Abstract
Olfactory receptors are G-protein-coupled chemoreceptors expressed on millions of olfactory sensory neurons within the nasal cavity. These receptors detect environmental odorants and signal the brain regarding the location of feed, potential mates, and the presence of possible threats (e.g., predators or chemical toxins). Olfactory receptors also are present in organs outside of the nasal cavity where they bind to molecules such as nutrients and metabolites from the animal’s internal environment to elicit physiological responses, including changes in gut motility, ventilation rate, and cellular migration. Recent evidence supports an additional role of olfactory receptors in the regulation of appetite in humans and rodents. In particular, genetic variation among individuals in specific odorant receptor genes has been linked to differences in their feeding behaviors, food choices, and the regulation of energy balance. This review provides a general overview of the olfactory receptors of vertebrates and their genetic variability and provides supporting evidence for a physiological role of olfactory receptors in appetite regulation of livestock. Basic research on olfactory receptors of livestock and their ligands should facilitate the development of novel odorant receptor agonists and identification of specific olfactory receptor variants that may be developed to enhance animal production efficiency.
Keywords: appetite regulation, feed intake, genetic variant, odorant, olfactory receptor
The first molecular characterization of the mammalian sense of smell by Buck and Axel (1991) earned the Nobel Prize in Physiology or Medicine in 2004. Since that time, olfactory receptor genes have been described and classified in livestock species including cattle, pigs, and chickens through comparative genomics. In addition, connections between the olfactory system and appetite have been investigated by neuroscientists and human nutritionists to gain a better understanding of human feeding behavior, food choice, and the regulation of energy balance, particularly to treat appetite-related disorders, such as obesity, bulimia nervosa, and anorexia nervosa (Ruijschop et al., 2009; Palouzier-Paulignan et al., 2012; Islam et al., 2015; Rolls, 2015). However, practically no research has been published on the links between the olfactory system and appetite regulation in production animals or how we might take advantage of these connections that have been demonstrated in humans and rodents to improve feed intake and intake-related traits (e.g., rate of gain and carcass composition) in livestock species. Therefore, the purpose of this review is to briefly describe the olfactory receptors of vertebrates and their genetic variability and to provide supporting evidence for a physiological role of olfactory receptors in appetite regulation of livestock. The goal is to promote interest in basic research on olfactory receptors of livestock species and their ligands in order to facilitate the development of novel odorant receptor (OR) agonists and genetic selection of particular receptor variants that enhance animal production.
STRUCTURE AND FUNCTION OF ORS
Two major regions of the nasal cavity of vertebrates function in odor perception. These are the olfactory epithelium and the vomeronasal organ, the latter of which may or may not be functional depending on the species (Spehr and Munger, 2009). These tissues contain different types of chemoreceptors, including ORs, vomeronasal receptors, and trace amine associate receptors that detect odorants, pheromones, and volatile amines, respectively, and transmit these chemical messages to the olfactory bulb (odorants and volatile amines) and accessory olfactory bulb (pheromones) of the brain (Buck, 2000; Liberles and Buck, 2006; Dalton and Lomvardas, 2015). The ORs, the focus of this review, are expressed on millions of olfactory sensory neurons within the olfactory epithelium, although each olfactory sensory neuron expresses only a single OR protein type on the cilia of its dendrites (Serizawa et al., 2004; Hayden and Teeling, 2014). However, each OR can bind and detect multiple odorant molecules, and conversely, each odorant can bind to multiple ORs with varying binding affinities, creating a distinct pattern of odorant binding and OR activation which enables animals to discriminate very diverse and complex odors (Malnic et al., 1999). These chemical messages are essential for animal survival by assisting animals in locating feed, detecting toxins in the environment, alerting them to the presence of predators, and identifying and selecting potential mates (Spehr and Munger, 2009). It also is becoming clearer that ORs may have an additional role in the regulation of appetite.
