Abstract
The objective of this study was to develop and validate a customized cost-effective single nucleotide polymorphism (SNP) panel for genetic improvement of feed efficiency in beef cattle. The SNPs identified in previous association studies and through extensive analysis of candidate genomic regions and genes, were screened for their functional impact and allele frequency in Angus and Hereford breeds used as validation candidates for the panel. Association analyses were performed on genotypes of 159 SNPs from new samples of Angus (n = 160), Hereford (n = 329), and Angus-Hereford crossbred (n = 382) cattle using allele substitution and genotypic models in ASReml. Genomic heritabilities were estimated for feed efficiency traits using the full set of SNPs, SNPs associated with at least one of the traits (at P ≤ 0.05 and P < 0.10), as well as the Illumina bovine 50K representing a widely used commercial genotyping panel. A total of 63 SNPs within 43 genes showed association (P ≤ 0.05) with at least one trait. The minor alleles of SNPs located in the GHR and CAST genes were associated with decreasing effects on residual feed intake (RFI) and/or RFI adjusted for backfat (RFIf), whereas minor alleles of SNPs within MKI67 gene were associated with increasing effects on RFI and RFIf. Additionally, the minor allele of rs137400016 SNP within CNTFR was associated with increasing average daily gain (ADG). The SNPs genotypes within UMPS, SMARCAL, CCSER1, and LMCD1 genes showed significant over-dominance effects whereas other SNPs located in SMARCAL1, ANXA2, CACNA1G, and PHYHIPL genes showed additive effects on RFI and RFIf. Gene enrichment analysis indicated that gland development, as well as ion and cation transport are important physiological mechanisms contributing to variation in feed efficiency traits. The study revealed the effect of the Jak-STAT signaling pathway on feed efficiency through the CNTFR, OSMR, and GHR genes. Genomic heritability using the 63 significant (P ≤ 0.05) SNPs was 0.09, 0.09, 0.13, 0.05, 0.05, and 0.07 for ADG, dry matter intake, midpoint metabolic weight, RFI, RFIf, and backfat, respectively. These SNPs contributed to genetic variation in the studied traits and thus can potentially be used or tested to generate cost-effective molecular breeding values for feed efficiency in beef cattle.
Keywords: association analysis, Beef cattle, candidate genes, feed efficiency
INTRODUCTION
Selecting beef cattle for improved feed efficiency or low residual feed intake (RFI) has two direct benefits: 1) reduced feed intake without compromising growth and product quality (Mao et al., 2013), and 2) reducing the environmental footprint, particularly greenhouse gas emissions, per animal (Basarab et al., 2005; Manafiazar et al., 2016). These benefits can increase profitability for producers. Therefore, it is important to identify efficient animals and utilize them for breeding and production purposes. A main challenge facing producers is to cost-effectively measure individual feed efficiency because performance testing is expensive and takes time to accumulate sufficient feed intake records to identify efficient animals.
Utilizing genomics offers a potential alternative with several benefits including the ability to immediately predict feed efficiency at a young age. In beef cattle, various studies have been used to identify genetic markers associated with feed efficiency including genome-wide association studies (GWAS) (Sherman et al., 2008a; Sherman et al., 2009; Lu et al., 2013; Saatchi et al., 2014a) and the candidate gene approach (Sherman et al., 2008b; Abo-Ismail et al., 2013; Karisa et al., 2013; Abo-Ismail et al., 2014). These and other studies have reported a large number of single nucleotide polymorphisms (SNPs) associated with feed efficiency and its components traits. It is important to validate the associations of these SNPs, genomic regions or candidate genes in other populations before they are considered for use in breeding programs.
The objectives of this study were to 1) identify SNPs located in genes within the regions reported to be associated with feed efficiency and to select SNPs with an increased likelihood of having a functional impact on the gene product or on gene expression; 2) to validate the association of SNPs with RFI and its component traits using genetically distinct populations of beef cattle; and 3) to measure the proportion of variance explained by these SNPs in order to develop a low cost SNP panel to select for feed efficiency.
MATERIALS AND METHODS
SNP Panel Development and Design
Previous work identified significant associations among genomic regions or SNPs with feed efficiency in “discovery populations” of mainly crossbred or hybrid cattle. The breed composition of the crossbred animals reported by Abo-Ismail et al. (2014) were estimated as follows; Angus (45.9%), Simmental (20.7%), Piedmontese (5%), Gelbvieh (4.2%), Charolais (2%), and Limousin (1.4%). The crossbred and hybrid animals reported by Karisa et al. (2013) were raised at the University of Alberta Roy Berg Kinsella Research Ranch. These steers were sired by Angus, Charolais, or University of Alberta hybrid bulls. Their dams were produced from crosses among three composite cattle lines: Beef Synthetic 1 (BS1), Beef Synthetic 2 (BS2), and Dairy × Beef Synthetic (DBS). Briefly, the BS1 is composed of about 33% each of Angus and Charolais, about 20% Galloway, with the remainder comprised of other beef breeds, while the BS2 is made up of approximately 60% Hereford and 40% other beef breeds. The DBS is composed of approximately 60% dairy breeds (Holstein, Brown Swiss, or Simmental) and approximately 40% beef breeds (Berg et al., 1990; Goonewardene et al., 2003; Karisa et al., 2013).
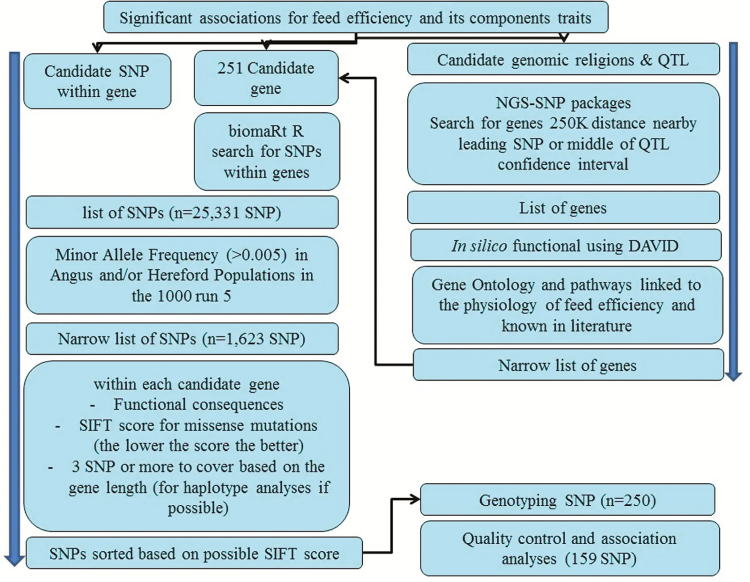
The workflow for the SNP panel development and design is illustrated in Fig. 1. Additionally, a comprehensive literature search was performed to identify additional genes or SNPs associated with feed efficiency traits and its components in previous studies (Connor et al., 2010; Rempel et al., 2012; Rolf et al., 2012; Serao et al., 2013; Serão et al., 2013; Yao et al., 2013; Zhang et al., 2013; de Oliveira et al., 2014; Saatchi et al., 2014a; Saatchi et al., 2014b; Lindholm-Perry et al., 2015). These SNPs were then combined with those identified by screening sequences generated from the 1,000 Bulls Genome Project and the Canadian Cattle Genome Project (CCGP) (Daetwyler et al., 2014; Stothard et al., 2015). These resources were mined in silico to further improve the panel by seeking candidate genes within previously reported quantitative trait loci (QTL) (Hu et al., 2013) and polymorphisms predicted to impact gene function or expression using NGS-SNP, a SNP annotation tool (Grant et al., 2011). In addition, we selected genes within each genomic region or confidence intervals for a QTL that had more than one candidate gene. In this case, these genes were filtered based on their in-silico biological background using bioinformatics tools such as DAVID (Huang et al., 2009) to refine the list of genes and to focus on those known to be involved in biological processes or pathways linked to feed efficiency. The impact of each polymorphism was assessed based on several criteria including sorting intolerant from tolerant (SIFT) scores to predict whether amino acid substitutions significantly affected protein function (Ng and Henikoff, 2003). The SNP was declared deleterious if the SIFT score was less than 0.05.
Figure 1.
The workflow for development of the marker SNP panel in the current study.
After initial filtering, we started with a set of 25,331 SNPs and focused on predicting functional variants in candidate genes that were segregating in Angus (AN) and Hereford (HH) cattle. Selected SNPs within genes known to be biologically linked to growth as well as lipid and energy metabolism were identified for consideration in the candidate SNP list. Allele frequency was also used to help select SNPs with minor allele frequency more than 0.005 based on 136 Angus and 31 Hereford individuals from the data from the Canadian bulls (CCGP) in the 1,000 Bulls genome Project (Daetwyler et al., 2014). By using minor allele frequency information from the previous step, the chance of detecting segregating SNP in the genotyping step is increased. This would reduce the cost of genotyping nonsegregating selected SNPs. The final selected list contained 250 SNPs used to optimize the multiplexes developed for this study. The number of multiplexes is a major factor in determining the final assay cost.
Blood samples were collected by jugular venipuncture into evacuated tubes containing ethylenediaminetetraacetate (Vacutainer, Becton Dickinson and Co., Franklin Lakes, NJ, USA) and stored at −80 °C until DNA extraction using the QiagenDNeasy 96 blood and tissue kit (Qiagen Sciences, Germantown, MD, USA) and was completed at Delta Genomics (Edmonton, Alberta, Canada). The resulting samples were then used to develop multiplex sets for the Sequenom Mass-Array platform (San Diego, CA, United States). The aim was to optimize the number of assays required to generate the maximum number of genotyped SNPs. The final panel design achieved by Sequenom resulted in assays for 216 SNPs. The panel was divided into five polymerase chain reaction-based assays or multiplexes in order to generate genotypes for the 216 SNPs by Delta Genomics.
Animals and Phenotypic Data
All animals in the current study were cared for according to the guidelines of the Canadian Council on Animal Care (1993). The protocols were approved by the University of Alberta Animal Use Committee. A set of animals born between 2002 and 2012 with accurate phenotypes for feed efficiency were identified (Crowley et al., 2014) from the Phenomic Gap project (PG1) which was initiated in 2008 primarily to generate phenotypic and genotypic information needed to discover and validate genome-wide selection methods and to help address the lack of data for difficult to measure traits in the Canadian beef cattle industry. Within the PG1 database, we selected 874 animals consisting of AN, HH, and their crossbreds (ANHH) which represent a population that was reasonably genetically distinct from the research populations used for the initial SNP association studies, as previously described (Karisa et al., 2013; Abo-Ismail et al., 2014). Additionally, inclusion of crossbreds was considered as a representative of the commercial beef industry, and to make the developed panel relevant for predicting feed efficiency for both purebred and commercial cattle. This assumption makes the selected population ideal for testing our hypothesis.
The validation population consisted of bulls (HH, n = 284), replacement heifers (HH, n = 25; AN, n = 88; ANHH, n = 185), and finishing heifers (AN, n = 1; ANHH, n = 14), and steers (HH, n = 23; AN, n = 71; ANHH, n = 183). The replacement heifers (n = 298) were born from 2004 to 2012 and tested from 2005 to 2013 whereas the finished heifers were born in 2002, 2003 and 2011 and tested in 2003, 2004 and 2012 at the Lacombe Research Center (LRC). A detailed description of the breeding and management for the replacement and finishing heifers were described in previous studies (Basarab et al., 2011; Manafiazar et al., 2015). The steers were born in 2002 to 2010 at LRC. Additional information on the breeding and management of the steers was reported by Basarab et al. (2007) and Basarab et al. (2012). Briefly, heifers and steer calves were placed into separate feedlot pens each fitted with eight GrowSafe (GrowSafe Systems Ltd., Airdrie, Alberta, Canada) feeding stations for the automatic monitoring of individual animal feed intake. The steers’ finishing diet consisted of an average of 1% alfalfa silage, 22% barley silage, 70% barley grain, and 7% supplement (as dry matter basis; Supplementary File 1) and was fed ad libitum. The HH bulls were born in 2012 and tested at Olds College (n = 165) and Cattleland Feedyards (n = 119) in 2012 and 2013. The bulls’ test diet consisted of ~50% roughage (silage and hay), and ~50% concentrate and vitamin/mineral supplement (as dry matter basis; Supplementary File 1). The feed intake testing protocol for the HH bulls was the same as in heifers and steer tests. Daily dry matter intake (DMI) was observed on all animal as well as frequent body weight measurements and ultrasound measurements at the start and end of test. The duration of the feed intake test ranged from 60 to 120 days. From the performance test data, animals were tested for the following phenotypes: average daily gain (ADG), average daily DMI, midpoint metabolic weight (i.e., the body weight at the middle of the performance testing period powered 0.75, MMWT), off test back fat (BFat), RFI, and RFI adjusted for back fat (RFIf) (Table 1). RFI estimated as the difference between observed DMI and expected DMI was modeled on ADG, MMWT, and BFat measured at end of test. In addition, RFI was adjusted for the effect of gender, herd of origin, birth year, diet, and management group. The ADG was calculated as the regression coefficients (i.e., the slope) of animal’s body weights over the performance testing period. The ADG with accuracy (R2) less than 90% were excluded. Animals with less than 3 recorded weights were excluded so each animal had from 3 to 10 weights over the performance testing period.
Table 1.
