SUMMARY
Yin yang 1 (YY1) is a ubiquitous transcription factor and mammalian polycomb group protein (PcG) with important functions to regulate embryonic development, lineage differentiation, and cell proliferation. YY1 mediates stable PcG-dependent transcriptional repression via recruitment of PcG proteins that catalyze histone modifications. Many questions remain unanswered regarding how cell- and tissue-specificity is achieved by PcG proteins. Here, we demonstrate that a conditional knockout of Yy1 in hematopoietic stem cells (HSCs) decreases long-term repopulating activity and ectopic YY1 expression expands HSCs. Although the YY1 PcG domain is required for Igk chain rearrangement in B cells, the YY1 mutant lacking the PcG domain retained the capacity to stimulate HSC self-renewal. YY1 deficiency deregulated the genetic network governing HSC cell proliferation and impaired stem cell factor/c-Kit signaling, disrupting mechanisms conferring HSC quiescence. These results reveal a mechanism for how a ubiquitously expressed transcriptional repressor mediates lineage-specific functions to control adult hematopoiesis.
Graphical Abstract
In Brief
Lu et al. investigate the function of the polycomb group (PcG) protein YY1 in hematopoietic stem cells. Independent of its REPO domain/PcG function, YY1 promotes hematopoietic stem cell selfrenewal and quiescence, suggesting that REPO domain/PcG function is not utilized in all contexts within the hematopoietic hierarchy.
INTRODUCTION
Many adult tissue-specific stem cells persist in a quiescent stage, which allows them to act as a dormant reserve to replenish tissues during homeostasis. Mammalian adult bone marrow contains resident hematopoietic stem cells (HSCs) that can proliferate to compensate for blood loss and to maintain homeostasis. HSCs are undifferentiated, long-lived cells that give rise to lineage-specific progenitors and retain their stem cell identity by undergoing self-renewal. Adult HSCs can remain in a quiescent state for a prolonged time, and quiescence is a fundamental characteristic of adult bone marrow-resident HSCs (Pietras et al., 2011). Thus, a precisely regulated cell cycle is critical for HSC-mediated generation of mature hematopoietic cells, while preventing stem cell exhaustion (Orford and Scadden, 2008).
HSC quiescence is regulated by both intrinsic and extrinsic signals (Morrison and Weissman, 1994; Suda et al., 1983). Cell-cycle regulators, transcription factors, as well as epigenetic modifications, have been identified as intrinsic regulators of HSC cell-cycle progression. Yin yang 1 (YY1) is a ubiquitous multifunctional zinc-finger transcription factor that has important roles in early embryo development, X chromosome inactivation, DNA repair, cell-cycle progression, apoptosis, and hematopoiesis. In addition to its function as a transcription factor, YY1 is a critical polycomb group (PcG) protein and is a founding member of a very limited cohort of mammalian PcG proteins with sequence-specific DNA binding (Atchison et al., 2003; Srinivasan and Atchison, 2004; Srinivasan et al., 2005). While non-stable transcriptional repression can involve direct competition for DNA binding by activators and repressors, recruitment of corepressors that deacetylate histones, or direct interference with the transcriptional machinery, stable PcG-dependent repression involves the hierarchical recruitment of PcG complexes and subsequent chromatin modifications (Wang et al., 2004). Studies in mice deficient for PcG genes revealed that PcG proteins serve important and diverse roles in HSC self-renewal and differentiation. The PRC1 protein BMI1 is required for HSC self-renewal (Iwama et al., 2004; Park et al., 2003; Rizo et al., 2009). CBX7 is selectively expressed in HSCs, and its overexpression enhances HSC self-renewal and induces leukemia. In contrast, CBX2, CBX4, or CBX8 overexpression induces HSC differentiation and exhaustion (Klauke et al., 2013). Overexpression of the PRC2 protein EZH2 in HSCs preserves stem cell potential and prevents HSC exhaustion after serial transplantations (Kamminga et al., 2006). The heterozygous Yy1 mutation in an Mpl null background reveals an exacerbated phenotype of thrombocytopenia and leukopenia. Competitive bone marrow transplantation of Yy1+/− Mpl+/+ fetal liver cells suggested that Yy1+/−cells might have a compromised capacity to maintain the stem cell pool (Majewski et al., 2010). However, correlations of epigenetic signatures are often not highly instructive, and the mechanistic implication of epigenetic signatures in HSC self-renewal is incompletely understood.
The mechanisms responsible for targeting mammalian PcG proteins to specific DNA regions are not elucidated, as the majority of PcG proteins do not directly bind to specific DNA sequences. However, PcG complexes must occupy specific DNA regions to function. YY1 can repress transcription in a PcG-dependent manner, can recruit other PcG proteins to specific DNA sequences and can increase H3K27me3 at the recruitment site (Atchison et al., 2003; Srinivasan and Atchison, 2004; Wilkinson et al., 2006). The ability of YY1 to selectively recruit PcG factors to specific chromatin sites suggests that YY1 may have a pivotal role in orchestrating the function of other PcG proteins in HSC selfrenewal. Our previous results demonstrated that YY1 PcG domain is required for Igk chain rearrangement in early B cell development (Pan et al., 2013). Here, we evaluated the role of YY1PcG domain in HSC self-renewal and tested whether the requirement for YY1 PcG domain is context-dependent in adult hematopoiesis. We utilized a YY1 REPO domain mutant (YY1ΔREPO). The small 25 amino acid REPO domain is necessary and sufficient for recruiting other PcG proteins to YY1-bound chromatin sites in Drosophila. While the REPO domain YY1 mutant (YY1ΔREPO) is competent for DNA binding, transcriptional activation, transient transcriptional repression, and interaction with transcriptional coregulators such as HDACs, YY1ΔREPO is defective in all YY1 PcG functions and unable to recruit other PcG proteins to DNA (Wilkinson et al., 2006). This mutant is therefore a powerful tool for dissecting mechanisms governing YY1 PcG domain- dependent versus -independent functions in HSCs.
Here, we demonstrate that the long-term self-renewal mechanism is defective in Yy1-deficient HSCs, and ectopic YY1 expression increases HSC expansion. Unexpectedly, the YY1 REPO domain mutant retained the capacity to stimulate HSC self-renewal. YY1 deficiency deregulated the genetic network governing HSC cell proliferation and disrupted mechanisms conferring HSC quiescence. Stem cell factor/c-Kit signaling is essential for adult HSC self-renewal and quiescence (Ho et al., 2017; Thorén et al., 2008; Zhang et al., 2017), and YY1 directly regulated c-Kit expression and function in HSCs.