Structurally, ORs are classified as G-protein-coupled receptors, which in classical models bind odorants within the nasal mucus and initiate nerve impulses that are carried to the brain (Purves et al., 2001). Specifically, odorant binding to the receptor causes activation of the G-protein, Golf, which then stimulates the conversion of ATP to cAMP via adenylate cyclase activation. The cAMP opens cyclic nucleotide-gated ion channels, allowing Ca2+ and Na+ ions to enter the cell and opening of Ca2+-gated Cl− channels that permit Cl− movement out of the cell. The movement of ions results in depolarization of the olfactory sensory neuron and creates an action potential that is carried along the axon to the brain. It is now understood that OR expression is not limited to the olfactory epithelium of the nasal cavity. There is also “ectopic” expression in many organs, such as liver (Wu et al., 2015), pancreas (Kang et al., 2015), and lung (An and Liggett, 2018) where ORs can detect the internal environment of the animal and impact a myriad of other physiological processes (reviewed by Kang and Koo, 2012 and Chen et al., 2018). These include chemotaxis of sperm (Flegel et al., 2016) and migration of muscle cells during differentiation (Pavlath, 2010), regulation of blood pressure via changes in circulating concentrations of VFA that bind ORs in the kidney and stimulate renin secretion (Natarajan and Pluznick, 2016), and changes in respiration rate (e.g., through binding of lactate during hypoxia to ORs expressed in the carotid artery to stimulate hyperventilation; Chang et al., 2015) as well as alteration of gut motility. For example, OR-expressing enterochromaffin cells in the gut are activated by compounds like eugenol to release serotonin that affects gut motility (Braun et al., 2007) and increases satiety (Voigt and Fink, 2015). Of interest, unlike the olfactory sensory neurons of the nasal cavity, more than one receptor type may be expressed per cell in these tissues. For instance, Flegel et al. (2016) detected approximately 90 different ORs expressed in human sperm cells which varied in their distribution among the acrosomal cap, head, and flagellar regions. These ORs present in the olfactory epithelium as well as other tissues detect a variety of chemical cues that may ultimately impact endocrine and metabolic centers, affecting feeding behavior or appetite as described in the following section.
EVIDENCE FOR FUNCTIONAL ROLE OF ORS IN APPETITE REGULATION
Experimental evidence indicates that specific odorants can activate ORs in the olfactory epithelium to influence animal appetite. The effects of these odorants appear to be mediated by changes in secretion of orexigenic or anorexigenic neuropeptides as well as activity of the gastric vagal nerves. For example, a 10-min exposure of rats to grapefruit oil or its primary odorant called limonene was shown to inhibit the activity of efferent vagal nerves innervating the stomach (Shen et al., 2005a), which should reduce gastric emptying. Furthermore, in the same study, a 15-min exposure to these compounds three times per week for 6 wk reduced food intake and body weight of rats. On the contrary, the same exposures to lavender oil or its main odorant linalool was shown to stimulate activity of gastric vagal nerves and increase rat food intake and BW (Shen et al., 2005b; Tanida et al., 2006). In each of these studies, the responses were abolished by local nasal mucosa anesthesia or anosmia induced by ZnSO4 treatment of the nasal cavity, indicating the necessity for olfactory stimulation in the observed responses. Furthermore, it was shown that a 10 min exposure to these compounds not only affects mRNA expression of the appetite stimulator neuropeptide Y in rat olfactory nerve cells (Rolf B1.T) and primary rat olfactory ensheathing cells but inhalation of essences of limonene and linalool also alters circulating concentrations of NPY in human subjects (Chen et al., 2012). That is, consistent with a stronger appetite induced by linalool, both NPY mRNA and serum NPY were increased in these studies, and the opposite was observed with limonene.