The descriptive statistics for feed efficiency traits and its components of Herford, Angus and their crossbred beef cattle
| Trait | N 1 | Mean | SD | Minimum | Maximum |
|---|---|---|---|---|---|
| Average daily gain, kg/d2 | 870 | 1.28 | 0.364 | 0.452 | 2.42 |
| Average daily dry matter intake, kg3 | 831 | 8.577 | 1.259 | 4.97 | 12.43 |
| Midpoint metabolic weight, kg4 | 822 | 87.77 | 9.037 | 64.72 | 114.81 |
| Residual feed intake, kg/d5 | 855 | 0.038 | 0.450 | -1.3 | 1.35 |
| Residual feed intake adjusted for fatness, kg/d6 | 852 | 0.013 | 0.437 | -1.25 | 1.26 |
| Back fat thickness, mm7 | 865 | 6.833 | 2.590 | 1 | 14.81 |
1 N = total number of animals used in the association analyses.
2ADG = average daily gain: recorded in kg per day from start to end of the finishing period.
3DMI = dry matter intake: recorded in kg per day from start to end of the finishing period.
4MMWT =midpoint metabolic weight: expressed in kg.
5RFI = residual feed intake: expressed in kg per day.
6RFIf = residual feed intake adjusted for backfat: expressed in kg per day;
BFat7= backfat: recorded as fat depth at the end of the finishing period in millimeters.
Genomic-based Breed Composition and Retained Heterozygosity
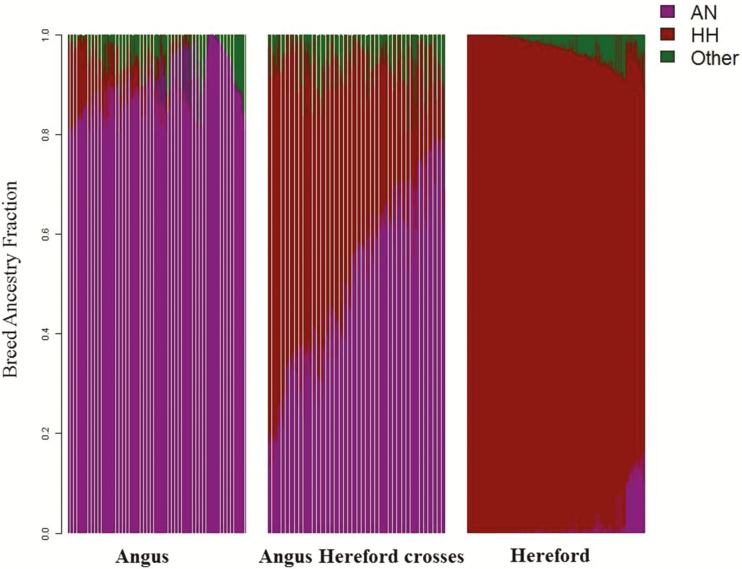
Breed composition was predicted using 43,172 SNPs distributed across the 29 autosomes from the Illumina Bovine 50K SNPs with ADMIXTURE software (Alexander et al., 2009) to account for stratification due to breed effects in the association analyses. A larger dataset (n = 7,845) of purebred animals of different breeds was used as a reference population. Predicted breed composition using the genotypes for all individuals in the study is illustrated in Fig. 2. Additionally, the heterosis effect was accounted for in the association analyses, by calculating the retained heterozygosity (RH) for each individual according to (Dickerson, 1973) as follows:
Figure 2.
The genomic based breed composition of all individuals in the study using BovineSNP50 chip. The breed composition of each individual of 871 animals was calculated using ancestry components in Admixture software on Illumina Bovine 50K SNPs genotypes (43,172 SNPs) across the 29 autosomes. The ancestor HH is Hereford, AN is Angus, and other is the sum of other fractions of other breeds and may be due to errors. Animals were grouped based on the breed fractions and considered purebred AN or HH if their breed fraction is ≥ 80%. An animal was considered ANHH crossbred if its breed fractions ≤ 80% of any of the AN or HH breeds. Each individual within a particular population is represented by a vertical bar which presents the ancestral populations.
| (1) |
where P is the predicted fraction of breed i from each of the n breeds. The predicted breed fractions using the genotypes and RH values were used as covariates in the following association analyses.
Association Analyses
Three models were used to evaluate SNP associations, including allele substitution effects, genotypic and additive/dominance models. Allele substitution effect is defined as the average change in phenotype value when the minor allele is substituted with the major allele. This effects included additive (a) and dominance (d) as in the following equation: 2pq[a + d(q-p)]2 where p and q are the alleles frequencies. The additive and dominance effect model adjusts the allele substitition effect with the dominance effect providing both effects and tests the significance of the a and d effects (Zeng et al., 2005). In addition, we used the genotypic model as provided by Fernando et al. (1998) which allows the mixed-model equations to be used to estimate the genotypes as fixed effects in order to identify the favorable genotype.
Allele substitution effect model.
In order to estimate allele substitution effects for each SNP, genotypes were coded as 0, 1, or 2 corresponding to the number of minor alleles present using PLINK (Purcell et al., 2007). A univariate mixed model was fitted where phenotypes were regressed on the number of copies of the minor allele (0, 1, or 2) using ASReml 4 software (Gilmour et al., 2009). The mixed model was applied as follows:
| (2) |
where Yijk is the trait measured in the kth animal of the jth contemporary group; µ is the overall mean for the trait; SNPi is the fixed effect of the ith genotype for the SNP considered; CGj is the fixed effect of the jth gender, herd of origin, birth year, diet and management group; β1 is the partial regression coefficient for age at the end of the test period (AET) of the kth animal; β2 is the partial regression coefficient for test duration length (TL) of the kth animal; β3 and β4 are the partial regression coefficients for the genomic-based breed proportion of AN and HH breeds in the kth animal; β5 is the regression coefficient of the linear regression on the percent of genomic-based retained heterozygosity of the kth animal; ak is the random additive genetic (polygenic) effect of the kth animal; and ejklm is the residual random effect associated with the kth animal record. Assumptions for this model are; ak: a ~ N (0, A σ2a) where A is a numerator relationship matrix, and σ2a is the additive genetic variance; and eijk: e ~ (0, I σ2e) where I is the identity matrix and σ2e is the error variance. The expectations are that E(ak) = 0; and E(eijk) = 0; and the variances are Var(ak) = σ2a; Var(eijk) = σ2e. Aσ2a is the covariance matrix of the vector of animal additive genetic effects and the relationship matrix (A). The number of individuals within each CG ranged from 4 to 74 animals. Any contemporary group level that had less than three animals was excluded from the analyses. Phenotypic outliers were identified by the Median Absolute Deviation method using R (Team, 2016) and excluded from the analysis.
Genotypic model.
This model included the same effects as those in the allele substitution effect model, except that the allele substitution effect was replaced with the genotypes as a class variable (e.g., AA and BB for homozygous genotypes and AB for the heterozygous genotype). The least square means for each genotypic class was estimated.
Additive and dominance effect model.
The additive and dominance effects of a SNP were estimated by fitting the substitution effect model in 2 above and adding a covariate to the model with zeros for homozygous genotypes (coded 0 and 2) and ones for heterozygous genotypes (coded as 1) (Zeng et al., 2005). Thus, the linear regression coefficient of the substitution effect is the additive effect and the linear regression coefficient of the added covariate is the dominance effect for the SNP. For a SNP to be associated with a particular trait, the significance threshold of SNP association was 5% absolute P-value. Setting a relaxed significance threshold (P < 0.05) allows the identification of markers with small effects on feed efficiency and its components traits to subsequently explain more genetic variance in these traits.
Gene Ontology and Pathway Enrichment Analyses
Enrichment analyses were performed to assign the associated candidate genes, i.e., those having at least one significant (P ≤ 0.05) SNP, to predefined gene ontology (GO) terms and pathways based on their functional characteristics using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (Huang et al., 2009). The absolute P-value < 0.05 was used to report the enriched GO terms and pathways. This relaxed threshold produces false positive results but may help in understanding the biological information about the candidate genes. To account for multihypotheses testing, the P-value of the enrichment analysis was adjusted using false discovery rate.
Heritability and Genetic Variance Explained by SNP Sets
Heritability was estimated using pedigree information and genomic-based methods. As the pedigree was available for all animals, the numerator relationship (A) matrix was constructed. The estimated breeding values (EBVs) for individuals and heritability of each trait were estimated using the univariate animal model using A matrix as the kernel in ASReml 4 software (Gilmour et al., 2009). To calculate the genomic based heritability, the genomic additive relationship matrix (G) was constructed following the formulas set out in (VanRaden, 2008) implemented in GVCBLUP software (Da et al., 2014). The genomic based heritability was calculated using the GREML method implemented in GVCBLUP software (Da et al., 2014) using different scenarios in terms of the number of SNPs; 1) all SNPs genotyped that passed quality control criteria in the small custom panel (n = 158 SNPs), 2) a set of associated (P < 0.05) SNPs with at least one of the feed efficiency traits (n = 63 SNPs within 43 genes), 3) a set of associated (P < 0.1) SNPs with at least one of the feed efficiency traits (n = 92 SNPs), and 4) a set of SNPs (n = 40465 SNPs) of the Illumina BovineSNP50 (50K SNP) panel that passed quality control criteria. The proportion of genetic variance explained by the full list of SNPs was calculated by dividing the heritability calculated from GVCBLUP by the heritability estimated from the animal model.
RESULTS AND DISCUSSION
Quality Control for the Developed Panel
A total of 874 animals were successfully genotyped for 216 SNPs. SNPs with call rates less than 70% (n = 11), minor allele frequencies less than 1% (n = 48), and excess of heterozygosity above 15%, were excluded from the analyses. Additionally, 20 animals with call rates less than 80% were excluded. Out of the initial 216 SNPs, 159 SNPs for 871 animals from AN (n = 160), HH (n = 329), and ANHH crossbred (n = 382) cattle were considered for association analyses (Supplementary File 2).
Association Analyses Using Allele Substitution Effect Model
A total of 54 markers within 34 genes were significantly (P ≤ 0.05) associated with at least one phenotypic trait using an allele substitution effect regression model (Table 2). Furthermore, significant effects were identified for both feed efficiency traits (i.e., RFI and/or RFIf) for 15 SNPs in 10 of the genes. The minor allele of SNPs within 8 of these genes (polycystin-2 (PKD2), calpastatin (CAST), calcium voltage-gated channel subunit alpha1 G (CACNA1G), occludin (OCLN), growth hormone receptor (GHR), proprotein convertase subtilisin/kexin type 6 (PCSK6), PAK1 interacting protein 1 (PAK1IP1), and phytanoyl-CoA 2-hydroxylase interacting protein like (PHYHIPL)) was associated with a negative, favorable, effect on RFI and/or RFIf (Table 2).
Table 2.
