ABSTRACT
The analysis of tumor growth curves is standard practice in experimental oncology including tumor immunology. In experimental oncology, cancer cells are inoculated into rodents (mostly mice) and their growth is monitored by measuring tumor diameter, surface or volume over time as a function of distinct treatments. Then, different groups of tumors/treatments are compared among each other for their evolution and possible responses to treatment. The R package TumGrowth has been created as a software tool allowing to carry out a series of statistical comparisons across or between groups of tumor growth curves obtained in a standard laboratory, for experimenters with limited knowledge in statistics. TumGrowth is freely available online at https://kroemerlab.shinyapps.io/TumGrowth/ and can be downloaded into any computer. It offers an exhaustive panoply of tools to visualize and analyze complex data sets including longitudinal, cross-sectional and time-to-endpoint measurements.
KEYWORDS: mice, statistical analyses, survival, tumor growth
Preclinical models of oncogenesis, tumor progression and treatment responses are essential for experimental cancer research. In a typical experiment, rodents (usually mice or rats) are inoculated subcutaneously with transplantable tumors that are either isogeneic (if the recipient is immunocompetent) or can be histo-incompatible or even xenogeneic (if the recipient is immunodeficient). Alternatively, cancer cells can be injected orthotopically, into the organ from which they originate, and tumor monitoring then requires image technologies based on fluorescence of luminescence signals (if the tumor has been transduced with appropriate biosensors) or radiological methods (such as micro-computer tomographic scans). Experimental cancers may also be induced by the regular administration of chemical carcinogens or genetic manipulations aiming at the activation of oncogenes.1
The scope of most experiments is to evaluate the capacity of treatments to prevent, reduce or abolish tumor growth, meaning that drugs or appropriate vehicle controls are administered to the animals and the growth of the tumor is recorded over time as a function of the treatment. Independently of the exact way how tumors are generated, treated and monitored, in a typical experimental setup, multiple measurements of tumor diameter, surface or volume are monitored, by calipers, in several groups of rodents at different time points. The analysis of such data, which often are non-linear in nature (due to the effects of treatments and the impact of increased or relaxed immunosurveillance) and frequently are afflicted by major inter-individual variations within the same treatment group, often trespasses the statistical competence of the experimenter, yielding suboptimal analyses or requiring the involvement of a professional biostatistician.
Driven by these premises, we decided to create an annotated software package that can be used by experimenters without programming knowledge to take advantage of the functionalities of the R environment coupled to the Shiny framework. The software entitled TumGrowth (see Fig. 1 for an overview) allows the user to upload data generated in conventional spreadsheet software, to verify the overall experimental design and data integrity, to visualize the entire experiments or selected groups alone or en masse, and then to perform three alternative, yet complementary statistical analyses, depending on three types of questions:
-
1.
Does a treatment affect the trajectory of tumor growth? For this, longitudinal analysis can be performed either on the entire tumor growth curve or on segments thereof, and the effects of the overall experimental intervention (over all group) can be analyzed statistically in the same way as groups can be subjected to pairwise comparisons.
-
2.
Does a treatment affect tumor size at a given time point chosen by the experimenter? For this, the visualization program facilitates the choice of the time point, and the software package yields statistical comparisons across and between groups.
-
3.
Does a variation in experimental conditions impact on the appearance (or disappearance) of tumors, the overall survival of animals or their survival up to a critical maximum tumor size?
Figure 1.
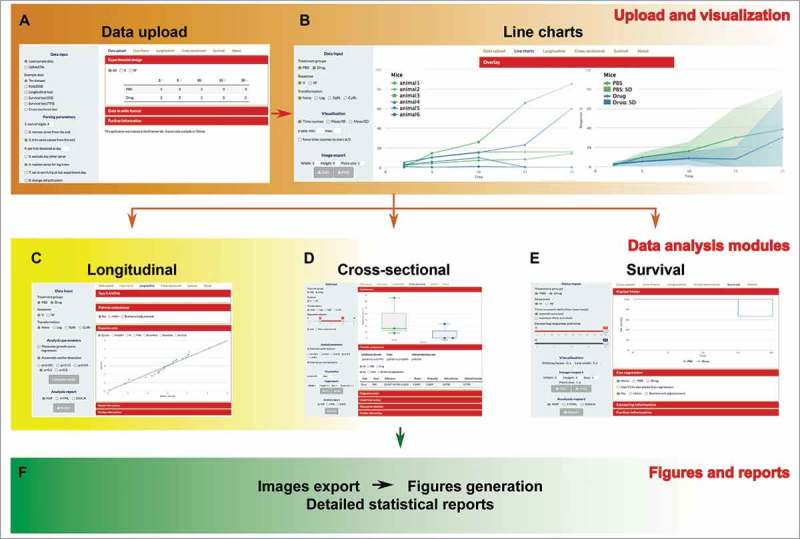
Overall workflow of the TumGrowth software. After data upload, the user verifies the integrity of the data file, visualizes the experimental outcome while allowing the investigator to choose among several possibilities, then performs several distinct statistical comparisons (longitudinal, cross-sectional, time-to-endpoint) and finally generates publication quality Figs., as well as detailed statistical reports.
TumGrowth application not only allows for exporting new generated graphs, for the inclusion in laboratory notebooks and scientific articles, but also it generates a full set of Materials and Methods (by automatically creating statistical reports that comprise experimental design, modeling settings, description of the statistical tests used for the calculation of p-value as well as diagnostic plots) to explain how each graph and each statistical comparison has been calculated.