There is also evidence that some ectopic ORs may play a role in appetite regulation, including mouse OR51E2. Fleischer et al. (2015) used transgenic mice that express green fluorescent protein on OR51E2 to demonstrate that this receptor is expressed on enteroendocrine L cells within the crypts of the colon and that all OR51E2-positive cells within the mouse colon coexpress the appetite-regulating gut hormone peptide YY (PYY). The authors interpreted this finding as a likely functional connection between OR51E2 and PYY release. Yet, only about a third of PYY-positive cells co-expressed OR51E2, indicating that regulation of PYY secretion likely involves additional mechanisms beyond the potential role of OR51E2. Additionally, a small percentage of OR51E2-positive cells in this study also co-expressed glucagon-like peptide 1 (GLP-1). This is of particular interest in the context of appetite regulation as both PYY and GLP-1 are believed to mediate what is known as the “ileal brake,” a feedback mechanism whereby the introduction of nutrients (especially energy-rich foods) to the lower gut results in PYY and GLP-1 release from L cells, slowing of gut emptying, reduced gut motility, and an increased sense of satiety (Spreckley and Murphy, 2015). It was further demonstrated by Fleischer et al. (2015) that propionate is an activator of OR51E2, providing evidence that this OR may be a critical link in appetite regulation; whereby, propionate derived from microbial digestion in the colon could activate OR51E2 and stimulate the release of PYY and GLP-1 from L cells as a satiety signal.
A very similar OR protein, OR51E1, was shown to be expressed throughout the gastrointestinal tract of pigs, with the highest concentration in the region connecting the stomach to the duodenum (Priori et al., 2015). Notably, this OR was shown to colocalize on enteroendocrine cells containing PYY and serotonin (Priori et al., 2015), both of which regulate gut motility and appetite (Braun et al., 2007; Spreckley and Murphy, 2015; Voigt and Fink, 2015). One of the ligands of this OR is butyrate (Adipietro et al., 2012), produced by microbes in the intestinal lumen. Indeed, there appears to be evidence for mechanistic links between ORs and appetite regulation not only in rodents and humans, but also in a livestock species.
Lastly, there are notable connections among nutrient-sensing mechanisms and hormones that regulate energy balance and appetite with the olfactory and central nervous systems which have been reviewed extensively by Palouzier-Paulignan et al. (2012) and Julliard et al. (2017). The notion is that activity of the olfactory system is heavily influenced by hormonal, nutritional, and metabolic cues of energy balance that impact animal odor-related behaviors like feed intake and food preference to assist in maintaining their energy homeostasis. Hormonal examples include orexins A and B, which stimulate food intake, and their receptors that are expressed at the transcript and protein levels in both olfactory sensory neurons and surrounding areas of the olfactory mucosa, as well as the hypothalamus (Caillol et al., 2003). Likewise, leptin, which suppresses appetite, and its receptors are expressed on olfactory receptor neurons and in the hypothalamus (Baly et al., 2007). These hormonal signals, along with activation of other nutrient receptors (e.g., members of the solute carrier transporter family; Julliard et al., 2017), can directly impact olfactory sensitivity and odorant detection through changes in the frequency and amplitude of neuronal firing as well as the expression of odorant binding proteins that present odorants to ORs in the olfactory mucosa (Palouzier-Paulignan et al., 2012). Namely, as demonstrated in rodents, fasting increases olfactory sensitivity and time spent exploring food-related odors, and satiety decreases them (Prud’homme et al., 2009), which ultimately could impact feeding behavior and feed intake. Clearly, expression of appetite-regulating peptides and their receptors in addition to nutrient receptors on olfactory neurons and their proximity to the hypothalamus provide opportunities for extensive cross talk between olfaction and appetite regulation begging further exploration in livestock species.