The P-values and effect estimate (SE) for the markers associated (P ≤ 0.05) with feed efficiency traits using allele substitution effect model
| Gene Name | rs#1 | MAF2 | ADG3 | DMI4 | MMWT5 | RFI6 | RFIf7 | BFat8 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | Estimate ± SE | P-value | Estimate ± SE | P-value | Estimate ± SE | P-value | Estimate ± SE | P-value | Estimate ± SE | P-value | Estimate ± SE | |||
| SMARCAL1 | rs109065702 | 0.368 | 0.038 | -0.048 ± 0.023 | ||||||||||
| SMARCAL1 | rs109808135 | 0.367 | 0.043 | -0.047 ± 0.023 | ||||||||||
| SMARCAL1 | rs110348122 | 0.367 | 0.038 | -0.048 ± 0.023 | ||||||||||
| SMARCAL1 | rs109382589 | 0.312 | 0.030 | 0.051 ± 0.024 | 0.036 | 0.048 ± 0.023 | ||||||||
| SMARCAL1 | rs208660945 | 0.417 | 0.009 | -0.024 ± 0.009 | 0.041 | -0.045 ± 0.022 | 0.05 | -0.042 ± 0.022 | ||||||
| LRRIQ3 | rs42417924 | 0.128 | 0.014 | -1.124 ± 0.45 | ||||||||||
| MGAM | rs110632853 | 0.076 | 0.032 | 0.17 ± 0.079 | ||||||||||
| DPP6 | rs110519795 | 0.388 | 0.030 | -0.651 ± 0.3 | ||||||||||
| DPP6 | rs132717265 | 0.468 | 0.002 | -0.267 ± 0.09 | ||||||||||
| CHADL | rs109499238 | 0.445 | 0.032 | -0.644 ± 0.3 | ||||||||||
| PPM1K | rs134225543 | 0.086 | 0.019 | 0.038 ± 0.016 | 0.006 | 1.402 ± 0.512 | 0.005 | -0.394 ± 0.14 | ||||||
| ABCG2 | rs110362902 | 0.102 | 0.016 | 1.224 ± 0.51 | ||||||||||
| PKD2 | rs29010894 | 0.207 | 0.044 | -0.055 ± 0.027 | ||||||||||
| PKD2 | rs43702346 | 0.114 | 0.022 | 1.085 ± 0.47 | 0.041 | 0.072 ± 0.035 | 0.024 | 0.078 ± 0.034 | 0.025 | -0.29 ± 0.13 | ||||
| EVC2 | rs207525537 | 0.174 | 0.041 | -0.026 ± 0.013 | ||||||||||
| CAST | rs137601357 | 0.376 | 0.048 | -0.047 ± 0.024 | 0.046 | -0.046 ± 0.023 | ||||||||
| CAST | rs210072660 | 0.379 | 0.019 | -0.052 ± 0.022 | 0.022 | -0.05 ± 0.022 | ||||||||
| CAST | rs384020496 | 0.096 | 0.021 | 0.032 ± 0.014 | 0.032 | 0.925 ± 0.43 | 0.009 | 0.317 ± 0.12 | ||||||
| CAST | rs133057384 | 0.107 | 0.044 | -0.072 ± 0.036 | 0.05 | -0.067 ± 0.035 | 0.005 | 0.366 ± 0.13 | ||||||
| CAST | rs110711318 | 0.087 | 0.031 | 1.087 ± 0.503 | 0.004 | 0.401 ± 0.14 | ||||||||
| CNTFR | rs137400016 | 0.417 | 0.015 | 0.023 ± 0.01 | ||||||||||
| ANXA2 | rs471723345 | 0.084 | 0.049 | 0.079 ± 0.04 | ||||||||||
| CNGA3 | rs43657898 | 0.405 | 0.039 | 0.02 ± 0.01 | ||||||||||
| AFF3 | rs42275280 | 0.080 | 0.007 | -1.105 ± 0.41 | 0.025 | -0.248 ± 0.11 | ||||||||
| ATP6V1E2 | rs43673198 | 0.278 | 0.002 | -0.034 ± 0.011 | 0.023 | -0.111 ± 0.049 | ||||||||
| MAPK15 | rs110323635 | 0.373 | 0.016 | 0.746 ± 0.309 | 0.020 | 0.2 ± 0.086 | ||||||||
| FAM135B | rs109575847 | 0.169 | 0.021 | -0.141 ± 0.061 | 0.037 | -0.902 ± 0.43 | ||||||||
| TG | rs133269500 | 0.133 | 0.006 | -0.038 ± 0.014 | 0.051 | -0.836 ± 0.43 | ||||||||
| TG | rs110547220 | 0.291 | 0.002 | -0.033 ± 0.011 | ||||||||||
| RB1CC1 | rs109800133 | 0.133 | 0.019 | -0.303 ± 0.129 | ||||||||||
| CNTN5 | rs42544329 | 0.431 | 0.022 | 0.052 ± 0.022 | ||||||||||
| ELMOD1 | rs42235500 | 0.164 | 0.018 | -0.189 ± 0.08 | ||||||||||
| HMCN1 | rs211555481 | 0.476 | 0.049 | 0.163 ± 0.083 | ||||||||||
| HMCN1 | rs381726438 | 0.295 | 0.033 | 0.024 ± 0.011 | ||||||||||
| HMCN1 | rs209012152 | 0.348 | 0.011 | 0.027 ± 0.01 | ||||||||||
| HMCN1 | rs41821600 | 0.049 | 0.021 | -0.117 ± 0.051 | 0.024 | -0.111 ± 0.049 | ||||||||
| HMCN1 | rs210494625 | 0.296 | 0.026 | 0.025 ± 0.011 | ||||||||||
| HMCN1 | rs209439233 | 0.305 | 0.033 | 0.023 ± 0.011 | ||||||||||
| CACNA1G | rs476872493 | 0.154 | 0.0001 | -0.124 ± 0.032 | 0.0004 | -0.109 ± 0.031 | ||||||||
| OCLN | rs134264563 | 0.133 | 0.05 | -0.066 ± 0.033 | 0.024 | -0.074 ± 0.033 | ||||||||
| IPO11 | rs207541156 | 0.018 | 0.041 | 0.177 ± 0.087 | ||||||||||
| GHR | rs385640152 | 0.157 | 0.05 | -0.06 ± 0.031 | 0.021 | -0.07 ± 0.03 | ||||||||
| OSMR | rs41947101 | 0.417 | 0.05 | 0.043 ± 0.022 | ||||||||||
| SLC45A2 | rs134604394 | 0.414 | 0.015 | 0.026 ± 0.011 | ||||||||||
| SLC45A2 | rs41946086 | 0.484 | 0.001 | -0.04 ± 0.012 | 0.016 | -0.13 ± 0.054 | 0.027 | -0.844 ± 0.38 | ||||||
| PCSK6 | rs43020736 | 0.451 | 0.016 | -0.109 ± 0.045 | 0.011 | -0.812 ± 0.32 | 0.048 | -0.046 ± 0.023 | 0.047 | -0.045 ± 0.023 | ||||
| TMEM40 | rs133838809 | 0.216 | 0.022 | 0.862 ± 0.375 | ||||||||||
| TMEM40 | rs132658346 | 0.216 | 0.022 | 0.862 ± 0.375 | ||||||||||
| PAK1IP1 | rs42342962 | 0.340 | 0.034 | -0.048 ± 0.023 | ||||||||||
| MKI67 | rs110216983 | 0.381 | 0.021 | 0.104 ± 0.045 | 0.030 | 0.693 ± 0.319 | 0.014 | 0.059 ± 0.024 | 0.025 | 0.052 ± 0.023 | ||||
| MKI67 | rs109930382 | 0.328 | 0.007 | 0.123 ± 0.045 | 0.024 | 0.727 ± 0.322 | 0.009 | 0.064 ± 0.025 | 0.016 | 0.058 ± 0.024 | ||||
| MKI67 | rs109558734 | 0.332 | 0.006 | 0.124 ± 0.045 | 0.020 | 0.748 ± 0.32 | 0.007 | 0.066 ± 0.024 | 0.013 | 0.059 ± 0.024 | 0.039 | 0.184 ± 0.089 | ||
| C27H8orf40 | rs135814528 | 0.054 | 0.002 | 2.015 ± 0.654 | ||||||||||
| PHYHIPL | rs209765899 | 0.312 | 0.003 | -0.076 ± 0.025 | 0.003 | -0.074 ± 0.024 | ||||||||
1rs# = a reference SNP ID number assigned by National Center for Biotechnology Information (NCBI).
2MAF = Minor allele frequency.
3ADG = average daily gain: recorded in kg per day from start to end of the finishing period.
DMI4 = dry matter intake: recorded in kg per day from start to end of the finishing period.
MMWT5 = midpoint metabolic weight.
6RFI = residual feed intake expressed kg per day.
7RFIf = residual feed intake adjusted for backfat
8BFat = backfat: recorded as fat depth at the end of the finishing period in millimeters.
The minor alleles of three SNPs, rs137601357, rs210072660 and rs133057384, located in CAST were associated with decreases in RFI and RFIf (favorable effect). In addition, SNP rs384020496 in CAST was associated with MMWT, ADG, and Bfat, whereas SNP rs110711318 was associated with an increase in MMWT and Bfat (Table 2). The association of SNP rs384020496 with RFI was reported previously (Karisa et al., 2013). SNP rs137601357 is located 12 bases from SNP rs109727850 which had an additive effect on RFI (Karisa et al., 2013). Thus, the current results provide evidence that polymorphisms within CAST have important potential effects on feed efficiency and its component traits. The CAST gene is known to be associated with inhibition of the normal post-mortem tenderization of meat (Schenkel et al., 2006; Li et al., 2010). Additionally, the CAST gene can also play an important role in the metabolism of the live animal. For example, a previous study reported that during nutrient intake restriction, activity of the calpain system is upregulated by decreasing the expression level of CAST gene in bovine skeletal muscles, whereas the activity of the calpain system in a fetus is downregulated through an increase in CAST expression maintaining fetal muscle growth during starvation (Du et al., 2004). Nonetheless, when selecting for favorable alleles for tenderness, this would be associated with higher protein metabolism (i.e., turnover) without negative effects on growth, efficiency, temperament, or carcass characteristics (Cafe et al., 2010).
The current study confirmed OCLN to be associated with RFI and RFIf. The SNP rs134264563 within OCLN was associated with RFI and RFIf. Another SNP rs109638814 within the OCLN gene was previously reported to be associated with RFI (Karisa et al., 2013), however, this was not the case in the current study. A previous study suggested an association between SNP rs134264563 and both cow as well as daughter conception rates in dairy cattle (Ortega et al., 2016). The SNP rs134264563 was reported to segregate in nine beef cattle breeds (Ortega et al., 2017). Although the SIFT values were 0.2 and 1 for rs134264563 and rs109638814, respectively, suggesting they are tolerated missense mutations, this may be in agreement with the hypothesis that when using causal or functional variants based selection (i.e., rs134264563), the SNP effect would be repeatable across different populations and breeds; this is in contrast to LD markers (i.e., rs109638814).
Our results indicated that a synonymous SNP rs110362902 within ABCG2 was associated with an increase in MMWT. The allele G of rs110362902 SNP was reported to be associated with increasing MMWT and decreasing intermuscular fat and marbling in beef cattle (Abo-Ismail et al., 2014). Additionally, SNP rs43702346 on BTA 6, within PKD2, was significantly associated with RFI, RFIf, and MMWT, whereas substitution with the minor allele was associated with an increase of RFI, RFIf and MMWT, as well as a decrease in Bfat. These findings agreed with previous results reported for rs43702346 (Abo-Ismail et al., 2014) where substitution with the minor allele was associated with an increase in MMWT and a decrease in intermuscular fat percentage. The PKD2 gene is involved in negative regulation of G1/S transition of mitotic cell cycle process. Gene PKD2 is located near an identified QTL for bone percentage, fat percentage, meat percentage, meat to bone ratio, moisture content, and subcutaneous fat (Gutiérrez-Gil et al., 2009). A SNP near to PKD2 (1063 Kbp) was associated with body weight in Australian Merino sheep (Al-Mamun et al., 2015). The results suggest that the SNP may be in linkage disequilibrium with a causative mutation associated with these traits.
Our findings indicated that the minor allele of tolerated missense mutations rs109065702, rs109808135, rs110348122, and rs208660945 within the SWI/SNF (SWItch/Sucrose Non Fermentable)-related matrix associated actin-dependent regulator (SMARCAL1) gene were significantly associated with a decrease in RFIf, whereas the minor allele of rs109382589, and having a deleterious mutation SIFT score = 0.02, was associated with an increase of RFI and RFIf. This study confirmed the significant association between rs109065702 (missense mutation) and RFI reported by Karisa et al. (2013). Furthermore, the minor allele of rs208660945 within SMARCAL1 was associated with a decrease in RFI and RFIf (favorable effect) and a decrease in ADG (unfavorable effect). The SMARCAL1 gene is involved in a network interacting with the Ubiquitin C (UBC) gene, which in turn, is involved in regulation of gene expression through DNA transcription, protein stability and degradation (Karisa et al., 2014).
The current results also revealed that the minor allele of the deleterious SNP rs476872493, within CACNA1G on BTA 19, was associated with decreasing RFI and RFIf. SNP rs476872493 is located close to (23,710 bases) a synonymous SNP, rs41914675, which was reported to be associated with RFI, DMI and FCR (Abo-Ismail et al., 2014). These results lend support to the relationship between CACNA1G and feed efficiency traits. Feed efficiency was also associated with a deleterious SNP (rs385640152), within the GHR, gene where the minor allele was associated with favorable effects by decreasing RFI and RFIf (Table 2). SNP rs385640152 is located close to (18,371 bases) to SNP rs209676814, which was previously reported to have an over-dominant effect on RFI (Karisa et al., 2013). Another SNP in the 4th intronic region was associated with RFI (Sherman et al., 2008b). The minor allele of the deleterious SNP rs43020736, within PCSK6, was associated with decreasing DMI, MMWT, RFI, and RFIf (Table 2). This SNP was previously reported to affect DMI and RFI where animals with the C allele have lower DMI and RFI (Abo-Ismail et al., 2014). The current result is in agreement with the physiological role of PCSK6 as it is involved in apoptosis and other physiological processes (Wang et al., 2014). The results indicated that the marker of the proliferation Ki-67 (MKI67) gene harbours three SNPs (rs110216983, rs109930382 and rs109558734), which were associated with MMWT, DMI, RFI, and RFIf (Table 2). The minor allele of these SNPs was associated with increasing MMWT, DMI, RFI and RFIf. Other studies suggested that polymorphisms within MKI67 were associated with meat tenderness and meat quality traits in Blonde d’Aquitaine cattle (Ramayo-Caldas et al., 2016). The findings in the current study are consistent with previous reports that RFI is phenotypically independent of growth, and body size, as well as fatness if phenotypes were adjusted for fat, but this is not the case from a genetic standpoint (Crowley et al., 2010; Ceacero et al., 2016). Pleiotropic loci affecting MMWT and RFI have previously been reported in other studies of beef cattle populations (Saatchi et al., 2014a).
In total, minor alleles of 5 SNPs were associated with a decrease in DMI, while minor alleles of four SNPs were associated with an increase in DMI (Table 2). A positive effect (i.e., decreased feed intake) of the minor allele provides the greatest opportunity for improvement. However, the value depends on the actual frequency in the population of interest and markers with a frequency less than 0.8 associated with reduced intake are still expected to be useful for improvement, especially when combined into a molecular breeding value (Uemoto et al., 2015).
Genotypic and Additive and Dominance Models
The genotypes of 46 SNPs within 32 genes were associated (P ≤ 0.05) with at least one feed efficiency trait or its component traits based on the genotypic model. Of these SNPs, 18 were associated with RFI and/or RFIf (Table 3). Four SNPs located in UMPS (rs110953962), SMARCAL1 (rs208660945), CCSER1 (rs41574929), and LMCD1 (rs208239648) genes showed significant overdominance effects on RFI and RFIf (Table 3). Other SNPs located in SMARCAL1 (rs109382589), ANXA2 (rs471723345), CACNA1G (rs476872493), and PHYHIPL (rs209765899) showed significant additive effects on RFI and RFIf (Table 3).
Table 3.