TumGrowth is freely available online at https://kroemerlab.shinyapps.io/TumGrowth/ (utilization limited to 25 hours per month) and can be downloaded into any computer to be used offline as well (without limitations to its use). As a free interactive web tool, TumGrowth should profit to technicians, students, post-docs and established investigators working in the area of experimental oncology. Examples of its recent use include articles in Cancer Cell,2 Immunity,3 OncoImmunology4,5 and Science.6,7
We anticipate that its use will improve the statistical analyses of experimental tumor growth curves in an ever-expanding number of laboratories. In principle, this statistical package might also be used to analyze growth curves of mice subjected to obesogenic diets.
Materials and methods
Data input
Growth data of each individual tumor (usually as raw data resulting from the measurement of two or three dimensions of the tumor surface/volume) can be uploaded as a tab-separated text file (.txt). Such text files can be exported from any standard spreadsheet software (e.g. .xls or .xlsx) or even text files (e.g. .doc), which are widely used for the documentation of tumor growth experiment in experimental oncology laboratories.
Analyses performed by TumGrowth
The data are subjected to interactive visualization using several distinct submenus (Line charts section) of the program allowing the experimenter to understand the biological outcome of the experiment. Moreover, tumor growth data are subjected to three different types of statistical comparison, and each of the three submenus (Longitudinal analysis, Cross-sectional analysis, Survival analysis) designed for this purpose contain text annotations to guide the user.
First, longitudinal analyses are performed over entire segments of the tumor growth curves that can be subjected to automatic analyses of breakpoints in the tumor growth characteristics and outlier detection on the basis of Bonferroni-corrected p-values calculated from the residuals (if required). In the next step, the tumor growth curves are subjected to type II ANOVA and pairwise comparisons across groups. The user can select the comparison of choice. Moreover, the software comprises a series of diagnostic plots, allowing the user to verify the adequacy of the statistical model that is applied.
Second, cross-sectional analyses allow for the comparison of tumor dimensions at one single time point that is chosen by the user upon interactive visualization of different options. Such data sets can again be subjected to outlier detection on the basis of Bonferroni-corrected p-values calculated from the residuals (if required), followed by comparison of groups. The software then calculates p-values via the Wilcoxon rank sum test that can be adjusted by the Holm method.
Third, time-to-endpoint data are visualized as Kaplan Meier plots and subjected to statistical comparisons by means of the Cox regression model, likelihood ratios, Wald and score tests. It is important to note that the program is flexible in specifying alternative censoring cut-offs (e.g. time of apparition of the tumor, maximum tumor size, survival of the animal, length of the experiment etc.). Estimation of hazard ratios is always possible, even in the absence of events, by means of the Firth correction.
Exporting results
Each graph can be exported as a .png or .svg file, knowing that .svg files are compatible with graphical software (such as Adobe Illustrator) that is widely used by the biomedical research community. Each of the statistical report can be exported as .doc, or .htlm or .pdf document. Such reports include plots of the transformed data, tables summarizing design and model specification, a description of the statistical methods used for intergroup comparisons, as well as diagnostic plots for statistical quality control.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Zitvogel L, Pitt JM, Daillere R, Smyth MJ, Kroemer G. Mouse models in oncoimmunology. Nat Rev Cancer. 2016;16:759–73. doi: 10.1038/nrc.2016.91. PMID:27687979. [DOI] [PubMed] [Google Scholar]
- 2.Pietrocola F, Pol J, Vacchelli E, Rao S, Enot DP, Baracco EE, Levesque S, Castoldi F, Jacquelot N, Yamazaki T, et al.. Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell. 2016;30:147–60. doi: 10.1016/j.ccell.2016.05.016. PMID:27411589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daillere R, Vetizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, et al.. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity. 2016;45:931–43. doi: 10.1016/j.immuni.2016.09.009. PMID:27717798. [DOI] [PubMed] [Google Scholar]
- 4.Baracco EE, Pietrocola F, Buque A, Bloy N, Senovilla L, Zitvogel L, Vacchelli E, Kroemer G. Inhibition of formyl peptide receptor 1 reduces the efficacy of anticancer chemotherapy against carcinogen-induced breast cancer. Oncoimmunology. 2016;5:e1139275. doi: 10.1080/2162402X.2016.1139275. PMID:27471610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang H, Ma Y, Chen G, Zhou H, Yamazaki T, Klein C, Pietrocola F,Vacchelli E, Souquere S, Sauvat A, et al.. Contribution of RIP3 and MLKL to immunogenic cell death signaling in cancer chemotherapy. Oncoimmunology. 2016;5:e1149673. doi: 10.1080/2162402X.2016.1149673. PMID:27471616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vacchelli E, Ma Y, Baracco EE, Sistigu A, Enot DP, Pietrocola F, Yang H, Adjemian S, Chaba K, Semeraro M, et al.. Chemotherapy-induced antitumor immunity requires formyl peptide receptor 1. Science. 2015;350:972–8. doi: 10.1126/science.aad0779. PMID:26516201. [DOI] [PubMed] [Google Scholar]
- 7.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al.. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–84. doi: 10.1126/science.aad1329. PMID:26541610. [DOI] [PMC free article] [PubMed] [Google Scholar]