GENETIC STRUCTURE OF ORS
The OR gene family comprises the largest gene family in mammals (Gilad et al., 2005; Fleischer et al., 2009). In humans, the OR family comprises ~30 Mb, or 1% of the genome, where OR genes are distributed across nearly all chromosomes, but often occur in clusters within a chromosome (Glusman et al., 2001). For example, 313 intact OR genes are located on human chromosome 11 alone, whereas nine other chromosomes have only one to five intact OR genes, and chromosomes 8, 20, and the Y chromosome are devoid of any OR genes (Malnic et al., 2004). Numerous OR pseudogenes (~300) also exist (Malnic et al., 2004) in which a mutation has occurred resulting in a nonfunctional OR protein. Genomic studies indicate that the number of OR genes is highly variable across species and may be related to each species’ ecology as well as its dependence on smell vs. other senses like vision for locating feed (Vandewege et al., 2016). For instance, bottlenose dolphins, which live in an aquatic environment and depend on echolocation for locating prey, have fewer than 30 OR genes of which only about half are functional, whereas cattle and rodents have between 1,100 and 1,600 functional OR genes (Hayden et al., 2010; Lee et al., 2013). Notably, the African elephant may have the greatest number of functional OR genes at about 2,000 which is believed to provide them with a very keen sense of smell (Hayden et al., 2010; Niimura et al., 2014). In fact, it has been proposed that African elephants have the ability to distinguish between ethnic groups of people based on smell. Specifically, exposure to odors of the Kenyan Maasai tribe who routinely hunt elephants elicit a greater fear response by African elephants than do odors of the Kamba tribe who lead an agricultural lifestyle and do not hunt elephants (Bates et al., 2007).
Extensive genetic variation exists in human OR genes which impacts olfactory function. For example, it is estimated that 66% of OR genes contain insertions/deletions, SNPs, and copy number variations (CNVs; Olender et al., 2012). CNVs are large segments of DNA (>1 Kb) that are repeated within the genome, and the number of repeats of this region varies from one individual to another (Freeman et al., 2006). The genetic variations can change the encoded AA, create nonsense codons in encoded ORs, or in the case of CNVs, differences in the expression level of a particular OR. Thus, genetic variation in ORs contributes to differences among individuals in their ability to smell, their sensitivity to different odors, which can differ by several orders of magnitude, as well as odor or food preferences, and feed intake (Keller et al., 2007; Hasin-Brumshtein et al., 2009; Choquette et al., 2012). For instance, SNPs in genes like OR11H7P and OR6A2 are associated with sensitivity to the sweaty smell of isovaleric acid (Menashe et al., 2007) and preference for the herb cilantro (Eriksson et al., 2012), respectively. An SNP in OR2J3 affects the ability to detect the grassy smell of cis-3-hexen-1-ol (McRae et al., 2012), and an SNP in OR5A1, which results in an AA change in one of the extracellular loops of the receptor, affects the preference for the floral scent of beta ionone and food choices (Jaeger et al., 2013). Lastly, SNPs in human OR7D4 affect the degree of aversion of individuals to the smell of androstenone, one of the compounds responsible for boar taint (Keller et al., 2007). Polymorphisms in this gene are also associated with human susceptibility to hunger and body mass index (Choquette et al., 2012), providing a possible link between OR variation and human appetite regulation.
Lastly, similar to humans, considerable variation has been shown to exist among OR genes of cattle and swine. For instance, 40% of OR genes sampled by Lee et al. (2013) exhibited CNVs across and within cattle breeds. Likewise, 66% of OR genes overlap CNV regions in the pig genome (Paudel et al., 2015). Thus, these CNVs within the OR gene loci provide substantial opportunity for animal-to-animal variation in OR function and impacts on phenotype, such as appetite or feed intake.