The least squares means (SE) and P-values for the markers associated (P ≤ 0.05) with residual feed intake using genotypic effect and additive and dominance models
| Gene Name | rs#1 | Genotype | Residual Feed Intake2 | Adjusted Back Fat Residual Feed Intake | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P-value | LSM ± SE | a3 ± SE | d4 ± SE | P Value | LSM ± SE | a ± SE | d ± SE | |||
| UMPS | rs110953962 | CC | 0.031 | 0.0164 ± 0.0281 | -0.0274 ± 0.028 | 0.0931 ± 0.035** | 0.035 | -0.0026 ± 0.028 | -0.0366 ± 0.028 | 0.0894 ± 0.034** |
| CT | 0.0821 ± 0.0262 | 0.0501 ± 0.026 | ||||||||
| TT | -0.0384 ± 0.0514 | -0.0759 ± 0.050 | ||||||||
| SMARCAL1 | rs109382589 | GG | 0.027 | 0.1569 ± 0.0477 | 0.0688 ± 0.026** | -0.0545 ± 0.035 | 0.018 | 0.1333 ± 0.046 | 0.0684 ± 0.025** | -0.0638 ± 0.034 |
| GT | 0.0337 ± 0.0277 | 0.0011 ± 0.027 | ||||||||
| TT | 0.0194 ± 0.0258 | -0.0035 ± 0.025 | ||||||||
| SMARCAL1 | rs208660945 | CC | 0.015 | 0.0274 ± 0.037 | -0.0369 ± 0.022 | -0.0648 ± 0.031* | 0.009 | 0.009 ± 0.036 | -0.0325 ± 0.022 | -0.0723 ± 0.030* |
| CT | -0.0005 ± 0.0264 | -0.0308 ± 0.026 | ||||||||
| TT | 0.1011 ± 0.0294 | 0.074 ± 0.029 | ||||||||
| CCSER1 | rs41574929 | GG | 0.003 | 0.0232 ± 0.021 | -0.0639 ± 0.057 | 0.1957 ± 0.067** | 0.005 | -0.0007 ± 0.021 | -0.0708 ± 0.055 | 0.1884 ± 0.065** |
| GT | 0.1551 ± 0.0411 | 0.1168 ± 0.040 | ||||||||
| TT | -0.1046 ± 0.1129 | -0.1424 ± 0.11 | ||||||||
| PKD2 | rs29010894 | CC | 0.0649 ± 0.0235 | -0.0154 ± 0.042 | -0.0528 ± 0.049 | 0.041 | 0.0422 ± 0.023 | -0.0099 ± 0.040 | -0.0717 ± 0.047 | |
| TC | -0.0032 ± 0.03 | -0.0393 ± 0.029 | ||||||||
| TT | 0.0342 ± 0.0813 | 0.0225 ± 0.079 | ||||||||
| PKD2 | rs43702346 | GG | 0.025 | 0.0202 ± 0.022 | -0.0171 ± 0.061 | 0.1250 ± 0.069 | 0.015 | -0.0079 ± 0.022 | -0.0102 ± 0.059 | 0.1232 ± 0.067 |
| GT | 0.1281 ± 0.0379 | 0.1051 ± 0.037 | ||||||||
| TT | -0.014 ± 0.1203 | -0.0283 ± 0.117 | ||||||||
| CAST | rs210072660 | AA | 0.046 | 0.0888 ± 0.0277 | -0.0470 ± 0.023* | -0.0264 ± 0.032 | 0.042 | 0.0617 ± 0.027 | -0.0437 ± 0.023 | -0.0317 ± 0.031 |
| AG | 0.0153 ± 0.0266 | -0.0137 ± 0.026 | ||||||||
| GG | -0.0053 ± 0.0404 | -0.0257 ± 0.039 | ||||||||
| ANXA2 | rs471723345 | AA | 0.3293 ± 0.1361 | 0.1491 ± 0.068* | -0.1008 ± 0.080 | 0.048 | 0.3314 ± 0.132 | 0.1622 ± 0.066* | -0.1384 ± 0.077 | |
| AG | 0.0794 ± 0.045 | 0.0308 ± 0.044 | ||||||||
| GG | 0.0311 ± 0.021 | 0.0069 ± 0.021 | ||||||||
| CNTN5 | rs42544329 | GG | 0.024 | -0.0197 ± 0.030 | 0.0464 ± 0.023* | 0.0459 ± 0.031 | -0.0333 ± 0.030 | 0.0372 ± 0.022 | 0.0343 ± 0.030 | |
| GT | 0.0726 ± 0.026 | 0.0382 ± 0.026 | ||||||||
| TT | 0.0732 ± 0.038 | 0.0411 ± 0.037 | ||||||||
| CACNA1G | rs476872493 | AA | 0.0004 | -0.2255 ± 0.094 | -0.1499 ± 0.048** | 0.0407 ± 0.055 | 0.001 | -0.24 ± 0.0908 | -0.1413 ± 0.046** | 0.0505 ± 0.054 |
| GA | -0.0349 ± 0.034 | -0.0481 ± 0.034 | ||||||||
| GG | 0.0742 ± 0.022 | 0.0426 ± 0.022 | ||||||||
| IPO11 | rs207541156 | CA | 0.041 | 0.184 ± 0.086 | ||||||
| CC | 0.0068 ± 0.020 | |||||||||
| GHR | rs385640152 | AA | 0.021 | 0.0425 ± 0.023 | -0.0148 ± 0.047 | -0.0825 ± 0.054 | ||||
| TA | -0.0548 ± 0.032 | |||||||||
| TT | 0.0129 ± 0.092 | |||||||||
| PCSK6 | rs43020736 | CC | 0.039 | 0.0667 ± 0.033 | -0.0500 ± 0.023* | 0.0503 ± 0.031 | 0.0507 ± 0.032 | -0.0467 ± 0.023* | 0.0217 ± 0.030 | |
| TC | 0.067 ± 0.0259 | 0.0257 ± 0.025 | ||||||||
| TT | -0.0333 ± 0.036 | -0.0427 ± 0.035 | ||||||||
| LMCD1 | rs208239648 | CC | 0.050 | 0.0382 ± 0.021 | -0.4254 ± 0.223 | 0.5324 ± 0.233* | 0.014 ± 0.021 | -0.4114 ± 0.217 | 0.4691 ± 0.226* | |
| TC | 0.1452 ± 0.071 | 0.0717 ± 0.069 | ||||||||
| TT | -0.8127 ± 0.446 | -0.8088 ± 0.433 | ||||||||
| MKI67 | rs110216983 | AA | 0.011 | 0.0136 ± 0.028 | 0.0720 ± 0.025** | -0.0559 ± 0.033 | 0.038 | -0.0125 ± 0.028 | 0.0613 ± 0.024* | -0.0400 ± 0.032 |
| GA | 0.0297 ± 0.026 | 0.0088 ± 0.026 | ||||||||
| GG | 0.1576 ± 0.044 | 0.11 ± 0.043 | ||||||||
| MKI67 | rs109930382 | CC | 0.025 | 0.0073 ± 0.026 | 0.0738 ± 0.028** | -0.0276 ± 0.035 | 0.043 | -0.0158 ± 0.026 | 0.0664 ± 0.027* | -0.0245 ± 0.034 |
| CT | 0.0535 ± 0.027 | 0.0261 ± 0.026 | ||||||||
| TT | 0.1549 ± 0.05 | 0.117 ± 0.05 | ||||||||
| MKI67 | rs109558734 | CC | 0.019 | 0.0055 ± 0.026 | 0.0760 ± 0.027** | -0.0291 ± 0.035 | 0.036 | -0.0172 ± 0.026 | 0.0676 ± 0.026* | -0.0257 ± 0.034 |
| GC | 0.0524 ± 0.026 | 0.0247 ± 0.026 | ||||||||
| GG | 0.1575 ± 0.050 | 0.118 ± 0.049 | ||||||||
| PHYHIPL | rs209765899 | AA | 0.010 | -0.0749 ± 0.051 | -0.0812 ± 0.028** | 0.0149 ± 0.036 | 0.011 | -0.0941 ± 0.049 | -0.0773 ± 0.027** | 0.0101 ± 0.034 |
| TA | 0.0211 ± 0.027 | -0.0067 ± 0.027 | ||||||||
| TT | 0.0874 ± 0.026 | 0.0606 ± 0.026 | ||||||||
1rs# = a reference SNP ID number assigned by National Center for Biotechnology Information (NCBI).
2Residual Feed Intake = residual feed intake expressed in kg per day.
3a = Additive effect of SNP expressed in kg per day.
4d = Dominance effect of SNP expressed in kg per day.
*is significant at P < 0.05; **is significant at P < 0.01.
Three SNPs within MKI67 showed strong additive effects on RFI (Table 3). In addition to the substantial effect reported previously, results characterized the effect of rs210072660 SNP located in CAST on RFI as significantly additive in decreasing RFI. The MKI67 and CAST genes have both been reported to affect meat quality traits, particularly meat tenderness (Schenkel et al., 2006; Ramayo-Caldas et al., 2016). The significant association between CAST and feed efficiency may explain the correlation between the selection of efficient animals (low RFI) and a negative effect on meat tenderness through the changes in calpastatin and myofibril fragmentation (McDonagh et al., 2001). Also, the significant association of MKI67 may explain the relationship between RFI and meat tenderness and related meat quality traits (Ramayo-Caldas et al., 2016) especially as these associations remained significant after adjusting RFI for fatness (i.e., RFIf) (Table 3). These associations support the link between body composition and the true energetic efficiency of efficient animals (Richardson et al., 2001).
The genotypes of nine SNPs within six genes were associated (P < 0.05) with DMI (Table 4). Genotypes of three SNPs located in MKI67 had significant additive effects on DMI (Table 4). Additionally, SNPs located in ERCC5 (rs133716845) and LMCD1 (rs208239648) showed significant dominance effects increasing DMI (Table 4). Other SNPs located in ACAD11 (rs210293774 and rs208270150) and SMARCAL1 (rs109382589) showed significant dominance effects decreasing DMI (Table 4). If SNP genotypes showed significant dominance or over-dominance effects, it should be taken into account in mating design or selection in planning a genetic improvement program for a crossbreeding system. For example, for rs210293774 and rs208270150 may be useful in crossbreeding animals to combine the different alleles to decrease DMI. In a previous study, the rs133716845 SNP located in ERCC5 showed significant effects on carcass and meat quality traits by increasing lean meat yield and decreasing fatness (Abo-Ismail et al., 2014). A study in mice selected for high muscle mass found that ERCC5 was located in a QTL for lean mass (Kärst et al., 2011). Another 16 SNPs were significantly associated with ADG and 12 SNPs showed additive effects (Table 4).
Table 4.