RESULTS
YY1 Controls Normal Adult Hematopoiesis by Regulating HSC Self-Renewal
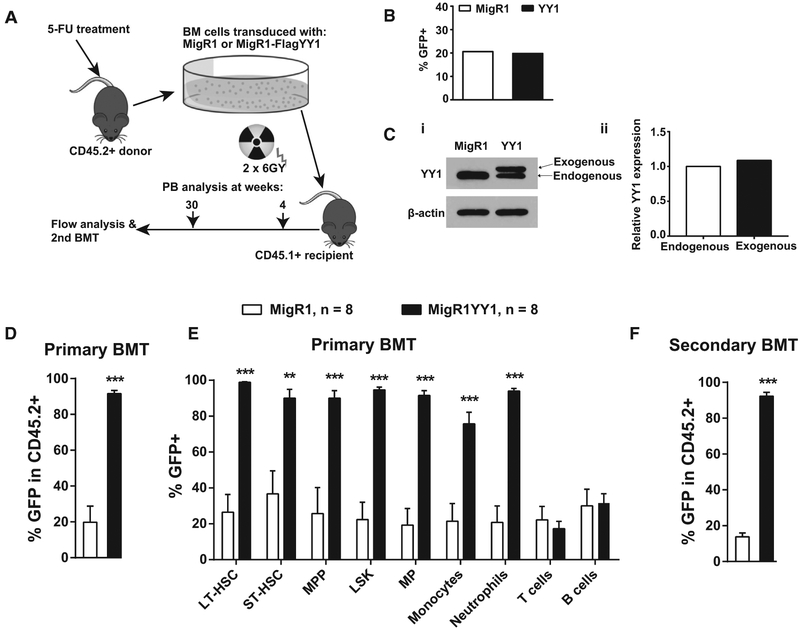
Our previous data demonstrated that ectopic YY1 expression increases HSCs and myeloid cells in adult bone marrow (Pan et al., 2012). We investigated whether ectopic YY1 expression impacts HSC long-term self-renewal. CD45.2+ wild-type bone marrow cells were retrovirally infected with either MigR1-FlagYY1 or MigR1 vector control and then transplanted to lethally irradiated (6 Gy ×2) congenic CD45.1+ mice (Figure 1A). Before bone marrow transplantation (BMT), the percentages of GFP+ cells in MigR1-Flag YY1 or MigR1- infected bone marrow were evaluated at 48 hr post-viral infection and were similar at 20% of infection rate (Figure 1B). At 4 weeks post-BMT, the exogenous Flag-tagged YY1 protein expression was compared with endogenous YY1 in the sorted GFP+ peripheral blood lymphocyte, and the total YY1 was twice that of the endogenous level (Figure 1C). Thus, the infection and reconstitution efficiencies were equivalent in recipient mice receiving MigR1-Flag YY1 or MigR1-infected BM cells, and the exogenous YY1 was expressed at a near-physiological level. At 30-weeks post-BMT, mice transplanted with MigR1-Flag YY1-infected bone marrow cells were nearly 100% GFP+ in donor-derived (CD45.2+) total bone marrow cells, in contrast to 20% GFP+ in mice receiving MigR1-infected bone marrow cells (Figure 1D). We compared the percentage of GFP+ in donor-derived long-term-hematopoietic stem cells (LT-HSCs) (CD45.2+ Lin−Sca1+ c-Kit+ CD48−CD150+), short-term-hematopoietic stem cells (ST-HSCs) (CD45.2+Lin−Sca1+c-Kit+CD48−CD150−), multipotent progenitor (MPP) (CD45.2+Lin−Sca1+ c-Kit+ CD48+CD150−), Lin-Sca1+Kit (LSK) (CD45.2+Lin−Sca1+ c-Kit+), myeloid progenitor (CD45.2+Lin−Sca1−c-Kit+), neutrophils (CD45.2+Mac1+Gr1+), monocytes (CD45.2+Mac1+Gr1−), T cells (CD45.2+CD19−Thy1.2+), and B cells (CD45.2+Thy1.2−CD19+). Whereas mice with ectopic YY1 expression had a significantly higher percentage of GFP+ cells in donor-derived bone marrow LT-HSC, ST-HSC, MPP, LSK, myeloid progenitor (MP), monocyte, and neutrophil populations, this increase was not detected in either B or T lymphocytes (Figure 1E). Consistently, the increase of GFP+ was associated with a decrease in the percentages of GFP− cells in mice with ectopic YY1 expression. As ectopic YY1 expression did not increase the percentage of GFP+ cells in lymphocytes, this suggested that the YY1-mediated HSC enrichment did not reflect a non-specific effect caused by retroviral integration. In secondary BMT, the bone marrow cellularity was similar in mice receiving MigR1-Flag YY1 or MigR1-infected bone marrow cells (Figure S1A). By 20 weeks post-secondary BMT, there was no obvious evidence of malignant transformation, as white blood cell (WBC) and red blood cell (RBC) numbers (Figure S1C) and spleen weight were normal (Figure S1B). CD45.2+ BM cells in secondary BMT mice with ectopic YY1 expression continue to maintain a high percentage of GFP+ (>90%) in comparison with a significantly low percentage of GFP+ (10%) in wild-type BMs (Figure 1F). Peripheral blood evaluation revealed that BMT mice with ectopic YY1 expression had a higher donor-derived percentage of monocyte and neutrophil populations (Figure S2). Thus, ectopic YY1 expression increased HSC long-term self-renewal and induced HSC expansion.
Figure 1. Ectopic YY1 Expression Promotes HSCs Long-Term Self-Renewal.
(A) Experimental strategy: MigR1 or MigR1-FlagYY1 transduced CD45.2+ wild-type bone marrow was transplanted into lethally irradiated CD45.1+ congenic mice. Bone marrow from reconstituted mice was subject to flow analyses at 30 weeks post BMT or harvested for the secondary BMT.
(B) Prior to BMT, flow analysis to evaluate the transduction efficiency of combined bone marrow cells from 8 donor mice infected with MigR1 or MigR1-Flag-YY1 at 48 hr post viral infection.
(C) (i) Western blot analysis to compare the exogenous Flag-YY1 expression to the endogenous YY1 in combined GFP+ peripheral lymphocytes sorted from 4 mice per cohort. (ii) Quantification of endogenous and exogenous YY1 protein expressions. (D and E) Evaluations of primary BMT mice.
(D) Flow analysis of % GFP+ in donor-derived total BM at 30 weeks post BMT.
(E) Flow analysis of % GFP+ of donor-derived bone marrow LSK, LT-HSC, ST-HSC, MPP, MP, neutrophils, monocytes, T cells, and B cells.
(F) Flow analysis of % GFP+ in donor-derived total BM cells at 16 weeks post-secondary BMTs. N represents the number of mice; graphs show means ± SEM; **p < 0.01, ***p < 0.001.
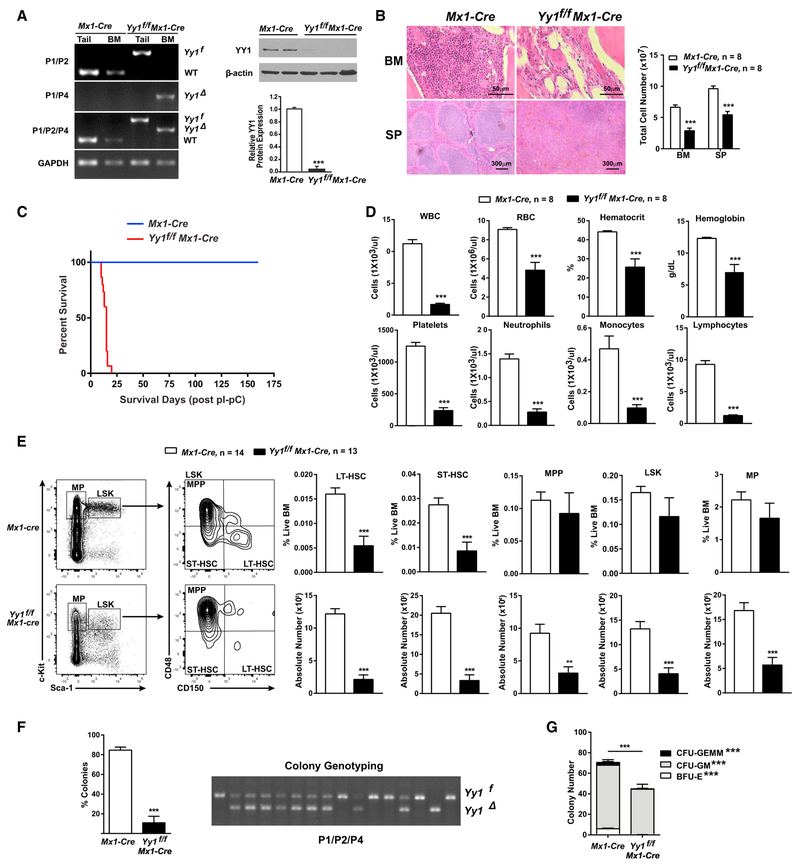
To test how loss-of-function of YY1 impacts hematopoiesis, we utilized a conditional Yy1 knockout allele Yy1f/f with loxP sites flanking the Yy1 promoter region and exon 1 (Liu et al., 2007) (Figure S3A). Yy1f/f mice were crossed to the either inducible Mx1-Cre or Vav-Cre. In Yy1f/f Mx1-Cre mice, YY1 deletion was achieved after treatment with the interferon alpha (IFN-a) stimulating polyinosinic:polycytidylic acid (pI-pC). Yy1f/f Mx1-Cre and Mx1-cre mice received 5 doses of pI-pC injections. At 7 days post-injections, PCR analysis failed to detect loxP-flanked Yy1f alleles in total BM cells of Yy1f/f Mx1-Cre mice (Figure 2A). In addition, there was a >90% reduction of YY1 protein levels in bone marrow in comparison with Mx1-Cre controls (Figure 2A). Yy1f/f Mx1-Cre mice died within 3 weeks post-pI-pC injection (Figure 2C), and Yy1f/f Vav-Cre mice died at the perinatal stage. Among 141 pups resulting from breeding Yy1f/f to Yy1f/+ Vav-Cre, only 7 were Yy1f/f Vav-Cre. All 7 pups died within 72 hr after birth (data not shown).
Figure 2. Yy1−/−Mice Are Pancytopenic with Decreased HSCs.
All Yy1f/f Mx1-cre and Mx1-Cre mice were treated with 5 doses of pI-pC and evaluated 7–10 days after the last injection.
(A) PCR and western blot to detect the deletion efficiency in total BM cells. Tail samples were used to show that Yy1 deletion was specific in the hematopoietic system. Mixed primers 1, 2, and 4 showed the equal primer efficiency.
(B) Total BM and spleen cell counts and representative images of H&E-stained BM (320) and spleen (34) sections.
(C) Kaplan-Meier survival curve.
(D) Complete blood count (CBC) analysis.
(E) Representative gating strategy, percentage, and absolute number of bone marrow LT-HSC, ST-HSC, MPP, LSK, and MP.
(F) Colony formation assays from sorted LT-HSCs. PCR detection of deletion efficiency of colonies growing from sorted LT-HSC of Yy1f/f Mx1-cre mice with mixed primers 1, 2, and 4.
