [Google Scholar]
- 10.Tugarinov V, Kay LE. 2003. Quantitative NMR studies of high molecular weight proteins: Application to domain orientation and ligand binding in the 723 residue enzyme malate synthase G. J Mol Biol 327(5):1121–1133. [DOI] [PubMed] [Google Scholar]
- 11.Wetzel R, Perry LJ, Baase WA, Becktel WJ. 1988. Disulfide bonds and thermal stability in T4 lysozyme. Proc Natl Acad Sci U S A 85(2):401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llinas M, Marqusee S. 1998. Subdomain interactions as a determinant in the folding and stability of T4 lysozyme. Protein Sci 7(1):96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uversky VN, Leontiev VV, Gudkov AT. 1992. Triple point mutation Asp10→His, Asn101→Asp, Arg148→Ser in T4 phage lysozyme leads to the molten globule. Protein Eng 5(8):781–783. [DOI] [PubMed] [Google Scholar]
- 14.Baase WA, Liu L, Tronrud DE, Matthews BW. Lessons from the lysozyme of phage T4. Protein Sci 19(4):631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MatsumuraM Becktel WJ,Matthews BW. 1988. Hydrophobic stabilization in T4 lysozyme determined directly by multiple substitutions of Ile 3. Nature 334(6181):406–410. [DOI] [PubMed] [Google Scholar]
- 16.Matsumura M, Wozniak JA, Sun DP, Matthews BW. 1989. Structural studies of mutants of T4 lysozyme that alter hydrophobic stabilization. J Biol Chem 264(27):16059–16066. [PubMed] [Google Scholar]
- 17.Chang M, Shi Y, Nail SL, HogenEsch H, Adams SB, White JL, Hem SL. 2001. Degree of antigen adsorption in the vaccine or interstitial fluid and its effect on the antibody response in rabbits. Vaccine 19(20–22):2884–2889. [DOI] [PubMed] [Google Scholar]
- 18.Heimlich JM, Regnier FE, White JL, Hem SL. 1999. The in vitro displacement of adsorbed model antigens from aluminium-containing adjuvants by interstitial proteins. Vaccine 17(22):2873–2881. [DOI] [PubMed] [Google Scholar]
- 19.Iyer S, HogenEsch H, Hem SL. 2003. Relationship between the degree of antigen adsorption to aluminum hydroxide adjuvant in interstitial fluid and antibody production. Vaccine 21(11–12):1219–1223. [DOI] [PubMed] [Google Scholar]
- 20.Morefield GL, Jiang D, Romero-Mendez IZ, Geahlen RL, Hogenesch H, Hem SL. 2005. Effect of phosphorylation of ovalbumin on adsorption by aluminum-containing adjuvants and elution upon exposure to interstitial fluid. Vaccine 23(12):1502–1506. [DOI] [PubMed] [Google Scholar]
- 21.Egan PM, Belfast MT, Gimenez JA, Sitrin RD, Mancinelli RJ.2009. Relationship between tightness of binding and immunogenicity in an aluminum-containing adjuvant-adsorbed hepatitis B vaccine. Vaccine 27(24):3175–3180. [DOI] [PubMed] [Google Scholar]
- 22.Hansen B, Belfast M, Soung G, Song L, Egan PM, Capen R, Hogenesch H, Mancinelli R, Hem SL. 2009. Effect of the strength of adsorption of hepatitis B surface antigen to aluminum hydroxide adjuvant on the immune response. Vaccine 27(6):888–892. [DOI] [PubMed] [Google Scholar]
- 23.Hansen B, Malyala P, Singh M, Sun Y, Srivastava I, Hogenesch H, Hem SL. Effect of the strength of adsorption of HIV 1 SF162dV2gp140 to aluminum-containing adjuvants on the immune response. J Pharm Sci 100(8):3245–3250. [DOI] [PubMed] [Google Scholar]
- 24.Hansen B, Sokolovska A, HogenEsch H, Hem SL. 2007. Relationship between the strength of antigen adsorption to an aluminum-containing adjuvant and the immune response. Vaccine 25(36):6618–6624. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, HogenEsch H, Hem SL. 2001. Change in the degree of adsorption of proteins by aluminum-containing adjuvants following exposure to interstitial fluid: Freshly prepared and aged model vaccines. Vaccine 20(1–2):80–85. [DOI] [PubMed] [Google Scholar]
- 26.McGuire J,Wahlgren M, Arnebrant T. 1995. Structural stability effects on the adsorption and dodecyltrimethylammonium bromide-mediated elutability of bacteriophage T4 lysozyme at silica surfaces. J Colloid Interface Sci 170. [Google Scholar]