The least squares means (SE) and P-values for the markers associated (P ≤ 0.05) with average daily gain and dry matter intake using genotypic effect and additive and dominance models
| Gene Name | rs#1 | Genotype | Average Daily Gain (kg) | Dry Matter Intake (kg) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P-value | LSM ± SE | a2 ± SE | d3 ± SE | P-value | LSM ± SE | a ± SE | d ± SE | |||
| ACAD11 | rs210293774 | CC | 0.004 | 1.405 ± 0.024 | 0.028 ± 0.012* | -0.047 ± 0.015** | 0.008 | 8.754 ± 0.048 | 0.114 ± 0.053* | -0.196 ± 0.067** |
| GC | 1.331 ± 0.016 | 8.444 ± 0.048 | ||||||||
| GG | 1.35 ± 0.015 | 8.526 ± 0.048 | ||||||||
| ACAD11 | rs208270150 | CC | 0.006 | 1.349 ± 0.015 | 0.028 ± 0.012* | -0.044 ± 0.015** | 0.013 | 8.526 ± 0.048 | 0.108 ± 0.054* | -0.187 ± 0.067** |
| CT | 1.333 ± 0.016 | 8.447 ± 0.048 | ||||||||
| TT | 1.405 ± 0.024 | 8.742 ± 0.048 | ||||||||
| SMARCAL1 | rs109065702 | CC | 0.05 | 1.354 ± 0.016 | 0.013 ± 0.01 | -0.032 ± 0.013* | 8.529 ± 0.048 | 0.039 ± 0.047 | -0.095 ± 0.058 | |
| CT | 1.335 ± 0.015 | 8.472 ± 0.048 | ||||||||
| TT | 1.38 ± 0.022 | 8.606 ± 0.048 | ||||||||
| SMARCAL1 | rs109808135 | CC | 0.049 | 1.38 ± 0.022 | 0.012 ± 0.011 | -0.032 ± 0.013* | 8.6 ± 0.048 | 0.035 ± 0.047 | -0.092 ± 0.058 | |
| TC | 1.336 ± 0.015 | 8.473 ± 0.048 | ||||||||
| TT | 1.356 ± 0.016 | 8.531 ± 0.048 | ||||||||
| SMARCAL1 | rs109382589 | GG | 1.382 ± 0.022 | 0.017 ± 0.011 | -0.029 ± 0.014* | 0.029 | 8.678 ± 0.048 | 0.082 ± 0.049 | -0.161 ± 0.062* | |
| GT | 1.336 ± 0.016 | 8.436 ± 0.048 | ||||||||
| TT | 1.348 ± 0.016 | 8.515 ± 0.048 | ||||||||
| SMARCAL1 | rs208660945 | CC | 0.024 | 1.311 ± 0.019 | -0.026 ± 0.009** | 0.011 ± 0.013 | 8.434 ± 0.048 | -0.067 ± 0.042 | -0.015 ± 0.056 | |
| CT | 1.348 ± 0.016 | 8.486 ± 0.048 | ||||||||
| TT | 1.362 ± 0.016 | 8.567 ± 0.048 | ||||||||
| LRRIQ3 | rs42417924 | CC | 1.35 ± 0.014 | -0.055 ± 0.024* | 0.056 ± 0.027* | 8.528 ± 0.048 | -0.067 ± 0.106 | -0.028 ± 0.116 | ||
| GC | 1.352 ± 0.019 | 8.433 ± 0.048 | ||||||||
| GG | 1.241 ± 0.049 | 8.394 ± 0.048 | ||||||||
| PPM1K | rs134225543 | CC | 0.028 | 1.338 ± 0.014 | 0.015 ± 0.024 | 0.038 ± 0.029 | 8.481 ± 0.048 | 0.019 ± 0.104 | 0.147 ± 0.128 | |
| TC | 1.391 ± 0.022 | 8.648 ± 0.048 | ||||||||
| TT | 1.368 ± 0.049 | 8.52 ± 0.048 | ||||||||
| CAST | rs137601357 | CC | 0.039 | 1.377 ± 0.021 | 0.006 ± 0.01 | -0.034 ± 0.013* | 8.505 ± 0.05 | -0.045 ± 0.046 | -0.079 ± 0.058 | |
| TC | 1.337 ± 0.016 | 8.471 ± 0.05 | ||||||||
| TT | 1.366 ± 0.017 | 8.594 ± 0.05 | ||||||||
| CAST | rs384020496 | AA | 0.013 | 1.444 ± 0.035 | 0.051 ± 0.017** | -0.045 ± 0.025 | 8.769 ± 0.048 | 0.14 ± 0.075 | -0.114 ± 0.109 | |
| GA | 1.348 ± 0.022 | 8.514 ± 0.048 | ||||||||
| GG | 1.343 ± 0.014 | 8.488 ± 0.048 | ||||||||
| CNTFR | rs137400016 | CC | 0.022 | 1.321 ± 0.017 | 0.021 ± 0.01* | 0.016 ± 0.013 | 8.418 ± 0.048 | 0.055 ± 0.044 | 0.09 ± 0.055 | |
| CT | 1.358 ± 0.015 | 8.562 ± 0.048 | ||||||||
| TT | 1.363 ± 0.019 | 8.527 ± 0.048 | ||||||||
| ATP6V1E2 | rs43673198 | CC | 0.008 | 1.365 ± 0.015 | -0.033 ± 0.013* | -0.002 ± 0.016 | 8.567 ± 0.048 | -0.091 ± 0.059 | -0.04 ± 0.07 | |
| CT | 1.33 ± 0.016 | 8.436 ± 0.048 | ||||||||
| TT | 1.299 ± 0.027 | 8.385 ± 0.048 | ||||||||
| ERCC5 | rs133716845 | CC | 1.343 ± 0.016 | -0.011 ± 0.011 | 0.024 ± 0.014 | 0.036 | 8.488 ± 0.048 | -0.083 ± 0.048 | 0.149 ± 0.061* | |
| TC | 1.356 ± 0.015 | 8.554 ± 0.048 | ||||||||
| TT | 1.321 ± 0.023 | 8.322 ± 0.048 | ||||||||
| TG | rs133269500 | AA | 0.011 | 1.231 ± 0.051 | -0.062 ± 0.025* | 0.032 ± 0.027 | 8.037 ± 0.048 | -0.244 ± 0.106* | 0.18 ± 0.116 | |
| GA | 1.324 ± 0.018 | 8.461 ± 0.048 | ||||||||
| GG | 1.354 ± 0.014 | 8.525 ± 0.048 | ||||||||
| TG | rs110547220 | CC | 0.005 | 1.289 ± 0.024 | -0.039 ± 0.012** | 0.017 ± 0.015 | 8.328 ± 0.047 | -0.118 ± 0.054* | 0.081 ± 0.064 | |
| GC | 1.345 ± 0.016 | 8.527 ± 0.047 | ||||||||
| GG | 1.367 ± 0.015 | 8.565 ± 0.047 | ||||||||
| HMCN1 | rs209012152 | AA | 0.037 | 1.385 ± 0.023 | 0.028 ± 0.011* | -0.004 ± 0.014 | 8.587 ± 0.048 | 0.046 ± 0.051 | -0.048 ± 0.061 | |
| GA | 1.353 ± 0.015 | 8.493 ± 0.048 | ||||||||
| GG | 1.329 ± 0.016 | 8.496 ± 0.048 | ||||||||
| SLC45A2 | rs134604394 | AA | 0.05 | 1.38 ± 0.021 | 0.027 ± 0.011* | -0.005 ± 0.013 | 8.606 ± 0.048 | 0.085 ± 0.049 | -0.008 ± 0.057 | |
| AT | 1.348 ± 0.015 | 8.513 ± 0.048 | ||||||||
| TT | 1.325 ± 0.018 | 8.436 ± 0.048 | ||||||||
| SLC45A2 | rs41946086 | AA | 0.003 | 1.388 ± 0.019 | -0.04 ± 0.012** | -0.009 ± 0.013 | 0.042 | 8.642 ± 0.048 | -0.128 ± 0.054* | -0.043 ± 0.058 |
| AG | 1.339 ± 0.015 | 8.472 ± 0.048 | ||||||||
| GG | 1.309 ± 0.02 | 8.387 ± 0.048 | ||||||||
| LMCD1 | rs208239648 | CC | 0.053 | 1.347 ± 0.014 | -0.216 ± 0.091* | 0.228 ± 0.095* | 0.042 | 8.517 ± 0.048 | -0.987 ± 0.395* | 0.916 ± 0.408* |
| TC | 1.359 ± 0.032 | 8.445 ± 0.048 | ||||||||
| TT | 0.916 ± 0.183 | 6.543 ± 0.048 | ||||||||
| MKI67 | rs110216983 | AA | 1.337 ± 0.016 | 0.013 ± 0.011 | -0.001 ± 0.013 | 0.039 | 8.441 ± 0.048 | 0.119 ± 0.047* | -0.063 ± 0.058 | |
| GA | 1.349 ± 0.016 | 8.496 ± 0.048 | ||||||||
| GG | 1.362 ± 0.021 | 8.678 ± 0.048 | ||||||||
| MKI67 | rs109930382 | CC | 1.335 ± 0.016 | 0.013 ± 0.011 | 0.01 ± 0.014 | 0.025 | 8.433 ± 0.048 | 0.131 ± 0.05** | -0.023 ± 0.062 | |
| CT | 1.357 ± 0.016 | 8.54 ± 0.048 | ||||||||
| TT | 1.36 ± 0.024 | 8.694 ± 0.048 | ||||||||
| MKI67 | rs109558734 | CC | 1.334 ± 0.016 | 0.014 ± 0.011 | 0.009 ± 0.014 | 0.02 | 8.431 ± 0.048 | 0.134 ± 0.05** | -0.029 ± 0.061 | |
| GC | 1.357 ± 0.016 | 8.536 ± 0.048 | ||||||||
| GG | 1.362 ± 0.023 | 8.7 ± 0.048 | ||||||||
1rs# = a reference SNP ID number assigned by National Center for Biotechnology Information (NCBI).
2a = Additive effect of SNP.
3d = Dominance effect of SNP.
*is significant at P < 0.05; **is significant at P < 0.01
Genotypes of nine SNPs located within nine genes had significant associations with MMWT (Table 5). Out of these SNPs, five showed significant additive effects. For example, SNP rs133269500 within the thyroglobulin precursor (TG) gene showed an additive effect on MMWT (Table 5). These findings are in agreement with TG gene biological role as the precursor for thyroid hormones which control fat and lean deposition. A previous study reported polymorphisms in the TG gene to have effects on growth and carcass composition (Zhang et al., 2015). Polymorphisms in TG were associated with marbling score (Gan et al., 2008) and one of the commercially available DNA markers known as GeneSTAR MARB for evaluating marbling in beef cattle is in TG (Rincker et al., 2006). Nonetheless, the DNA marker in TG5 in GeneSTAR was independently validated by the National Beef Cattle Evaluation Consortium (NBCEC, www.NBCEC.org). The results of this validation study reported that TG5 did not have a significant association on marbling score but the favorable allele of TG5 showed a tendency for association with increasing quality grade (Van Eenennaam et al., 2007). The current results also found that rs110519795 SNP, a missense mutation, located in DPP6, showed a significant additive effect on MMWT, whereas SNP rs132717265 showed a significant additive effect on back fat (Table 5). The current associations are in agreement with the physiological role of DPP6 as the latter is involved in ion and cation transport, and which is reported to contribute to variation in feed efficiency (Richardson and Herd, 2004; Herd and Arthur, 2009). In a previous GWAS study in Angus and Simmental, as well as their crosses, an intronic SNP (rs110787048) located in DPP6, was reported to affect the efficiency of gain (i.e., residual ADG) (Serão et al., 2013). In another GWAS in Canchim beef cattle, DPP6 was reported to affect birth and weaning weights (Buzanskas et al., 2014). SNPs located in the C27H8orf40 (rs135814528), ELMOD1 (rs42235500), MAPK15 (rs110323635), AFF3 (rs42275280), and PPM1K (rs134225543) genes all showed significant additive effects on backfat (Table 5).
Table 5.
The least squares means (SE) and P-values for the markers associated (P ≤ 0.05) with midpoint metabolic weight and back fat using genotypic effect and additive and dominance models
| Gene Name | rs#1 | Genotype | Midpoint Metabolic Weight (kg) | Back fat (mm) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P-value | LSM ± SE | a2 ± SE | d3 ± SE | P-value | LSM ± SE | a1 ± SE | d2 ± SE | |||
| RRP1B | rs43285609 | AA | 85.739 ± 0.381 | -0.135 ± 0.321 | 0.312 ± 0.388 | 0.05 | 7.022 ± 0.183 | 0.019 ± 0.089 | 0.254 ± 0.109* | |
| RRP1B | rs43285609 | GA | 86.185 ± 0.381 | 7.257 ± 0.137 | ||||||
| RRP1B | rs43285609 | GG | 86.008 ± 0.381 | 6.984 ± 0.151 | ||||||
| GALNT13 | rs438856835 | AA | 85.883 ± 0.38 | 0.196 ± 0.962 | 0.828 ± 1.063 | 0.035 | 7.094 ± 0.13 | -0.486 ± 0.274 | 0.77 ± 0.304* | |
| GALNT13 | rs438856835 | CA | 86.908 ± 0.38 | 7.378 ± 0.191 | ||||||
| GALNT13 | rs438856835 | CC | 86.275 ± 0.38 | 6.122 ± 0.555 | ||||||
| SMARCAL1 | rs208660945 | CC | 85.764 ± 0.382 | -0.172 ± 0.297 | 0.219 ± 0.391 | 0.049 | 7.177 ± 0.174 | 0.111 ± 0.083 | 0.203 ± 0.109 | |
| SMARCAL1 | rs208660945 | CT | 86.155 ± 0.382 | 7.269 ± 0.141 | ||||||
| SMARCAL1 | rs208660945 | TT | 86.109 ± 0.382 | 6.955 ± 0.148 | ||||||
| LRRIQ3 | rs42417924 | CC | 0.043 | 86.331 ± 0.38 | -0.857 ± 0.742 | -0.368 ± 0.806 | 7.171 ± 0.132 | -0.013 ± 0.211 | -0.187 ± 0.231 | |
| LRRIQ3 | rs42417924 | GC | 85.107 ± 0.38 | 6.971 ± 0.17 | ||||||
| LRRIQ3 | rs42417924 | GG | 84.618 ± 0.38 | 7.145 ± 0.427 | ||||||
| DPP6 | rs110519795 | AA | 0.05 | 86.44 ± 0.381 | -0.725 ± 0.308* | 0.446 ± 0.395 | 7.178 ± 0.148 | -0.062 ± 0.086 | 0.017 ± 0.112 | |
| DPP6 | rs110519795 | AG | 86.161 ± 0.381 | 7.133 ± 0.142 | ||||||
| DPP6 | rs110519795 | GG | 84.99 ± 0.381 | 7.054 ± 0.18 | ||||||
| DPP6 | rs132717265 | AA | 85.689 ± 0.382 | -0.393 ± 0.313 | -0.015 ± 0.381 | 0.007 | 6.895 ± 0.164 | -0.264 ± 0.086** | -0.067 ± 0.107 | |
| DPP6 | rs132717265 | GA | 86.068 ± 0.382 | 7.092 ± 0.138 | ||||||
| DPP6 | rs132717265 | GG | 86.476 ± 0.382 | 7.423 ± 0.159 | ||||||
| PPM1K | rs134225543 | CC | 0.018 | 85.78 ± 0.383 | 1.013 ± 0.727 | 0.673 ± 0.886 | 0.015 | 7.198 ± 0.129 | -0.521 ± 0.207* | 0.214 ± 0.253 |
| PPM1K | rs134225543 | TC | 87.465 ± 0.383 | 6.891 ± 0.194 | ||||||
| PPM1K | rs134225543 | TT | 87.806 ± 0.383 | 6.156 ± 0.424 | ||||||
| CAST | rs384020496 | AA | 87.727 ± 0.377 | 0.973 ± 0.518 | -0.124 ± 0.751 | 0.014 | 7.465 ± 0.31 | 0.203 ± 0.15 | 0.277 ± 0.214 | |
| CAST | rs384020496 | GA | 86.631 ± 0.377 | 7.539 ± 0.199 | ||||||
| CAST | rs384020496 | GG | 85.782 ± 0.377 | 7.058 ± 0.13 | ||||||
| CAST | rs133057384 | AA | 87.548 ± 0.38 | 0.827 ± 0.731 | -0.102 ± 0.846 | 0.018 | 7.665 ± 0.42 | 0.307 ± 0.207 | 0.088 ± 0.24 | |
| CAST | rs133057384 | GA | 86.619 ± 0.38 | 7.447 ± 0.179 | ||||||
| CAST | rs133057384 | GG | 85.894 ± 0.38 | 7.052 ± 0.13 | ||||||