(G) Colony formation assays from total bone marrow cells. N represents the number of mice; graphs show means ± SEM; **p < 0.01, ***p < 0.001.
Yy1−/− spleen and bone marrow were hypocellular with decreased total cell numbers (Figure 2B). Yy1−/− were pancytopenic with significantly decreased WBC, hemoglobin, and platelet counts (Figure 2D). In addition, the percentage and absolute numbers of LT-HSCs and ST-HSCs in Yy1−/− bone marrow decreased significantly (Figure 2D). Yy1−/− HSCs had reduced colony forming activity. Single LT-HSC cells (Lin−Sca1+c-Kit+CD48−CD150+) sorted from Yy1−/− or Yy1+/+ BM were plated into Methocult M03434 complete medium. The colony forming activity (total colony number/total cell number plated) from Yy1−/− was reduced significantly in comparison with control (85% versus 10%) (Figure 2F). Quantitation of deletion efficiency revealed the majority of colonies generated by LT-HSCs from Yy1f/f Mx1-Cre mice had either no, or heterozygous YY1 deletion (Figure 2F). To assess whether YY1 deficiency impacts lineage differentiation, total bone marrow cells were plated in Methocult M03434 complete medium. Yy1−/− had significantly reduced CFU-GEMM, CFU-GM, and BFU-E (Figure 2G). Thus, YY1 controls hematopoiesis by regulating HSC numbers and colony-forming activity.
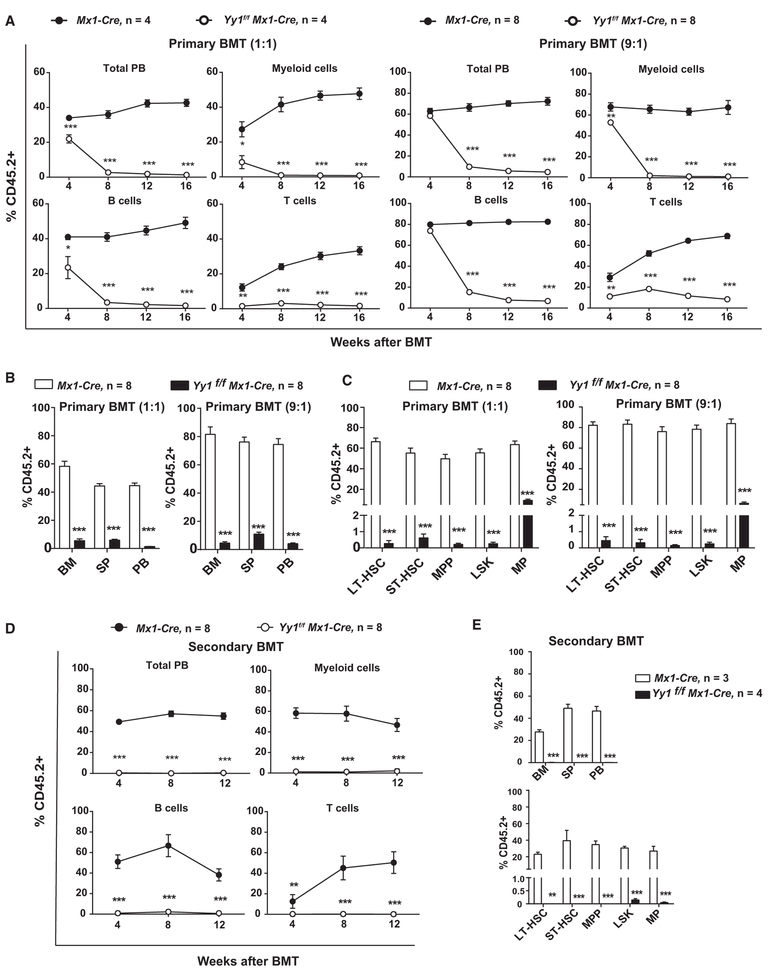
To assess whether YY1-deficiency impacts HSC long-term self-renewal, serial BMTs were conducted. Bone marrows from Yy1f/f Mx1-Cre or Mx1-Cre were mixed with freshly isolated CD45.1+ bone marrow cells at ratio of 1:1 or 9:1 and then transplanted into lethally irradiated CD45.1+ recipients. Recipient mice were treated with 5 doses of pI-pC 4 weeks post-transplantation to delete endogenous YY1. At week 16 post-BMT, Yy1−/− peripheral blood had a significantly reduced percentage of CD45.2+ in every cell lineage, including myeloid (Thy1.2−CD19−), T cells (Thy1.2+CD19−), and B cells (Thy1.2−CD19+). This reduction occurred even under very stringent conditions when using 9 times more Yy1f/f Mx1-Cre bone marrow (9:1 ratio) (Figure 3A). Total peripheral blood, spleen, and bone marrow from mice transplanted with Yy1−/− bone marrows had only ~5% CD45.2+ in comparison with 40%–80% CD45.2+ in mice transplanted with Mx1-cre bone marrow (Figure 3B). As Yy1−/− donor cells failed to give rise to myeloid and lymphoid lineages, we tested whether the YY1 deficiency impacts HSCs. Indeed, there was a significant reduction in the percentage of CD45.2+ cells in bone marrow LT-HSC (Lin−Sca1+c-Kit+ CD48−CD150+), ST-HSC (Lin−Sca1+c-Kit+CD48−CD150−), MPP (Lin−Sca1+c-Kit+ CD48+CD150−), LSK (Lin−Sca1+c-Kit+), and myeloid progenitor (Lin−Sca1−c-Kit+) populations (Figure 3C). Furthermore, in secondary BMT, no CD45.2+ cells were detected in peripheral blood, spleen, and bone marrow in mice transplanted with Yy1−/− bone marrows, in comparison to 30%–50% CD45.2+ in mice transplanted with wild-type bone marrow (Figures 3D and 3E). A terminal evaluation of secondary BMT mice at 16 weeks revealed no CD45.2+ cells in Yy1−/− LT-HSC, STHSC, and MPP (Figure 3E). Our data prove that Yy1−/− HSCs are defective in long-term self-renewal and are prone to stem cell exhaustion.
Figure 3. Yy1 Deficiency Results in Reduced HSC Long-Term Repopulating Activity in Competitive Serial Transplantations.
Mx1-cre or Yy1f/f Mx1-cre bone marrow was mixed with CD45.1+ bone marrow competitor cells in a 1:1 or 9:1 ratio and transplanted into lethally irradiated CD45.1+ recipients. Four weeks after transplantation, recipient mice received 5 doses of pI-pC to delete the endogenous YY1.
(A) Lineage evaluations of donor-derived contribution in primary total peripheral blood cell, myeloid, T cells, and B cells at 4, 8, 12, and 16 weeks post BMTs.
(B) Donor-derived contribution in primary total bone marrow, spleen, and peripheral blood cells.
(C) Donor-derived contribution in primary bone marrow LSK, LT-HSC, ST-HSC, MPP, and MP.
(D) Donor-derived contribution in secondary total peripheral blood, myeloid, T and B cell.