ORS VARIATION ASSOCIATED WITH LIVESTOCK PRODUCTION TRAITS
To date, a few genome-wide association studies have reported associations between OR genetic variants and feed intake or intake-related traits of livestock, such as rate of gain, carcass composition, and residual feed intake (RFI), defined as the difference between observed and expected feed intake based on production level proposed by Koch et al. (1963). For example, Veerkamp et al. (2012) identified 500 genes that were located near SNPs significantly associated with the traits of BW, DMI, or BCS in European first-parity Holstein dairy cows. These genes were enriched in olfactory, taste, and pheromone receptors, suggesting a functional role for variation in OR genes in regulation of feed intake of dairy cows. Similarly, a genome-wide association study of pigs identified 25 OR genes located near SNPs significantly associated with RFI (Do et al., 2014), and another study in beef cattle identified OR9Q2 as a positional candidate gene contributing to RFI in the SimAngus breed (Seabury et al., 2017). Thus, olfactory transduction may be an important biological pathway contributing to variation in feed conversion efficiency measured as RFI. Furthermore, a study of Nellore cattle identified three OR genes (OR2D3, OR2D2, and OR6A2) within the 46 Mb region of BTA 15 associated with DMI, two OR (OR52J3 and OR51A7) in the 50 Mb region of BTA 15 associated with ADG, and OR9A4 in the 105.9 Mb region of BTA 4 associated with RFI (Olivieri et al., 2016). A second study in Nellore cattle also identified a cluster of OR within the 31 to 32 Mb region of BTA 5 associated with carcass marbling (Magalhães et al., 2016). Of interest, a study of beef steers of various breeds exhibiting divergent residual weight gain also showed differential mRNA expression in ileum of LOC618173, an OR similar to OR52K1 that was identified in a previous genome-wide association study as associated with weight gain in crossbred beef cattle (Lindholm-Perry et al., 2015). However, differences in ileal expression of this transcript could not be confirmed by the authors within a separate beef cattle population differing in ADG and ADFI. Finally, our laboratory recently completed a genome-wide analysis of CNVs and their association with production traits of Holstein dairy cows including DMI and RFI and identified significant CNVs associated with both traits overlapping OR genes (Zhou et al., unpublished data): first, OR2A2 gene, located on BTA and a second CNV region associated with RFI that includes two other ORs, OR2T12 and OR2AK2. Collectively, these results indicate that multiple ORs are positional candidate genes contributing to differences in feeding and appetite-related traits of livestock and may play an important role in appetite regulation.
MANIPULATING APPETITE VIA ORS
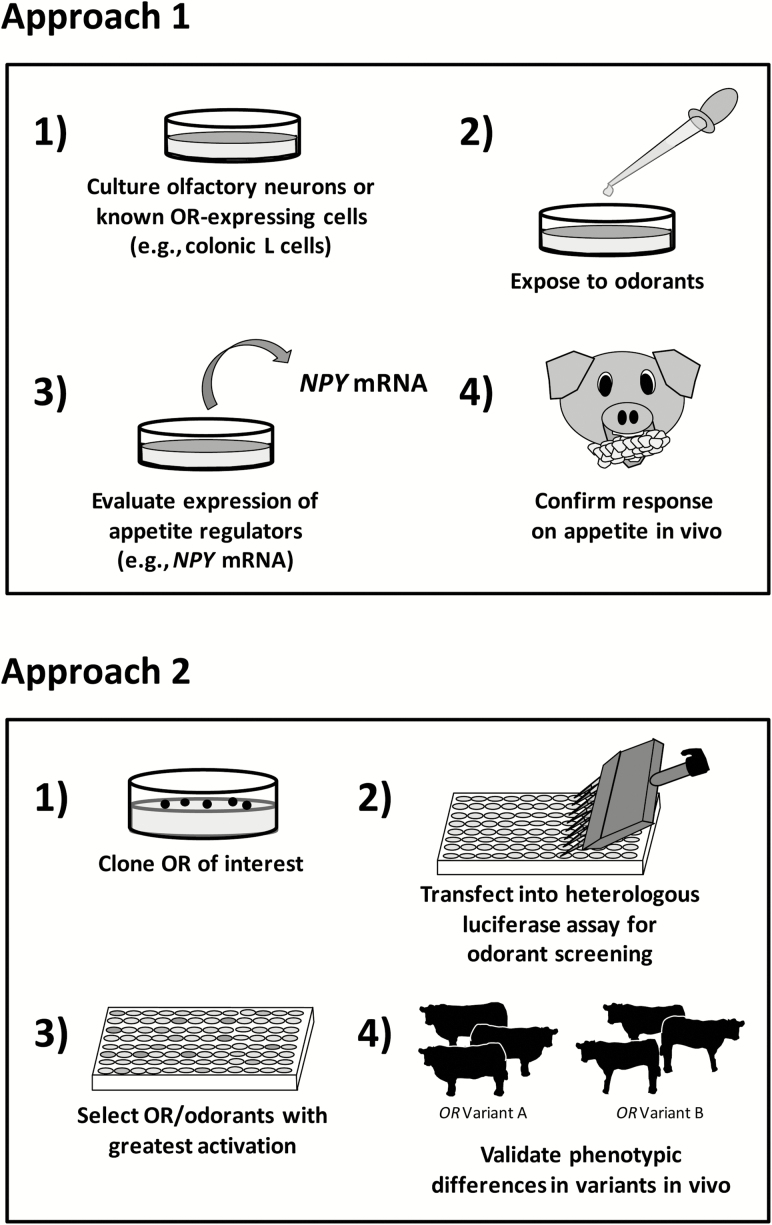
Overall, there appears to be reasonable evidence to support a functional link between ORs and appetite regulation in production animals and the potential to manipulate appetite and feed intake via direct OR activation or selection for particular OR genetic variants. Herein, we propose two approaches by which specific ORs or odorants could be identified and studied for targeted manipulation in livestock species based on prior techniques described for rodents and humans (Figure 1). First, one could isolate and culture-specific OR-expressing cells such as primary colonic L