| CAST | rs110711318 | CC | 85.843 ± 0.38 | 1.273 ± 0.831 | -0.272 ± 0.968 | 0.017 | 7.058 ± 0.131 | 0.37 ± 0.233 | 0.045 ± 0.272 | |
| CAST | rs110711318 | TC | 86.843 ± 0.38 | 7.473 ± 0.188 | ||||||
| CAST | rs110711318 | TT | 88.388 ± 0.38 | 7.797 ± 0.472 | ||||||
| AFF3 | rs42275280 | CC | 0.011 | 84.288 ± 0.382 | -0.992 ± 0.417* | -2.594 ± 1.999 | 6.691 ± 0.242 | -0.238 ± 0.112* | -0.271 ± 0.577 | |
| AFF3 | rs42275280 | CT | 82.687 ± 0.382 | 6.658 ± 0.576 | ||||||
| AFF3 | rs42275280 | TT | 86.272 ± 0.382 | 7.167 ± 0.13 | ||||||
| ERCC5 | rs133716845 | CC | 0.031 | 85.967 ± 0.38 | -0.598 ± 0.34 | 1.054 ± 0.419* | 7.098 ± 0.144 | 0.019 ± 0.096 | 0.066 ± 0.119 | |
| ERCC5 | rs133716845 | TC | 86.423 ± 0.38 | 7.182 ± 0.141 | ||||||
| ERCC5 | rs133716845 | TT | 84.771 ± 0.38 | 7.135 ± 0.205 | ||||||
| MAPK15 | rs110323635 | AA | 0.047 | 85.592 ± 0.382 | 0.811 ± 0.33* | -0.228 ± 0.403 | 6.957 ± 0.148 | 0.185 ± 0.091* | 0.055 ± 0.114 | |
| MAPK15 | rs110323635 | GA | 86.175 ± 0.382 | 7.196 ± 0.138 | ||||||
| MAPK15 | rs110323635 | GG | 87.213 ± 0.382 | 7.326 ± 0.192 | ||||||
| TG | rs133269500 | AA | 0.02 | 82.153 ± 0.379 | -2.021 ± 0.731** | 1.59 ± 0.798* | 6.736 ± 0.437 | -0.214 ± 0.212 | 0.073 ± 0.232 | |
| TG | rs133269500 | GA | 85.765 ± 0.379 | 7.024 ± 0.165 | ||||||
| TG | rs133269500 | GG | 86.195 ± 0.379 | 7.165 ± 0.131 | ||||||
| ELMOD1 | rs42235500 | AA | 85.947 ± 0.382 | -0.039 ± 0.287 | 0.502 ± 1.095 | 0.043 | 6.751 ± 0.192 | -0.2 ± 0.081* | 0.25 ± 0.301 | |
| ELMOD1 | rs42235500 | GA | 86.488 ± 0.382 | 7.202 ± 0.314 | ||||||
| ELMOD1 | rs42235500 | GG | 86.025 ± 0.382 | 7.151 ± 0.129 | ||||||
| UGT3A1 | rs42345570 | AA | 85.329 ± 0.381 | -0.278 ± 0.455 | 0.747 ± 0.512 | 0.027 | 7.009 ± 0.255 | 0.001 ± 0.124 | 0.292 ± 0.143* | |
| UGT3A1 | rs42345570 | CA | 86.354 ± 0.381 | 7.3 ± 0.142 | ||||||
| UGT3A1 | rs42345570 | CC | 85.884 ± 0.381 | 7.006 ± 0.14 | ||||||
| SLC45A2 | rs134604394 | AA | 86.476 ± 0.382 | 0.406 ± 0.348 | 0.081 ± 0.397 | 7.012 ± 0.185 | -0.05 ± 0.097 | 0.128 ± 0.112 | ||
| SLC45A2 | rs134604394 | AT | 86.151 ± 0.382 | 7.191 ± 0.137 | ||||||
| SLC45A2 | rs134604394 | TT | 85.664 ± 0.382 | 7.113 ± 0.161 | ||||||
| PCSK6 | rs43020736 | CC | 0.027 | 86.725 ± 0.38 | -0.824 ± 0.319* | 0.336 ± 0.383 | 7.09 ± 0.16 | -0.041 ± 0.09 | 0.183 ± 0.109 | |
| PCSK6 | rs43020736 | TC | 86.237 ± 0.38 | 7.232 ± 0.142 | ||||||
| PCSK6 | rs43020736 | TT | 85.078 ± 0.38 | 7.009 ± 0.169 | ||||||
| C27H8orf40 | rs135814528 | AA | 0.009 | 85.856 ± 0.379 | 1.856 ± 2.012 | 0.175 ± 2.098 | 0.036 | 7.117 ± 0.128 | -1.284 ± 0.564* | 1.505 ± 0.589* |
| C27H8orf40 | rs135814528 | GA | 87.887 ± 0.379 | 7.338 ± 0.215 | ||||||
| C27H8orf40 | rs135814528 | GG | 89.569 ± 0.379 | 4.549 ± 1.132 | ||||||
1rs# = a reference SNP ID number assigned by National Center for Biotechnology Information (NCBI).
2a = Additive effect of SNP.
3d = Dominance effect of SNP.
*is significant at P < 0.05; **is significant at P < 0.01.
Gene Ontology and Pathways Enrichment Analyses
Gland development.
The gene set enrichment analysis suggested that the biological process of gland development (GO:0048732) was significantly enriched (P = 0.0016) by the MKI67, PKD2, TG, and RB1CC1 genes (Table 6). Additionally, MKI67, PKD2, and RB1CC1 genes were each significantly (P < 0.05) over-represented in liver development (GO:0001889) and mechanisms in the hepaticobiliary system (GO:0061008). The importance of these genes in organ development were presented in a study by Saatchi et al. (2014b) where the study identified eight pleiotropic QTLs affecting body weights and carcass traits, and having genes involved in tissue development. The PKD2 gene was identified in the confidence interval of across-breed and breed-specific pleiotropic QTL for carcass and meat quality traits and was over-represented in the tissue development process (GO:0009888) (Saatchi et al., 2014b). Liver development and tissue development are part of the anatomical structure development biological process (GO:0048856).
Table 6.
The enriched (at P ≤ 0.05) gene ontology terms and biological pathways having genes associated with feed efficiency and its components traits
| Category1 | Term | P-Value2 | Gene Names |
|---|---|---|---|
| BP | GO:0001889~liver development | 0.010& | MKI67, PKD2, RB1CC1 |
| BP | GO:0034220~ion transmembrane transport | 0.011& | DPP6, CNGA3, PKD2, ATP6V1E2, ANXA2, CACNA1G |
| BP | GO:0061008~hepaticobiliary system development | 0.011& | MKI67, PKD2, RB1CC1 |
| BP | GO:0055085~transmembrane transport | 0.012& | DPP6, CNGA3, PKD2, ATP6V1E2, ANXA2, CACNA1G, ABCG2 |
| BP | GO:0006812~cation transport | 0.018 | DPP6, CNGA3, PKD2, ATP6V1E2, ANXA2, CACNA1G |
| BP | GO:0098655~cation transmembrane transport | 0.018 | DPP6, CNGA3, PKD2, ATP6V1E2, ANXA2 |
| BP | GO:0006811~ion transport | 0.024 | DPP6, CNGA3, PKD2, ATP6V1E2, ANXA2, TG, CACNA1G |
| BP | GO:0070509~calcium ion import | 0.031 | PKD2, ANXA2, CACNA1G |
| BP | GO:0030001~metal ion transport | 0.034 | DPP6, CNGA3, PKD2, ANXA2, CACNA1G |
| BP | GO:0015672~monovalent inorganic cation transport | 0.036 | DPP6, CNGA3, PKD2, ATP6V1E2 |
| BP | GO:0006813~potassium ion transport | 0.040 | DPP6, CNGA3, PKD2 |
| BP | GO:0048732~gland development | 0.040 | MKI67, PKD2, TG, RB1CC1 |
| MF | GO:0008324~cation transmembrane transporter activity | 0.009& | SLC45A2, CNGA3, PKD2, ATP6V1E2, ANXA2, CACNA1G |
| MF | GO:0004896~cytokine receptor activity | 0.017& | CNTFR, OSMR, GHR |
| MF | GO:0005262~calcium channel activity | 0.023 | PKD2, ANXA2, CACNA1G |
| MF | GO:0022890~inorganic cation transmembrane transporter activity | 0.023 | CNGA3, PKD2, ATP6V1E2, ANXA2, CACNA1G |
| MF | GO:0005261~cation channel activity | 0.028 | CNGA3, PKD2, ANXA2, CACNA1G |
| MF | GO:0015085~calcium ion transmembrane transporter activity | 0.029 | PKD2, ANXA2, CACNA1G |
| MF | GO:0022843~voltage-gated cation channel activity | 0.038 | CNGA3, PKD2, CACNA1G |
| MF | GO:0072509~divalent inorganic cation transmembrane transporter activity | 0.049 | PKD2, ANXA2, CACNA1G |
| KEGG | bta04630:Jak-STAT signaling pathway | 0.027 | CNTFR, OSMR, GHR |
1Category = gene ontology (GO) and pathway categories where BP is biological process, MF is molecular function and KEGG is the Kyoto Encyclopedia of Genes and Genomes pathway.
2 P-Value is the absolute P-Value; &P-value is significant at less than 20% false discovery rate.
Ion transport (GO:0034220).
The current results highlighted the importance of ion transport as a mechanism for controlling feed efficiency traits where it was promoted by DPP6, CNGA3, PKD2, ATP6V1E2, ANXA2, TG, and CACNA1G genes (Table 6). Previous studies have emphasized the importance of ion transport as part of the metabolic processes controlling variation in feed efficiency (Herd et al., 2004). Metabolism was reported to account for 42% variation in observed RFI (Herd and Arthur 2009).
Jak-STAT signaling pathway (bta04630).
In the current study, JAK-STAT signaling was identified as a key pathway contributing to variation in feed efficiency traits. This pathway was enriched by the CNTFR, OSMR, and GHR genes (Table 6). Growth hormone binds its receptors (GHR) to activate the Janus kinases (Jaks) signal transduction pathway affecting important processes such as lipid metabolism and the cell cycle (Richard and Stephens, 2014). The mRNA expression of GHR is greater in the muscle and liver of efficient animals when compared to nonefficient animals (Chen et al., 2011; Kelly et al., 2013) where RFI was negatively associated with GHR expression (r = −0.5) (Kelly et al., 2013). The JAK-STAT pathway mediates several biological mechanisms including lipid and glucose metabolism, insulin signaling, development, and adipogenesis regulation (Richard and Stephens, 2014). Other studies suggested that the GHR and OSMR genes repress adipocyte differentiation through an antiadipogenic activity of STAT5 in different model systems (Richard and Stephens, 2014). This might explain the relationship between variation in RFI and body composition, especially body fat (Richardson et al., 2001; Richardson and Herd, 2004; Herd and Arthur, 2009).
Pedigree and Genomic Heritability and Genetic Variance Explained by SNP Panel
The pedigree-based heritability (hp2) estimates for feed efficiency traits in the current population were moderate to high, and ranged from 0.25 to 0.69 (Table 7). In general, the hp2 for the studied traits were in agreement with published values for Hereford and Angus populations (Schenkel et al., 2004). Generally, the estimated heritability for RFI (0.25) and RFIf (0.27) are within the reported range of 0.16 to 0.45 (Herd and Bishop, 2000; Crowley et al., 2010) in British Hereford and Irish beef cattle breeds. Also, the heritability (0.69) for MMWT agreed with that reported by Crowley et al. (2010). For DMI, heritability (0.49) was within previous estimates ranging from 0.31 to 0.49 (Herd and Bishop, 2000; Crowley et al., 2010). The fact that the heritability estimates calculated for feed efficiency traits were consistent with previously documented values support that the standard error and level of significance are appropriately estimated in the current population because the uncertainty of heritability has a large influence on SNP significance tests (Hassen et al., 2009).
Table 7.
Heritability values estimated using the different SNP sets
| Trait | h2p ± SE1 | h250k ± SE2 | h2full ± SE3 | h2sig10 ± SE4 | h2sig5 ± SE5 |
|---|---|---|---|---|---|
| ADG, kg/d6 | 0.276 ± 0.083 | 0. 254 ± 0.080 | 0.078 ± 0.030 | 0.089 ± 0.030 | 0.072 ± 0.027 |
| DMI, kg7 | 0.499 ± 0.095 | 0.513 ± 0.077 | 0.079 ± 0.031 | 0.089 ± 0.032 | 0.077 ± 0.029 |
| MMWT, kg8 | 0.690 ± 0.090 | 0.572 ± 0.072 | 0.126 ± 0.037 | 0.111 ± 0.036 | 0.076 ± 0.03 |
| RFI, kg/d9 | 0.247 ± 0.078 | 0.213 ± 0.066 | 0.038 ± 0.020 | 0.048 ± 0.021 | 0.047 ± 0.021 |
| RFIf, kg/d10 | 0.273 ± 0.080 | 0.240 ± 0.069 | 0.044 ± 0.021 | 0.053 ± 0.022 | 0.053 ± 0.022 |
| BFat, mm11 | 0.446 ± 0.093 | 0.369 ± 0.073 | 0.037 ± 0.024 | 0.064 ± 0.027 | 0.067 ± 0.028 |
1h2p = Heritability estimate from using the pedigree information.
2h250k = Heritability estimate from using the 50k panel (n = 40465 SNP).
3h2full = Heritability estimate using the full SNPs set (n = 159 SNP).
4h2sig10 = Heritability estimate from using the significant (P < 0.10) SNPs set (n = 92 SNP).
5h2sig5 = Heritability estimate from using the significant (P ≤ 0.05) SNPs set (n = 63 SNP).
6ADG = average daily gain: recorded in kg per day from start to end of the finishing period.
7DMI = dry matter intake: recorded in kg per day from start to end of the finishing period.
8MMWT = midpoint metabolic weight.
9RFI = residual feed intake expressed kg per day.
10RFIf = residual feed intake adjusted for backfat.
11BFat = backfat: recorded as fat depth at the end of the finishing period in millimeters.