(E) Donor-derived contribution in secondary total bone marrow, spleen, peripheral blood cells, bone marrow LSK, LT-HSC, ST-HSC, MPP, and MP. N represents the number of mice; graphs show means ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
PcG Domain-Independent YY1 Function Confers HSC Long-Term Self-Renewal
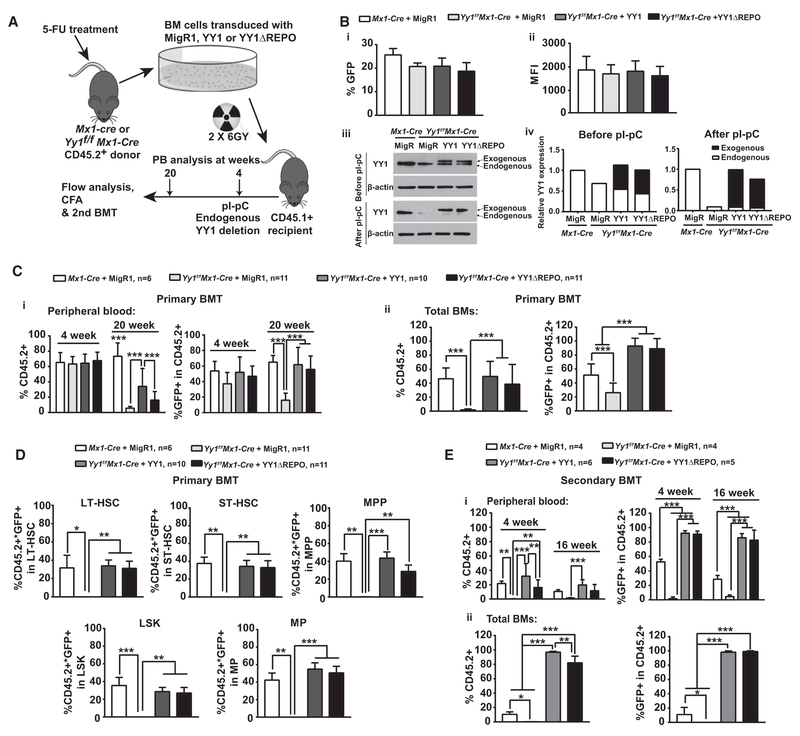
The YY1 REPO domain is necessary and sufficient for YY1 PcG function in Drosophila. As YY1 REPO domain/PcG function is required for early B cell development (Pan et al., 2013), we tested whether YY1 REPO domain/PcG function is also required for HSC self-renewal. Bone marrow cells from Yy1f/f Mx1-Cre mice were transduced retrovirally with MigR1-FlagYY1, MigR1-Flag REPO, or MigR1 vector and transplanted into lethally irradiated CD45.1+ mice. In addition, Mx1-Cre bone marrow cells infected with MigR1 vector were used as the wild-type control and were transplanted into CD45.1+ recipient mice (Figure 4A). Prior to BMT, viral transduction efficiency was assessed 48 hr post-infection. MigR1-FlagYY1, MigR1-FlagYY1ΔREPO, or MigR1 had a similar infection rate (% GFP+) and a similar GFP median fluorescence intensity (MFI) (Figure 4B, i and ii). Before pI-pC injections, GFP+ lymphocytes were sorted from the recipient mice peripheral blood at 4 weeks post BMT. Western blot analysis revealed that the exogenous Flag-tagged YY1 or YY1 REPO was expressed at level equivalent to endogenous YY1. Upon pI-pC injection 4 weeks post-transplantation, endogenous YY1 was deleted, and YY1 function beyond this stage was dependent upon the exogenous YY1 constructs transduced into the cells. Western blot analysis revealed that >90% of endogenous YY1 was deleted in GFP+ peripheral blood lymphocytes. Exogenous YY1 and YY1ΔREPO were expressed at a similar level 7 days post-pI-pC injection (Figure 4B, iii and iv). Peripheral blood or bone marrow cells were analyzed by gating CD45.2+ donor-derived cells and further gating for GFP+ cells within LT-HSC (CD45.2+Lin−Sca1+c-Kit+CD48−CD150+), ST-HSC (CD45.2+Lin−Sca1+c-Kit+CD48−CD150−), MPP (CD45.2+Lin−Sca1+c-Kit+CD48+CD150−), LSK (CD45.2+Lin−Sca1+c-Kit+), and myeloid progenitor (CD45.2+Lin−Sca1−c-Kit+) populations (Figure S4A). Prior to the endogenous YY1 deletion 4 weeks post-transplantation, peripheral blood from all cohorts: Yy1+/+ + MigR1, Yy1−/− + MigR1, Yy1−/− + MigR1-Flag YY1, and Yy1−/− + MigR1-Flag REPO had an equivalent percentage of CD45.2+ and percentage of GFP+ within donor-derived cells. However, a 20-week peripheral blood evaluation revealed that mice rescued with wild-type YY1 or YY1ΔREPO had a significantly higher percentage of GFP+ within donor-derived cells in comparison with mice transplanted with Yy1−/− + MigR1 bone marrow cells (Figure 4C, i). In the bone marrow, both YY1- and ΔREPO-rescued mice were nearly 100% GFP+ in donor-derived cells, in contrast to low percentage of GFP+ cells in mice transplanted with MigR1-infected Yy1−/− bone marrow cells (Figure 4C, ii). The YY1- or YY1ΔREPO-rescued mice had a similar percentage of GFP+ donor-derived cell (% GFP+ CD45.2+) in bone marrow LT-HSC, ST-HSC, MPP, LSK, and MP populations in comparison with the wild-type control (Yy1+/+ + MigR1), and the percentage was significantly higher in comparison with mice transplanted with Yy1-deficient bone marrow cells (Yy1−/−+ MigR1) (Figure 4D). Clearly, both full-length YY1 and YY1ΔREPO were competent to rescue the Yy1−/− HSC phenotype. HSCs rescued with YY1 or YY1ΔREPO had an indistinguishable capacity to form colonies (Figure S4B). To assess YY1REPO domain/PcG function in HSC long-term self-renewal, a secondary BMT was conducted. Mice rescued with full-length YY1 or YY1ΔREPO had a similarly high percentage of CD45.2+ and GFP+ cells in peripheral blood and bone marrow (Figure 4E). Thus, YY1-dependent HSC long-term self-renewal does not require REPO domain/PcG function.
Figure 4. YY1 Regulation of HSC Long-Term Self-Renewal Is Independent on its PcG Function.
(A) Experimental strategy: Yy1f/f Mx1-Cre bone marrow cells transduced with MigR1, MigR1-Flag-YY1, or MigR1-Flag-ΔREPO, or Mx1-Cre bone marrow cells transduced with MigR1, was injected into lethally irradiated CD45.1+ congenic mice. Five doses of pI-pC injections were given at 4 weeks post BMTs. Bone marrows from reconstituted mice were harvested for analyses or for secondary BMTs at 20 weeks post transplantation.
(B) (i and ii) Prior to BMT, flow analysis of % GFP+ and MFI of transduced BM cells at 48 hr post viral infection. (iii) Western blot to detect exogenous Flag-YY1, Flag-ΔREPO, and endogenous YY1 in GFP+ peripheral lymphocytes before and after pI-pC injections. (iv) Quantification of exogenous Flag-YY1, Flag-REPO, and endogenous YY1 protein expressions.
(C) Primary BMT evaluations of donor-derived contribution and % GFP+ in donor-derived peripheral blood (i) and total bone marrow (ii).
(D) Primary BMT evaluation of % GFP+ donor-derived LSK, LT-HSC, ST-HSC, MPP, and MP.
(E) Secondary BMT evaluation of donor-derived contribution and % GFP+ in donor-derived peripheral blood (i) and total bone marrow (ii). N represents the number of mice; graphs show means ± SEM; **p < 0.01, ***p < 0.001.
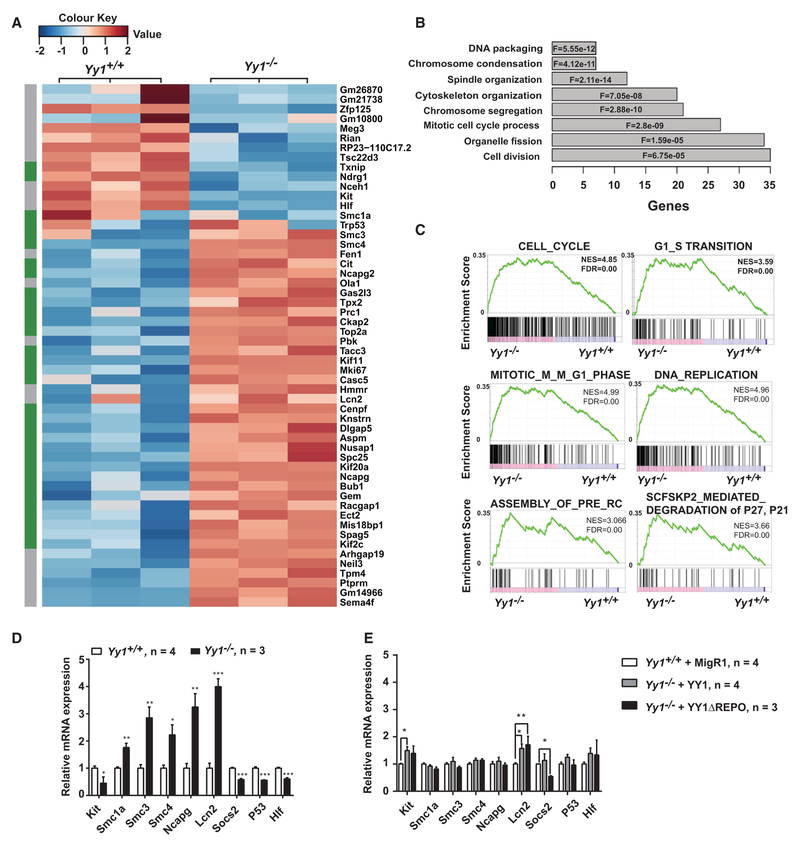
YY1-Deficient HSCs Have an Aberrant Genetic Network Linked to Cell Proliferation
In principle, the Yy1-dependent genetic network should reveal clues regarding the mechanism of YY1-mediated HSC self-renewal. We use fluorescence-activated cell sorting (FACS) to isolate bone marrow Lin−c-Kit+Sca-1+CD48− cells, which include LT-HSCs (Lin−Sca1+c-Kit+CD48−CD150+) and ST-HSCs (Lin−Sca1+c-Kit+CD48−CD150−) and used RNA sequencing (RNA-seq) to establish Yy1+/+ and Yy1−/− transcrip tomes. This analysis identified a small cohort of 100 significantly upregulated and 49 downregulated genes in Yy1−/− HSCs (false discovery rate [FDR] <0.05, logFC R1). Among them, many genes involved in cell cycle (highlighted in green) were deregulated in Yy1−/−HSCs (Figure 5A). Gene ontology analysis of the upregulated cohort revealed a significant enrichment of genes involved in cell cycle, cell division, chromosome condensation, and segregation, cytoskeleton, and spindle organizations (Figure 5B). Gene Set Enrichment Analysis (GSEA) revealed cell-cycle gene sets associated with pre-replication complex (RC) assembly, degradation of p27, DNA replication, G1 to S phase, and M to G1 phase transitions were significantly enriched (Figure 5C). The entry of quiescent cells into G1-phase is a prolonged process involving the accumulation of CDC6, CDT1, MCM and the assembly of pre-RCs, decreasing levels of p27 and the synthesis of D-type cyclin. The RNA-seq analysis revealed that YY1-deficient HSCs had an aberrant genetic network associated with G0 to G1 phase entry. Quantitative real-time PCR (real-time qPCR) validated the RNA-seq results, confirming that the cell-cycle genes Smc1a, Smc3, Smc4, and Ncapg were upregulated, and the HSPC-regulatory gene Kit was downregulated at the mRNA levels in Yy1−/− Lin− bone marrow cells (Figure 5D). To evaluate whether YY1 regulated cell-cycle genes and Kit via its REPO domain/PcG function, we sorted Lin− GFP+ bone marrow cells from mice rescued with YY1 or REPO (Figure 4A). Consistent with our conclusion that wild-type YY1 and REPO were competent to rescue the HSC self-renewal defect in Yy1−/− mice, Smc1a, Smc3, Smc4, Ncapg, and Kit mRNA expression were similar to the wild-type level in YY1-and YY1ΔREPO-rescued Lin−GFP+ bone marrow cells. The REPO domain/PcG dependency is gene-specific, as YY1-DREPO failed to rescue expression of Socs2, a member of the suppressor of cytokine signaling, while YY1 rescued Socs2 expression (Figure 5E). Thus, YY1 regulates important cell-cycle genes and Kit in a REPO domain/PcG function-independent manner.
Figure 5. Yy1−/−HSCs Exhibit an Aberrant Genetic Network with Corruption of Genes Involved in Cell-Cycle Regulation $$PARABREAKHERE$$RNA-seq-based comparison of gene expression in Yy1+/+ and Yy1−/−sorted HSCs.
(A) Heatmap depicting statistically significant, differentially expressed upregulated and downregulated genes.
(B) Gene Ontology analysis of genes upregulated in Yy1−/−HSCs. The most enriched biological processes are shown with corresponding FDR values. F, FDR.
(C) GSEA shows that genes involved in cell-cycle progression and G0 to G1 phase entry are enriched in Yy1−/−HSCs.
(D) Relative mRNA expression of genes in Lin−GFP+ bone marrow cells of Yy1+/+ versus Yy1−/−.
(E) Relative mRNA expressions of genes in Lin−GFP+ bone marrow cells of Yy1+/+ + MigR1, Yy1−/−+ YY1, or Yy1−/−+ ΔREPO. Graphs show means ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
YY1-Deficiency Impairs Mechanisms Governing HSC Quiescence
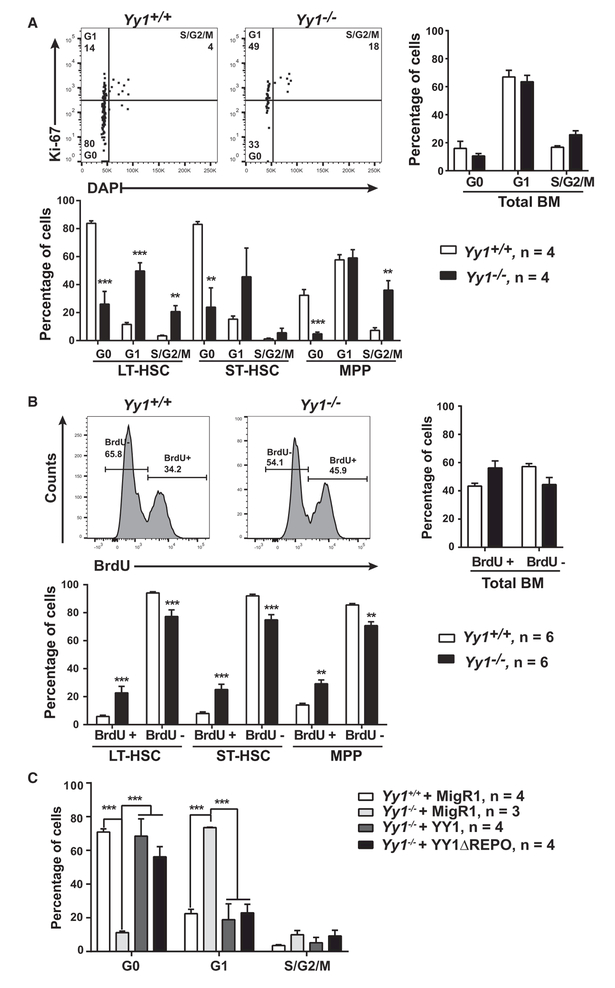
HSC quiescence is associated with its long-term self-renewal function. As the RNA-seq analysis revealed an aberrant genetic network involving cell-cycle and G1 phase entry, we hypothesized that YY1 deficiency may disrupt mechanisms involved in establishing and/or maintaining HSC quiescence. We analyzed cell-cycle status using Ki-67/DAPI staining. Within bone marrow LT-HSC, ST-HSC, and MPP populations, we gated cells in G1(Ki-67+DAPI−), S/G2/M (Ki-67+DAPI+), or G0 (Ki-67−DAPI−) phase (Figure 6A). Yy1−/− HSCs and earlier progenitors exhibited significantly reduced G0 phase cells. The reduction of quiescent cells included HSCs and early progenitors, as Yy1−/− total bone marrow had a normal G0 percentage (Figure 6A). Cell cycle was further tested using a BrdU incorporation assay. Consistent with the Ki-67/DAPI results, Yy1−/−LT-HSC and ST-HSC failed to establish normal quiescence with an increased proportion of BrdU+ cells (Figure 6B). As YY1 can regulate apoptosis (Affar et al., 2006; Chen et al., 2016; Sui et al., 2004), we tested whether YY1 deficiency impacted HSC apoptosis. Early and late apoptotic cells were stained with AnnexinV/DAPI. YY1 deficiency did not induce apoptosis in LSK, LT-HSC, ST-HSC, or MPP (Figure S5). These results indicate that YY1 controls HSC cell-cycle regulation and is an important determinant of HSC quiescence. To test whether YY1 function in HSC quiescence is REPO domain/PcG function-dependent, we conducted cell-cycle analysis using Ki-67/DAPI staining in GFP+ bone marrow HSC (Lin−c-Kit+Sca-1+CD48−) from YY1- or ΔREPO-rescued mice (Figure 4A). Wild-type YY1 and YY1ΔREPO were competent to restore the HSC cell-cycle (Figure 6C), indicating that YY1 regulation of HSC quiescence is REPO domain/PcG function-independent.
Figure 6. Yy1-Deficient HSCs Exhibit Loss of Quiescence and Increased Proliferation.
(A) Ki67/DAPI cell-cycle analysis of total bone marrow, LT-HSC, ST-HSC, and MPP of Yy1+/+ and Yy1−/−.
(B) BrdU incorporation assay of total bone marrow, LT-HSC, ST-HSC, and MPP of Yy1+/+ and Yy1−/−.