The genomic heritability using the different SNP sets ranged from 0.037 to 0.13 (Table 7). The associated (P < 0.05) SNPs list explained 19.4% of the genetic variance of RFI and RFIf with genomic heritability of 0.05. Up to 32%, 18%, 18%, 19.4%, 19.4%, and 15% of the genetic variance, relative to that calculated from pedigree information (i.e., A matrix) in ADG, DMI, midpoint metabolic weight, RFI, RFI adjusted for back fat, and BFat, respectively, were explained by the full set of the tested SNPs (n = 159) or subsets of these SNPs. About 16% of the genetic variance of the DMI was explained by the full set of SNPs (n = 159). Interestingly, the highest genomic heritability for the full set of the developed markers (n = 159) was for MMWT (0.13). This might support the link between candidate genes and the tissue development and energy maintenance mechanisms discussed previously. The population size used (n = 871) in the current study was relatively low and the accuracy of prediction may improve as the number of individuals in the reference population increases (Goddard, 2009; VanRaden et al., 2009; Zhang et al., 2011). Candidate genes explained up to 19.4 % in genetic variance in feed efficiency (RFI and RFIf). Thus, using the SNP panel in marker assisted selection could be effective. Nonetheless, feed efficiency is a complex trait affected by many genes, and adding more informative SNPs to this panel would be needed to achieve the same proportion of genetic variance explained by a larger panel such as 50K SNP which explained 87% of genetic variance in feed efficiency in this study.
This study sought to generate and validate a set of SNPs selected to have a high chance of being causative mutations, or closely linked to such mutations (i.e., in linkage disequilibrium), which could have an effect on feed efficiency. Such SNPs would likely be useful for genetic improvement of feed efficiency across different populations of cattle or for selection in commercial crossbred populations which are prevalent in Canada. The results obtained are in good agreement with those from previous studies including those describing the roles of these genes and pathways in traits related to feed efficiency and its component traits. Generally, to develop a SNP panel as a selection tool, Crews et al. (2008) suggested it would be necessary to explain at least 10% to 15% of the genetic variation in order to be cost effective. The current markers explained up to 19.5% of the genetic variance, which is equal to 0.11 Beef Improvement Federation (BIF) accuracy (Bullock et al., 2012). This BIF accuracy (i.e., 0.11) corresponds to testing 5 and 3 progeny (i.e., equivalent progeny number; EPN) for medium to high heritability traits such as RFI and MMWT, respectively. This value is estimated in for the current population, and it may decrease when applied to independent populations. Nonetheless, if we assumed that genomic data are the only source of information for feed efficiency or its components traits (i.e., an animal with 0 accuracy expected progeny difference, EPD), a small panel with a cost of approximately $12 will potentially save the producer the cost of progeny testing of around $48–96 per animal which represents a significant return. Although, the collection of data for correlated traits provides some accuracy this is often very limited. In addition, including the molecular breeding value (MBV) estimated using the SNP panel as a correlated trait (Kachman, 2008) or blending information from genotypes (G matrix) with pedigree (A matrix) could potentially increase the accuracy of selection and subsequently increase the rate of genetic change. The ability of forming G using a very small number of SNP depends on linkage disequilibrium (LD) and minor allele frequency (MAF) (Rolf et al., 2010). When fewer number of SNPs (less than 2,500 SNP) were randomly selected, the MBV estimates were sensitive to the number of SNPs and based on LD and MAF. The correlations between G estimates calculated using 384 and 1,536 randomly selected SNPs from the 50k SNP panel and G estimates calculated from 50K ranged from 0.61 to 0.69 and from 0.85 to 0.88 for 384 and 1,536 SNP, respectively. They also used a developed panel of 384 SNPs strongly associated with feed intake and correlation between G estimated from 384 SNP set and 50k SNP set was 0.62 which was slightly lower than using 384 SNPs randomly selected. This study reported that a small panel selected based on significant associations could be used for the estimation of genomic relationship coefficients and generation of molecular breeding values. Genomic selection is potentially cheaper than phenotypic selection especially if the number of SNPs on the panel is small and limited to only those with the largest effect (Zhang et al., 2011). Reducing the cost of the DNA test in this way could increase the uptake of the genomic testing significantly (Rolf et al., 2014). In addition, using a small DNA test panel with BIF accuracy 0.11 is better than just selecting animals based on unrelated indicators such as appearance with accuracy zero. Thus, we have developed this panel to be used for commercial animal selection (e.g., commercial heifer selection). The SNP panel can also be used with such panels for other traits by combining them into a larger panel for selection.
More recently, it has been shown that including causative mutations or functional annotations of polymorphisms, can potentially improve the performance of genomic prediction (e.g., see (MacLeod et al., 2016). Thus, the current study incorporated biological information by selecting genes based on gene expression analyses, enriched data, and identified causal variants in other studies, to improve the power and precision of genomic prediction, including for crossbred or less related cattle populations. The current study also supports the value of incorporating variants from candidate genes reported in previous studies and known to be related to feed efficiency.
CONCLUSION
An informative cost-effective SNP marker panel was developed that predicted a useful proportion of variation in important feed efficiency traits for cattle. The study identified 63 SNPs associated with substantial variation (19.4%) in feed efficiency which can subsequently be used in practice by the beef industry. Such a panel with a small set of SNPs may be useful to generate molecular breeding values for feed efficiency at relatively low cost. Further testing in other populations including a wider variety of crossbred cattle is warranted. Some of the SNPs within the UMPS, SMARCAL, CCSER1, and LMCD1 genes showed significant over-dominance effects, whereas other SNPs located in the SMARCAL1, ANXA2, CACNA1G, and PHYHIPL genes showed additive effects on RFI and RFIf. These results need to be taken into account in any cross breeding system to optimize useful allele combinations. Gland development, ion, and cation transport were important physiological mechanisms contributing to variation in the feed efficiency traits. Finally, the study revealed the effect of a Jak-STAT signaling pathway on feed efficiency through the CNTFR, OSMR, and GHR genes which could be useful for genetic selection for feed efficiency.
Supplementary Material
Footnotes
The authors gratefully acknowledge funding support from Alberta Livestock and Meat Agency Ltd., Alberta Innovates Bio Solutions, and Alberta Agriculture and Forestry (previously Agriculture and Rural Development [ARD]) and in-kind contribution in animals, facilities and staff received from Agriculture and Agri-Food Canada, Lacombe Research Centre. We would also like to thank Steve Miller (when at University of Guelph) for his support of the background studies and helping obtain funding for this study.
LITERATURE CITED
- Abo-Ismail M. K., Kelly M. J., Squires E. J., Swanson K. C., Bauck S., and Miller S. P.. 2013. Identification of single nucleotide polymorphisms in genes involved in digestive and metabolic processes associated with feed efficiency and performance traits in beef cattle. J. Anim. Sci. 91:2512–2529. [DOI] [PubMed] [Google Scholar]
- Abo-Ismail M. K., Vander Voort G., Squires J. J., Swanson K. C., Mandell I. B., Liao X., Stothard P., Moore S., Plastow G., and Miller S. P.. 2014. Single nucleotide polymorphisms for feed efficiency and performance in crossbred beef cattle. BMC Genet. 15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mamun H. A., Kwan P., Clark S. A., Ferdosi M. H., Tellam R., and Gondro C.. 2015. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. 47:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D. H., Novembre J., and Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basarab J., Baron V., López-Campos Ó., Aalhus J., Haugen-Kozyra K., and Okine E.. 2012. Greenhouse gas emissions from calf-and yearling-fed beef production systems, with and without the use of growth promotants. Animals 2:195–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basarab J., Colazo M., Ambrose D., Novak S., McCartney D., and Baron V.. 2011. Residual feed intake adjusted for backfat thickness and feeding frequency is independent of fertility in beef heifers. Can. J. Anim. Sci. 91:573–584. [Google Scholar]
- Basarab J. A., Okine E. K., Baron V. S., Marx T., Ramsey P., Ziegler K., and Lyle K.. 2005. Methane emissions from enteric fermentation in Alberta’s beef cattle population. Can. J. Anim. Sci. 85:501–512. [Google Scholar]
- Basarab J. A., McCartney D., Okine E. K., and Baron V. S.. 2007. Relationships between progeny residual feed intake and dam productivity traits. Can. J. Anim. Sci. 87:489–502. [Google Scholar]
- Berg R. T., Makarechian M., and Arthur P.. 1990. The University of Alberta beef breeding project after 30 years—A review. Univ. Alberta Annu. Feeders’ Day Rep. 69:65–69. [Google Scholar]
- Bullock D., Spangler M., Van Eenennaam A., and Weaber R.. 2012. Delivering genomics technology to the beef industry National Beef Cattle Evaluation Consortium White Paper.〈 http://www.nbcec.org/topics/WhitePaperGenomicsTechnology.pdf/〉(Accessed 15 October 2013).
- Buzanskas M. E., Grossi D. A., Ventura R. V., Schenkel F. S., Sargolzaei M., Meirelles S. L., Mokry F. B., Higa R. H., Mudadu M. A., da Silva M. V.,. et al. 2014. Genome-wide association for growth traits in Canchim beef cattle. PLOS ONE 9:e94802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafe L. M., McIntyre B. L., Robinson D. L., Geesink G. H., Barendse W., and Greenwood P. L.. 2010. Production and processing studies on calpain-system gene markers for tenderness in Brahman cattle: 1. Growth, efficiency, temperament, and carcass characteristics1. J. Anim. Sci. 88:3047–3058. [DOI] [PubMed] [Google Scholar]
- Canadian Council on Animal Care 1993. Guide to the care and use of experimental animals. Vol. 1 E. D. Olfert B. M. Cross, and A. A. McWilliams, editors. Ottawa, ON: CCAC. [Google Scholar]
- Ceacero T. M., Mercadante M. E., Cyrillo J. N., Canesin R. C., Bonilha S. F., and de Albuquerque L. G.. 2016. Phenotypic and genetic correlations of feed efficiency traits with growth and carcass traits in Nellore cattle selected for postweaning weight. PLoS ONE 11:e0161366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Gondro C., Quinn K., Herd R. M., Parnell P. F., and Vanselow B.. 2011. Global gene expression profiling reveals genes expressed differentially in cattle with high and low residual feed intake. Anim. Genet. 42:475–490. [DOI] [PubMed] [Google Scholar]
- Connor E. E., Kahl S., Elsasser T. H., Parker J. S., Li R. W., Van Tassell C. P., Baldwin R. L. 6th, and Barao S. M.. 2010. Enhanced mitochondrial complex gene function and reduced liver size may mediate improved feed efficiency of beef cattle during compensatory growth. Funct. Integr. Genomics 10:39–51. [DOI] [PubMed] [Google Scholar]
- Crews D., Moore S., and Enns R.. 2008. Optimizing traditional and marker assisted evaluation in beef cattle. Proceedings of the 40th Beef Improvement Federation, Calgary, Alberta: 44–49. [Google Scholar]
- Crowley J. J., McGee M., Kenny D. A., Crews D. H. Jr, Evans R. D., and Berry D. P.. 2010. Phenotypic and genetic parameters for different measures of feed efficiency in different breeds of Irish performance-tested beef bulls. J. Anim. Sci. 88:885–894. [DOI] [PubMed] [Google Scholar]
- Crowley J. J., Stothard P., Basarab J. A., Miller S. P., Li C., Wang Z., Plastow G., De Pauw E., Moore S. S., and Lu D.. 2014. Collation of Data and Genetic Parameter Estimation in Different Experimental Canadian Beef Cattle Populations Measured for Feed Efficiency 10th World Congress on Genetics Applied to Livestock Production (WCGALP). p Paper 117, Vancouver, Canada. [Google Scholar]
- Da Y., Wang C., Wang S., and Hu G.. 2014. Mixed model methods for genomic prediction and variance component estimation of additive and dominance effects using SNP markers. PLOS ONE 9:e87666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Capitan A., Pausch H., Stothard P., van Binsbergen R., Brøndum R. F., Liao X., Djari A., Rodriguez S. C., Grohs C.,. et al. 2014. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46:858–865. [DOI] [PubMed] [Google Scholar]
- de Oliveira P. S., Cesar A. S., do Nascimento M. L., Chaves A. S., Tizioto P. C., Tullio R. R., Lanna D. P., Rosa A. N., Sonstegard T. S., Mourao G. B.,. et al. 2014. Identification of genomic regions associated with feed efficiency in Nelore cattle. BMC Genet. 15:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson G. E. 1973. Inbreeding and heterosis in animals. J. Anim. Sci. 1973:54–77. [Google Scholar]
- Du M., Zhu M. J., Means W. J., Hess B. W., and Ford S. P.. 2004. Effect of nutrient restriction on calpain and calpastatin content of skeletal muscle from cows and fetuses1. J. Anim. Sci. 82:2541–2547. [DOI] [PubMed] [Google Scholar]
- Fernando R. L., Stricker C., and Wang T.. 1998. Detection and utilization of single genes without DNA assays. J. Dairy Sci. 81:64–75. [DOI] [PubMed] [Google Scholar]