(C) Ki67/DAPI cell-cycle analysis of HSCs (Lin−Sca1+c-Kit+ CD48−CD150 and Lin−Sca1+c-Kit+CD48−CD150−) of Yy1+/+ + MigR1, Yy1−/−+ MigR1, Yy1−/−+ YY1, and Yy1−/−+ ΔREPO. N represents the number of mice; graphs show means ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
YY1 c-Kit Axis in Adult HSC
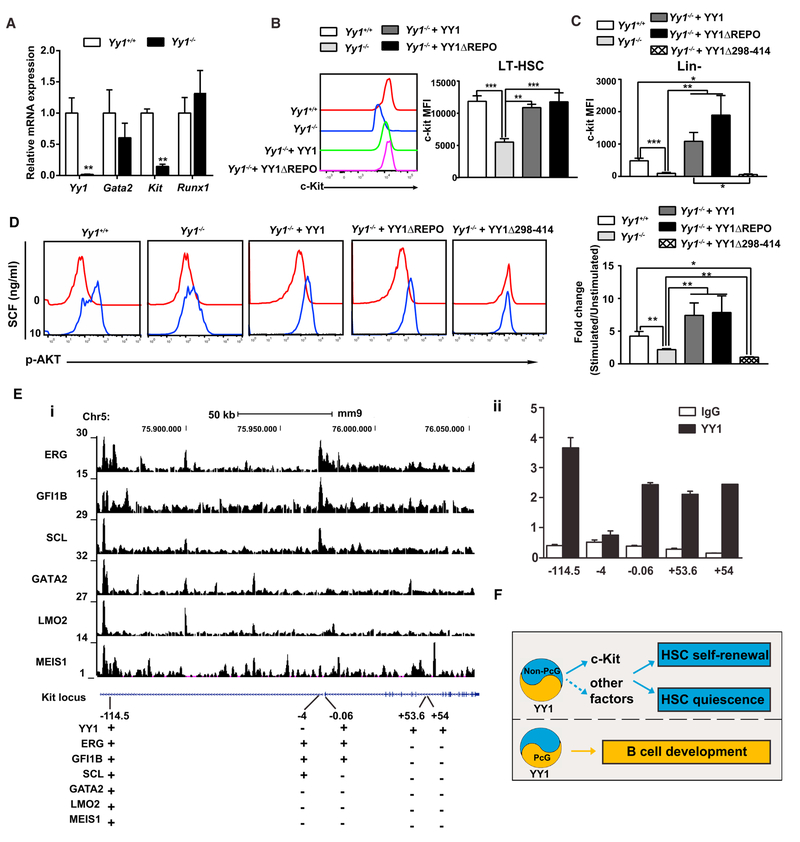
The c-Kit receptor tyrosine kinase is an essential regulator for HSPCs. Moderately decreased c-Kit signaling impairs HSC self-renewal and can yield anemia (Ho et al., 2017; Miller et al., 1996; Thorén et al., 2008; Zhang et al., 2017). Gain-of-function mutations have been detected in human leukemia patients (Cairoli et al., 2006; Malaise et al., 2009). SCF/c-Kit signaling controls HSC quiescence (Thorén et al., 2008). HSCs from W41/W41 mice, with a partial Kit loss-of-function, have decreased long-term reconstitution activity, and this defect correlates with a decreased proportion of cells in G0 phase (Miller et al., 1996). Although YY1 directly regulates c-Kit transcription in human melanocytes by occupying intron 7 (Li et al., 2012), its involvement in controlling Kit in HSCs has not been described. We demonstrate that Kit mRNA levels were significantly lower in Yy1−/− bone marrow. YY1 regulation of Kit was specific, as Gata2 and Runx1 mRNA expression were not significantly altered (Figure 7A). The MFI of c-Kit in LT-HSC (Lin−Sca1+c-Kit+ CD48−CD150+) cells was significantly lower in Yy1−/−mice (Figure 7B) and higher in mice with ectopic YY1 expression as described in Figure 1 (Figure S6). Ectopic expression of wild-type YY1 or ΔREPO in Yy1−/−mice rescued c-Kit MFI back to the wild-type level (Figure 7B). To test whether YY1 deficiency influences c-Kit function, we analyzed c-Kit-dependent phosphorylation of the downstream target, AKT in Lin−c-Kit+ cells of Yy1−/−, Yy1−/−+ YY1, and Yy1−/−+ REPO mice. While SCF induced AKT phosphorylation in wild-type Lin−c-Kit+ bone marrow cells, YY1-deficient Lin−c-Kit+ cells failed to respond to SCF. However, wild-type YY1- and YY1ΔREPO-rescued Lin−c-Kit+ bone marrow cells regained the capacity to respond to SCF (Figure 7D). These results indicate that YY1 deficiency reduced c-Kit cell surface expression and signaling in Lin−c-Kit+ bone marrow cells, and YY1 activity to regulate c-Kit expression is independent on its REPO domain/PcG function. In addition to its function as a critical Polycomb protein, YY1 is a multifunctional transcription factor that can bind specific DNA sites with its zinc finger DNA binding domain (YY1298–414) (Bushmeyer et al., 1995; Zaprazna and Atchison, 2012). We analyzed YY1 occupancy at Kit using mouse chromatin immunoprecipitation sequencing (ChIP-seq) datasets from HPC-7 (Wilson et al., 2010) and G1E cells (Hewitt et al., 2015; Jing et al., 2008). These data from hematopoietic cell lines included six regulators of hematopoietic stem/progenitor cells (ERG, GFI1B, SCL, GATA-2, LMO2, and MEIS1). YY1 occupied intron 7 sites (+53.6 and +54 from TSS) as reported previously in human melanocytes (Li et al., 2012). Our data also reveal YY1 co-occupancy at 114 and the Kit promoter with the additional six transcription factors, including GATA-2 (Figure 7E). GATA-2 is a key regulator of the SCF/c-Kit signaling pathway in erythroid maturation (Hewitt et al., 2015), and the 114 position of the Kit locus is implicated as a potential GATA-2-regu-lated enhancer (Jing et al., 2008). To test whether YY1 activity to regulate c-Kit expression and signaling requires its DNA binding function, we have prepared a DNA binding domain deletion mutant (YY1Δ298–414) (Zaprazna and Atchison, 2012). The YY1Δ298–414 protein was structurally stable and expressed at a near-physiological level in mice transplanted with bone marrow cells infected with YY1Δ298–414 (Figure S7). Compared with wild-type YY1 or YY1ΔREPO, YY1Δ298–414 was unable to rescue c-Kit expression and signaling in Lin−bone marrow cells (Figures 7C and 7D). Thus, YY1 regulates c-Kit surface expression and signaling in HSCs, and the YY1-c-Kit axis requires its DNA binding activity.
Figure 7. YY1 Promotes SCF/c-Kit Signaling.
(A) Quantitative real-time PCR analysis of Yy1, Gata2, Kit, and Runx1 transcript levels in total bone marrow cells.
(B) Evaluation of c-Kit MFI in LT-HSC.
(C) Evaluation of c-Kit MFI in Lin−bone marrow cells.
(D) Phosphorylated flow analysis of AKT.
(E) YY1 occupancy at Kit. (i) “+” and “− ” indicate positive versus negative occupancy of transcription factors at Kit based on ChIP-seq (GEO: GSE22178) or ChIP-qPCR, respectively. (ii) YY1 occupancy at Kit in HPC-7 cells detected by ChIP-qPCR and normalized to input.
(F) The model assumes that YY1 directly regulates SCF/c-Kit signaling in HSCs and promotes HSC long-term self-renewal and quiescence without a requirement for the REPO domain/PcG function. By contrast, YY1 REPO domain/ PcG function is required for Ig rearrangement in early B cells. N represents the number of mice; graphs show means ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
DISCUSSION
Multiple transcription factors and epigenetic regulators function as intrinsic regulators of HSC quiescence. YY1, a multifunctional transcription factor and a PcG protein, plays critical roles in B and T lymphocyte development (Chen et al., 2016; Kleiman et al., 2016; Liu et al., 2007; Zaprazna and Atchison, 2012). While many PcG proteins have important roles in HSC self-renewal and lineage differentiation, YY1 is one of a few mammalian PcG proteins with intrinsic sequence-specific DNA binding activity (Atchison et al., 2003; Srinivasan and Atchison, 2004). YY1 recruits other PcG proteins, including EZH2, to specific DNA sequences, leading to catalysis of H3K27me3 at the recruitment site (Srinivasan and Atchison, 2004; Wilkinson et al., 2006). YY1 activity to recruit other PcG factors to specific DNA sites suggests that YY1 has a pivotal role in orchestrating the function of various HSC-regulatory factors.
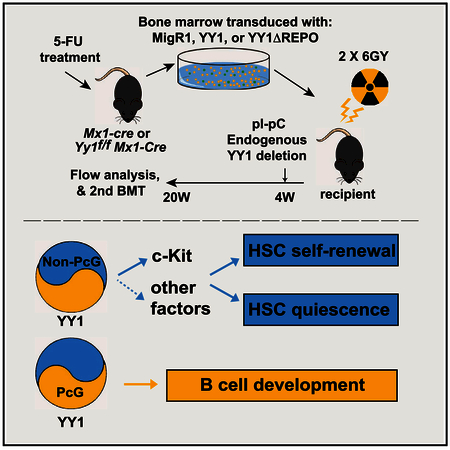
Our results described here demonstrate that YY1 promotes HSC long-term self-renewal and quiescence (Figures 3 and 6). By introducing a YY1 mutant (ΔREPO) into an YY1 null genetic background, we dissected YY1 REPO domain/ PcG function in different hematopoietic compartments (Pan et al., 2013) (Figure 4). While YY1 REPO domain/ PcG function is required for Igk rearrangement in early B cell development (Pan et al., 2013), YY1 control of HSC self-renewal and quiescence is insensitive to YY1 PcG domain deletion. By contrast, the genetic network governing cell-cycle progression is disrupted in Yy1-deficient HSCs. Genes required for cell-cycle G1 phase entry, G1 to S transition, and mitosis were deregulated. Many key regulators of HSC proliferation and quiescence, including c-Kit, are significantly deregulated in Yy1−/−HSCs and YY1 regulatory function of cell-cycle genes is also independent on its REPO/ PcG function (Figure 7F).
Intriguingly, YY1 utilizes distinct mechanisms in different hematopoietic compartments. Although YY1 is a ubiquitously expressed transcription factor with a high expression level in LT-HSC, ST-HSC, MPP, myeloid progenitor, CLP, and lymphocytes (https://gexc.riken.jp/models/3/genes/Yy1; Figure S3B), mechanisms underlying YY1 function in lymphocytes appear to be distinct. While a high YY1 level promotes HSC self-renewal (Figure 1), high YY1 induces B cell apoptosis and downregulates anti-apoptotic genes in vitro (Pan et al., 2012). The negative impact of high YY1 expression on B lymphocyte could also involve mechanisms beyond cell survival control. High YY1 expression may disrupt the stoichiometry of YY1-PcG complexes. While YY1 establishes HSC quiescence independent on its REPO domain/PcG function, YY1 PcG domain is required for proper Ig rearrangement and early B cell development (Pan et al., 2013). B cell development is delineated by the rearrangement status of the Ig heavy and light chain genes. The Ig loci are 2.4–3.2 Mb, and for rearrangement of distal variable region genes to occur, the loci must undergo a physical contraction process. PcG proteins mediate long-range interactions between their binding sites in polycomb response elements (PREs) and promoter DNA sequences (Lanzuolo et al., 2007). YY1 plays critical roles in IgH locus contraction in B cell development (Liu et al., 2007) and YY1 physically co-localizes with PcG protein EZH2 and condensin proteins across the Igk locus (Pan et al., 2013). Because YY1 recruits PcG proteins to DNA (Srinivasan and Atchison, 2004; Wilkinson et al., 2006), the recruitment of EZH2 to multiple sites across the Igk locus could assist in long-distance interactions leading to locus contraction and rearrangement in Igk locus. Further study of mechanisms underlying the context-dependent requirement of YY1 and its PcG domain for activation or repression will ultimately be important.
c-Kit is an important determinant of HSC self-renewal and quiescence (Ho et al., 2017; Thorén et al., 2008; Zhang et al., 2017). Our study established that YY1 regulates c-Kit surface expression and signaling in HSCs, and the YY1-c-Kit axis requires its DNA binding activity. YY1 overlaps with other hematopoietic regulators, including GATA-2 and SCL at the Kit locus and regulates c-Kit cell surface expression and function (Figure 7). GATA-2 promotes the SCF/c-Kit signaling pathway in erythroid maturation (Hewitt et al., 2015). Although Gata2 expression was not altered in Yy1−/−HSCs (Figure 7A), YY1 co-localized with GATA-2 at the Kit-114 site in the multi-potent HPC7 cell line (Figure 7D). Furthermore, YY1 regulates cohesin (Smc1a and Smc3) and condesin (Smc4 and Ncapg) complex proteins in a REPO domain/PcG-independent manner (Figures 5D and 5E), and those genes play critical roles in spindle dynamics, centrosome separation, and centromere-kinetochore complex formation. Clearly, the genetic network linked to cell proliferation is disrupted at multiple levels in Yy1−/−HSCs. YY1 function to control the HSC self-renewal and quiescence likely involves a multi-component mechanism and would not be expected to solely involve the regulation of c-Kit signaling. For instance, HSC and progenitor cells undergo both symmetric and asymmetric division to maintain proper stem cell pools and functions (Ting et al., 2012; Wu et al., 2007; Zimdahl et al., 2014). YY1 physically interacts and co-localizes with chromo-some condensin complex proteins SMC4 and SMC2 at the Igk locus (Pan et al., 2013). Yy1 deficiency might negatively impact proper mitotic spindle formation and positioning during either symmetric or asymmetric cell division, and the mouse model and genomic insights derived from our study will facilitate the subsequent testing of this potential mechanism.
EXPERIMENTAL PROCEDURES
Mice
Yy1f/f mice (Affar et al., 2006; Liu et al., 2007) in which the Yy1 promoter region and exon 1 are flanked by loxP sites, were crossed to Mx1-Cre mice to generate heterozygous Yy1f/+ Mx1-cre mice. Yy1f/+ Mx1-cre mice were then subsequently bred with Yy1f/f mice to generate homozygous Yy1f/f Mx1-cre mice. To induce the expression of Cre recombinase, 6- to 8-week-old mice were injected with 200 mg of polyinosinic-polycytidylic acid (pI-pC; Sigma-Aldrich) every other day for 5 doses. Approximately equal portions of mice of both genders were used, and aggregated data are presented because gender specific differences were not found. All experiments described in this manuscript were performed 7–10 days after the last injection of pI-pC unless stated otherwise. All experiments involving mice were approved by the Institutional Laboratory Animal Care and Use Committee of the University of Wisconsin-Madison and conform to the appropriate regulatory standards.
BMTs
For retroviral BMT, bone marrow cells were harvested from 6- to 8-week-old C57BL/6 (CD45.2+) mice 4 days after receiving a single intravenous injection of 5 mg 5-fluorouracil (5-FU, APP Pharmaceuticals) and transduced as previously described (Pan et al., 2012, 2013). More experimental details are described in the Supplemental Information.
Flow Cytometric Analysis and Cell Sorting
Directly conjugated or biotin-conjugated antibodies specific for the following surface antigens were purchased from eBioscience: CD3 (145–2C11), CD4 (RM4–5), CD8 (53–6.7), B220 (RA3–6B2), TER119 (TER-119), Gr-1 (RB6–8C5), IgM (eB121–15F9), CD19 (eBio1D3), IL-7Ra (A7R34), CD45.2 (104), CD45.1 (A20), Sca-1 (D7), c-Kit (2B8), Mac1 (M1/70), Gr1 (RB6–8C5), Thy1.2 (53–2.1), and CD19 (eBio1D3). CD48 (103427) and CD150 (TC15–12F12.2) were purchased from Biolegend. Acquisitions were performed on a LSR II or LSR II Fortessa (BD Biosciences). Data were analyzed using BD FlowJo v10.0.7 software. For cell sorting, Lin−c-Kit+Sca-1+CD1502212HSCs or Lin−GFP+ were sorted from freshly prepared single BM suspension by the University of Wisconsin-Madison Carbon Cancer Center Flow Cytometry and Cell Sorting Facility.
Ki67/DAPI and BrdU Incorporation Cell-Cycle Analyses
Bone marrow cells were fixed with 4% PFA, permeabilized with 0.1% saponin in PBS,and stained with FITC-conjugated Ki67(BD Biosciences) and DAPI(Thermo Fisher) in addition to HSC, LSK, MP, and MPP markers described above. A single dose 0.5 mg of BrdU (BD Biosciences) was injected intraperitoneally 24 hr prior to the evaluation. BrdU-labeled BM cells were prepared by using the FITC BrdU Flow Kit (BD Biosciences) following the manufacturer’s instructions.
Flow Cytometry Analysis of Phosphorylated AKT
Total bone marrow cells were deprived of serum and cytokines for 2 hr and then stimulated with murine SCF at 37°C for 10 min. Those stimulated cells were fixed with paraformaldehyde at a final concentration of 2% (Electron Microscopy Sciences) at 37°C for 10 min. Surface proteins were detected with FITC-conjugated CD3 (145–2C11), CD4 (RM4–5), CD8 (53–6.7), B220 (RA3–6B2), TER119 (TER119), Gr1 (RB6–8C5), and PE-conjugated c-Kit (2B8) antibodies. Phosphorylated AKT was detected by a primary antibody against p-AKT (Ser473; Cell Signaling Technology), followed by APC-conjugated donkey anti-rabbit F (ab′) 2 fragment (Jackson ImmunoResearch).
ChIP Assays
The ChIP-seq data were compiled from the UCSC website (http://genome.ucsc.edu/). ChIP-qPCR was carried out according to the Upstate Biotechnology protocol. Approximately 10 × 106 HPC7 cells were used for each immunoprecipitation. Anti-YY1 (H414, Santa Cruz Biotechnology) antibody was used followed by protein A conjugated bead precipitation. Pre-immune controls used normal rabbit IgG. DNA samples were analyzed by real-time PCR using a Roche Lightcycler 96. Relative enrichments for each region were presented as the percentage of input. The primers for specific DNA sequences are listed in Table S1.
RNA-Seq and Gene Set Enrichment Analysis
A total of 6 samples were chosen for RNA-seq, 3 from Yy1+/+ and 3 from Yy1−/−mice. Total RNA was purified from bone marrow HSCs (Lin−Sca1+c-Kit+CD48−CD150+and Lin−Sca1+c-Kit+CD48−CD150−) sorted from Yy1−/−versus Yy1+/+ mice using RNeasy Micro kit (QIAGEN). Sequencing libraries were prepared by using Ovation RNA-Seq System V2 (NuGen) according to the manufacturer’s specifications and were sequenced by an Illumina HiSeq 2000 at the University of Wisconsin Biotechnology Center Gene Expression Center. Gene set enrichment analysis (GSEA) was performed by calculating a ranked vector as sign (foldChange)* 1/p value. More experimental details are described in the Supplemental Information.