- Gan Q. F., Zhang L. P., Li J. Y., Hou G. Y., Li H. D., Gao X., Ren H. Y., Chen J. B., and Xu S. Z.. 2008. Association analysis of thyroglobulin gene variants with carcass and meat quality traits in beef cattle. Appl. Genet. 49:251–255. [DOI] [PubMed] [Google Scholar]
- Gilmour A., Gogel B. J., Cullis B., and Thompson R.. 2009. ASReml User Guide Release 3.0. HP1 1ES, UK: VSN International Ltd Hemel Hempstead. [Google Scholar]
- Goddard M. 2009. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136:245–257. [DOI] [PubMed] [Google Scholar]
- Goonewardene L., Wang Z., Price M., Yang R.-C., Berg R., and Makarechian M.. 2003. Effect of udder type and calving assistance on weaning traits of beef and dairy× beef calves. Livest. Prod. Sci. 81:47–56. [Google Scholar]
- Grant J. R., Arantes A. S., Liao X., and Stothard P.. 2011. In-depth annotation of SNPs arising from resequencing projects using NGS-SNP. Bioinformatics 27:2300–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Gil B., Williams J. L., Homer D., Burton D., Haley C. S., and Wiener P.. 2009. Search for quantitative trait loci affecting growth and carcass traits in a cross population of beef and dairy cattle. J. Anim. Sci. 87:24–36. [DOI] [PubMed] [Google Scholar]
- Hassen A., Avendano S., Hill W. G., Fernando R. L., Lamont S. J., and Dekkers J. C.. 2009. The effect of heritability estimates on high-density single nucleotide polymorphism analyses with related animals1. J. Anim. Sci. 87:868–875. [DOI] [PubMed] [Google Scholar]
- Herd R., and Arthur P.. 2009. Physiological basis for residual feed intake. J. Anim. Sci. 87:E64–E71. [DOI] [PubMed] [Google Scholar]
- Herd R., Oddy V., and Richardson E.. 2004. Biological basis for variation in residual feed intake in beef cattle. 1. Review of potential mechanisms. Animal Prod. Sci. 44:423–430. [Google Scholar]
- Herd R. M., and Bishop S. C.. 2000. Genetic variation in residual feed intake and its association with other production traits in British Hereford cattle. Livest. Prod. Sci. 63:111–119. [Google Scholar]
- Hu Z.-L., Park C. A., Wu X.-L., and Reecy J. M.. 2013. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acid Res. 41:D871–D879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., and Lempicki R. A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4:44–57. [DOI] [PubMed] [Google Scholar]
- Kachman S. D. 2008. Incorporation of marker scores into national genetic evaluations. In: 9th genetic prediction workshop; Kansas City, Missouri: p 92–98. [Google Scholar]
- Karisa B., Moore S., and Plastow G.. 2014. Analysis of biological networks and biological pathways associated with residual feed intake in beef cattle. J. Anim. Sci. 85:374–387. [DOI] [PubMed] [Google Scholar]
- Karisa B. K., Thomson J., Wang Z., Stothard P., Moore S. S., and Plastow G. S.. 2013. Candidate genes and single nucleotide polymorphisms associated with variation in residual feed intake in beef cattle. J. Anim. Sci. 91:3502–3513. [DOI] [PubMed] [Google Scholar]
- Kärst S., Cheng R., Schmitt A. O., Yang H., de Villena F. P., Palmer A. A., and Brockmann G. A.. 2011. Genetic determinants for intramuscular fat content and water-holding capacity in mice selected for high muscle mass. Mamm. Genome. 22:530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A. K., Waters S. M., McGee M., Browne J. A., Magee D. A., and Kenny D. A.. 2013. Expression of key genes of the somatotropic axis in longissimus dorsi muscle of beef heifers phenotypically divergent for residual feed intake. J. Anim. Sci. 91:159–167. [DOI] [PubMed] [Google Scholar]
- Li J., Zhang L.-P., Gan Q.-F., Li J.-Y., Gao H.-J., Yuan Z.-R., Gao X., Chen J.-B., and S.-Z Xu. 2010. Association of CAST gene polymorphisms with carcass and meat quality traits in Chinese commercial cattle herds. Asian-Australas J. Anim. Sci. 23:1405–1411. [Google Scholar]
- Lindholm-Perry A. K., Kern R. J., Kuehn L. A., Snelling W. M., Miles J. R., Oliver W. T., and Freetly H. C.. 2015. Differences in transcript abundance of genes on BTA15 located within a region associated with gain in beef steers. Gene 572:42–48. [DOI] [PubMed] [Google Scholar]
- Lu D., Miller S., Sargolzaei M., Kelly M., Vander Voort G., Caldwell T., Wang Z., Plastow G., and Moore S.. 2013. Genome-wide association analyses for growth and feed efficiency traits in beef cattle1. J. Anim. Sci. 91:3612–3633. [DOI] [PubMed] [Google Scholar]
- MacLeod I. M., Bowman P. J., Vander Jagt C. J., Haile-Mariam M., Kemper K. E., Chamberlain A. J., Schrooten C., Hayes B. J., and Goddard M. E.. 2016. Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Genet. 17:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manafiazar G., Basarab J., Baron V., McKeown L., Doce R., Swift M., Undi M., Wittenberg K., and Ominski K.. 2015. Effect of post-weaning residual feed intake classification on grazed grass intake and performance in pregnant beef heifers. Can. J. Anim. Sci. 95:369–381. [Google Scholar]
- Manafiazar G., Zimmerman S., and Basarab J.. 2016. Repeatability and variability of short-term spot measurement of methane and carbon dioxide emissions from beef cattle using GreenFeed emissions monitoring system. Can. J. Anim. Sci. 97:118–126. [Google Scholar]
- Mao F., Chen L., Vinsky M., Okine E., Wang Z., Basarab J., Crews D. H. Jr, and Li C.. 2013. Phenotypic and genetic relationships of feed efficiency with growth performance, ultrasound, and carcass merit traits in Angus and Charolais steers. J. Anim. Sci. 91:2067–2076. [DOI] [PubMed] [Google Scholar]
- McDonagh M., Herd R., Richardson E., Oddy V., Archer J., and Arthur P.. 2001. Meat quality and the calpain system of feedlot steers following a single generation of divergent selection for residual feed intake. Animal Prod. Sci. 41:1013–1021. [Google Scholar]
- Ng P. C., and Henikoff S.. 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acid Res. 31:3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega M. S., Denicol A. C., Cole J. B., Null D. J., and Hansen P. J.. 2016. Use of single nucleotide polymorphisms in candidate genes associated with daughter pregnancy rate for prediction of genetic merit for reproduction in Holstein cows. Anim. Genet. 47:288–297. [DOI] [PubMed] [Google Scholar]
- Ortega M. S., Denicol A. C., Cole J. B., Null D. J., Taylor J. F., Schnabel R. D., and Hansen P. J.. 2017. Association of single nucleotide polymorphisms in candidate genes previously related to genetic variation in fertility with phenotypic measurements of reproductive function in Holstein cows. J. Dairy Sci. 100:3725–3734. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J.,. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramayo-Caldas Y., Renand G., Ballester M., Saintilan R., and Rocha D.. 2016. Multi-breed and multi-trait co-association analysis of meat tenderness and other meat quality traits in three French beef cattle breeds. Genet. Sel. Evol. 48:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel L., Casas E., Shackelford S., and Wheeler T.. 2012. Relationship of polymorphisms within metabolic genes and carcass traits in crossbred beef cattle. J. Anim. Sci. 90:1311–1316. [DOI] [PubMed] [Google Scholar]
- Richard A. J., and Stephens J. M.. 2014. The role of JAK-STAT signaling in adipose tissue function. Biochim. Biophys. Acta. 1842:431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson E., Herd R., Oddy V., Thompson J., Archer J., and Arthur P.. 2001. Body composition and implications for heat production of Angus steer progeny of parents selected for and against residual feed intake. Animal Prod. Sci. 41:1065–1072. [Google Scholar]
- Richardson E. C., and Herd R. M.. 2004. Biological basis for variation in residual feed intake in beef cattle. 2. Synthesis of results following divergent selection. Aust. J. Exp. Agric. 44:431–440. [Google Scholar]
- Rincker C. B., Pyatt N. A., Berger L. L., and Faulkner D. B.. 2006. Relationship among GeneSTAR marbling marker, intramuscular fat deposition, and expected progeny differences in early weaned Simmental steers. J. Anim. Sci. 84:686–693. [DOI] [PubMed] [Google Scholar]
- Rolf M. M., Taylor J. F., Schnabel R. D., McKay S. D., McClure M. C., Northcutt S. L., Kerley M. S., and Weaber R. L.. 2012. Genome‐wide association analysis for feed efficiency in Angus cattle. Anim. Genet. 43:367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolf M. M., Decker J. E., McKay S. D., Tizioto P. C., Branham K. A., Whitacre L. K., Hoff J. L., Regitano L. C., and Taylor J. F.. 2014. Genomics in the United States beef industry. Livest. Sci. 166:84–93. [Google Scholar]
- Rolf M. M., Taylor J. F., Schnabel R. D., McKay S. D., McClure M. C., Northcutt S. L., Kerley M. S., and Weaber R. L.. 2010. Impact of reduced marker set estimation of genomic relationship matrices on genomic selection for feed efficiency in Angus cattle. BMC Genet. 11:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatchi M., Beever J. E., Decker J. E., Faulkner D. B., Freetly H. C., Hansen S. L., Yampara-Iquise H., Johnson K. A., Kachman S. D., Kerley M. S.,. et al. 2014a. QTLs associated with dry matter intake, metabolic mid-test weight, growth and feed efficiency have little overlap across 4 beef cattle studies. BMC Genet. 15:1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatchi M., Schnabel R. D., Taylor J. F., and Garrick D. J.. 2014b. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genomics 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel F. S., Miller S. P., Jiang Z., Mandell I. B., Ye X., Li H., and Wilton J. W.. 2006. Association of a single nucleotide polymorphism in the calpastatin gene with carcass and meat quality traits of beef cattle. J. Anim. Sci. 84:291–299. [DOI] [PubMed] [Google Scholar]
- Schenkel F. S., Miller S. P., and Wilton J. W.. 2004. Genetic parameters and breed differences for feed efficiency, growth, and body composition traits of young beef bulls. Can. J. Anim. Sci. 84:177–185. [Google Scholar]
- Serão N. V., González-Peña D., Beever J. E., Bollero G. A., Southey B. R., Faulkner D. B., and Rodriguez-Zas S. L.. 2013. Bivariate genome-wide association analysis of the growth and intake components of feed efficiency. PloS One 8:e78530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serão N. V., González-Peña D., Beever J. E., Faulkner D. B., Southey B. R., and Rodriguez-Zas S. L.. 2013. Single nucleotide polymorphisms and haplotypes associated with feed efficiency in beef cattle. BMC Genet. 14:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman E., Nkrumah J., Murdoch B., and Moore S.. 2008a. Identification of polymorphisms influencing feed intake and efficiency in beef cattle. Anim. Genet. 39:225–231. [DOI] [PubMed] [Google Scholar]
- Sherman E. L., Nkrumah J. D., Li C., Bartusiak R., Murdoch B., and Moore S. S.. 2009. Fine mapping quantitative trait loci for feed intake and feed efficiency in beef cattle1. J. Anim. Sci. 87:37–45. [DOI] [PubMed] [Google Scholar]
- Sherman E. L., Nkrumah J. D., Murdoch B. M., Li C., Wang Z., Fu A., and Moore S. S.. 2008b. Polymorphisms and haplotypes in the bovine neuropeptide Y, growth hormone receptor, ghrelin, insulin-like growth factor 2, and uncoupling proteins 2 and 3 genes and their associations with measures of growth, performance, feed efficiency, and carcass merit in beef cattle1. J. Anim. Sci. 86:1–16. [DOI] [PubMed] [Google Scholar]
- Stothard P., Liao X., Arantes A. S., De Pauw M., Coros C., Plastow G. S., Sargolzaei M., Crowley J. J., Basarab J. A., Schenkel F.,. et al. 2015. A large and diverse collection of bovine genome sequences from the Canadian Cattle Genome Project. GigaScience 4:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R. C. 2016. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- Uemoto Y., Sasaki S., Kojima T., Sugimoto Y., and Watanabe T.. 2015. Impact of QTL minor allele frequency on genomic evaluation using real genotype data and simulated phenotypes in Japanese Black cattle. BMC Genet. 16:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eenennaam A., Li J., Thallman R., Quaas R., Dikeman M., Gill C., Franke D., and Thomas M. . 2007. Validation of commercial DNA tests for quantitative beef quality traits. J. Anim. Sci. 85:891–900. [DOI] [PubMed] [Google Scholar]
- VanRaden P. M., Van Tassell C. P., Wiggans G. R., Sonstegard T. S., Schnabel R. D., Taylor J. F., and Schenkel F. S.. 2009. Invited review: reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 92:16–24. [DOI] [PubMed] [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang X.-H., Fan D.-X., Zhang Y., Li M.-Q., Wu H. -X., and Jin L.-P.. 2014. PCSK6 regulated by LH inhibits the apoptosis of human granulosa cells via activin A and TGFβ2. J. Endocrinol. 222:151–160. [DOI] [PubMed] [Google Scholar]
- Yao C., Spurlock D., Armentano L., Page C., VandeHaar M., Bickhart D., and Weigel K.. 2013. Random Forests approach for identifying additive and epistatic single nucleotide polymorphisms associated with residual feed intake in dairy cattle. J. Dairy Sci. 96:6716–6729. [DOI] [PubMed] [Google Scholar]
- Zeng Z.-B., Wang T., and Zou W.. 2005. Modeling quantitative trait loci and interpretation of models. Genetics 169:1711–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Gan Q., Hou G., Gao H., Li J., and Xu S.. 2015. Investigation of TG gene variants and their effects on growth, carcass composition, and meat quality traits in Chinese steers. Genet. Mol. Res. 14:5320–5326. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ding X., Liu J., Zhang Q., and de Koning D.-J.. 2011. Accuracy of genomic prediction using low-density marker panels. J. Dairy Sci. 94:3642–3650. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang W.. Li J. Li J. Zhong Z. Zhao, and Huang J.. 2013. Novel splice variants of the bovine PCK1 gene. Genet. Mol. Res. 12:4028–4035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.