Statistics
Results are expressed as mean ± SEM from at least three independent experiments using three or more animals per group. Differences between two groups were analyzed using a two-tailed Student’s t test, whereas differences among three or more groups were analyzed using two-way ANOVA followed by Tukey’s correction for multiple comparisons. GraphPad Prism 7 was used for all statistical analyses, and p values are indicated as follows: *p < 0.05, **p < 0.01, and ***p < 0.001.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq data reported in this paper is GEO: GSE108672. The accession number for ERG, GFI1B, SCL, GATA2, LMO2, and MEIS1 ChIP-seq in HPC7 cells reported in this paper is GEO: GSE22178. The DOI number for Mendeley data is https://doi.org/10.17632/swk6j63kft.1.
Supplementary Material
Highlights.
YY1 promotes adult HSC self-renewal and quiescence
PcG domain is selectively utilized at distinct contexts in hematopoiesis
YY1 regulates SCF/c-Kit signaling in HSCs
ACKNOWLEDGMENTS
We would like to thank the University of Wisconsin Carbone Comprehensive Cancer Center (UWCCC) for use of its Shared Services (Flow Cytometry Core, Cancer Informatics Shared Resources, and Histology Lab), which are supported by UWCCC grant P30 CA014520, to complete this research. This work was supported by startup funds from UW-Madison 101A-87–0641, NIH K01OD020153–01A1 (to X.P.), NIH R01CA152108, NIH R01HL113066, a Scholar Award from the Leukemia & Lymphoma Society (to J.Z.), and DK68634 (to E.H.B.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Affar B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, and Shi Y (2006). Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol 26, 3565–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison L, Ghias A, Wilkinson F, Bonini N, and Atchison ML (2003). Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 22, 1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushmeyer S, Park K, and Atchison ML (1995). Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem 270, 30213–30220. [DOI] [PubMed] [Google Scholar]
- Cairoli R, Beghini A, Grillo G, Nadali G, Elice F, Ripamonti CB, Colapietro P, Nichelatti M, Pezzetti L, Lunghi M, et al. (2006). Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood 107, 3463–3468. [DOI] [PubMed] [Google Scholar]
- Chen L, Foreman DP, Sant’Angelo DB, and Krangel MS (2016). Yin yang 1 promotes thymocyte survival by downregulating p53. J. Immunol 196, 2572–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt KJ, Kim DH, Devadas P, Prathibha R, Zuo C, Sanalkumar R, Johnson KD, Kang YA, Kim JS, Dewey CN, et al. (2015). Hematopoietic signaling mechanism revealed from a stem/progenitor cell cistrome. Mol. Cell 59, 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CCM, Chhabra A, Starkl P, Schnorr PJ, Wilmes S, Moraga I, Kwon HS, Gaudenzio N, Sibilano R, Wehrman TS, et al. (2017). Decoupling the functional pleiotropy of stem cell factor by tuning c-Kit signaling. Cell 168, 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, Ema H, Kamijo T, Katoh-Fukui Y, Koseki H, et al. (2004). Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 21, 843–851. [DOI] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, and Blobel GA (2008). Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol. Cell 29, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, Dontje B, and de Haan G (2006). The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood 107, 2170–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke K, Radulovic, V, Broekhuis M, Weersing E, Zwart E, Olthof S, Ritsema M, Bruggeman S, Wu X, Helin K, et al. (2013). Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat. Cell Biol. 15, 353–362. [DOI] [PubMed] [Google Scholar]
- Kleiman E, Jia H, Loguercio S, Su AI, and Feeney AJ (2016). YY1 plays an essential role at all stages of B-cell differentiation. Proc. Natl. Acad. Sci. USA 113, E3911–E3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, and Orlando V (2007). Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 9, 1167–1174. [DOI] [PubMed] [Google Scholar]
- Li J, Song JS, Bell RJ, Tran TN, Haq R, Liu H, Love KT, Langer R, Anderson DG, Larue L, and Fisher DE (2012). YY1 regulates melanocyte development and function by cooperating with MITF. PLoS Genet. 8, e1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, and Shi Y (2007). Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 21, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski IJ, Ritchie ME, Phipson B, Corbin J, Pakusch M, Ebert A, Busslinger M, Koseki H, Hu Y, Smyth GK, et al. (2010). Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood 116, 731–739. [DOI] [PubMed] [Google Scholar]
- Malaise M, Steinbach D, and Corbacioglu S (2009). Clinical implications of c-Kit mutations in acute myelogenous leukemia. Curr. Hematol. Malig. Rep 4, 77–82. [DOI] [PubMed] [Google Scholar]
- Miller CL, Rebel VI, Lemieux ME, Helgason CD, Lansdorp PM, and Eaves CJ (1996). Studies of W mutant mice provide evidence for alternate mechanisms capable of activating hematopoietic stem cells. Exp. Hematol 24, 185–194. [PubMed] [Google Scholar]
- Morrison SJ, and Weissman IL (1994). The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity 1, 661–673. [DOI] [PubMed] [Google Scholar]
- Orford KW, and Scadden DT (2008). Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet 9, 115–128. [DOI] [PubMed] [Google Scholar]
- Pan X, Jones M, Jiang J, Zaprazna K, Yu D, Pear W, Maillard I, and Atchison ML (2012). Increased expression of PcG protein YY1 negatively regulates B cell development while allowing accumulation of myeloid cells and LT-HSC cells. PLoS ONE 7, e30656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Papasani M, Hao Y, Calamito M, Wei F, Quinn Iii WJ, Basu A, Wang J, Hodawadekar S, Zaprazna K, et al. (2013). YY1 controls Igk repertoire and B-cell development, and localizes with condensin on the Igk locus. EMBO J. 32, 1168–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, and Clarke MF (2003). Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305. [DOI] [PubMed] [Google Scholar]
- Pietras EM, Warr MR, and Passegué E (2011). Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195, 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo A, Olthof S, Han L, Vellenga E, de Haan G, and Schuringa JJ (2009). Repression of BMI1 in normal and leukemic human CD34(+) cells impairs self-renewal and induces apoptosis. Blood 114, 1498–1505. [DOI] [PubMed] [Google Scholar]
- Srinivasan L, and Atchison ML (2004). YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 18, 2596–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan L, Pan X, and Atchison ML (2005). Transient requirements of YY1 expression for PcG transcriptional repression and phenotypic rescue. J. Cell. Biochem 96, 689–699. [DOI] [PubMed] [Google Scholar]
- Suda T, Suda J, and Ogawa M (1983). Single-cell origin of mouse hemopoietic colonies expressing multiple lineages in variable combinations. Proc. Natl. Acad. Sci. USA 80, 6689–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui G, Affar B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, and Shi Y (2004). Yin Yang 1 is a negative regulator of p53. Cell 117, 859–872. [DOI] [PubMed] [Google Scholar]
- Thorén LA, Liuba K, Bryder D, Nygren JM, Jensen CT, Qian H, Anton-chuk J, and Jacobsen SE (2008). Kit regulates maintenance of quiescent hematopoietic stem cells. J. Immunol 180, 2045–2053. [DOI] [PubMed] [Google Scholar]
- Ting SB, Deneault E, Hope K, Cellot S, Chagraoui J, Mayotte N, Dorn JF, Laverdure JP, Harvey M, Hawkins ED, et al. (2012). Asymmetric segregation and self-renewal of hematopoietic stem and progenitor cells with endocytic Ap2a2. Blood 119, 2510–2522. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, and Jones RS (2004). Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14, 637–646. [DOI] [PubMed] [Google Scholar]
- Wilkinson FH, Park K, and Atchison ML (2006). Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. USA 103, 19296– 19301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte €J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. (2010). Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7, 532–544. [DOI] [PubMed] [Google Scholar]
- Wu M, Kwon HY, Rattis F, Blum J, Zhao C, Ashkenazi R, Jackson TL, Gaiano N, Oliver T, and Reya T (2007). Imaging hematopoietic precursor division in real time. Cell Stem Cell 1, 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaprazna K, and Atchison ML (2012). YY1 controls immunoglobulin class switch recombination and nuclear activation-induced deaminase levels. Mol. Cell. Biol 32, 1542–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu P, Zhou Y, Sheng Y, Hong Y, Xiang D, Qian Z, Mosenson J, and Wu WS (2017). A novel slug-containing negative-feedback loop regulates SCF/c-Kit-mediated hematopoietic stem cell self-renewal. Leukemia 31, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimdahl B, Ito T, Blevins A, Bajaj J, Konuma T, Weeks J, Koechlein CS, Kwon HY, Arami O, Rizzieri D, et al. (2014). Lis1 regulates asymmetric division in hematopoietic stem cells and in leukemia. Nat. Genet 46, 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.