cells or olfactory neurons from the species of interest and then quantify their mRNA expression or protein production of known appetite regulators (e.g., NPY or PYY) in response to exposure to various odorants. Then, odorants with the desired effects in vitro could be evaluated in live animals to determine whether they produce the desired effects on appetite and feed intake. As described earlier, this approach was employed by Chen et al. (2012) who exposed Rolf B2.T cells or cultured rat olfactory ensheathing cells to differing concentrations of linalool or limonene for various time periods to assess the cellular responses in NPY mRNA expression. The NPY mRNA expression concentrations served to indicate the appetite-regulating potential of these compounds and provided a model system to screen additional odorants of interest.
Figure 1.
Proposed approaches by which specific ORs or odorants could be identified and studied for targeted manipulation in livestock species based on prior techniques described for rodents and humans.
Second, specific ORs from the species of interest (including different genetic variants of a particular receptor) could be cloned, transfected into a heterologous luciferase assay, and used as a high-throughput screening tool to evaluate receptor reactivity to various odorants (Zhuang and Matsunami, 2008; Mainland et al., 2014). Specifically, different OR clones could be arrayed into a 96-well (or greater) assay plate and exposed to putative appetite-altering odorants of choice to determine the receptors (and variants) with the greatest activation potentials based on luciferase assay. Alternatively, a single OR of interest could be screened against multiple appetite-altering odorants and concentrations to identify the odorant with the greatest stimulating ability. Ultimately, selected OR–odorant combinations could be targeted for further evaluation in live animals to confirm anticipated effects on DMI or feed preference as well as OR variant associations with other appetite-related production traits such as ADG or carcass marbling.
SUMMARY AND CONCLUSIONS
In summary, there appears to be reasonable evidence suggesting a link between olfactory receptors and appetite regulation, and variation in these receptors could contribute to differences in individuals in terms of feed intake, weight gain, and body composition. There are practical applications for developing novel compounds to activate various receptors to manipulate these processes in production animals or to select for particular OR genetic variants within the population to support desired outcomes on livestock production. Basic research focused on olfactory receptors of production animals and their ligands to regulate appetite has yet to be explored and provides an enormous opportunity to enhance appetite-related traits, such as feed intake, weight gain, and carcass composition for greater production efficiency.
ACKNOWLEDGMENTS
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
Footnotes
Present address: Key Laboratory of Agricultural Animal Genetics, Breeding, and Reproduction, Education Ministry of China, Huazhong Agricultural University, Wuhan, China.
LITERATURE CITED
- Adipietro K. A., Mainland J. D., and Matsunami H.. 2012. Functional evolution of mammalian odorant receptors. PLoS Genet. 8:e1002821. doi:10.1371/journal.pgen.1002821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S. S., and Liggett S. B.. 2018. Taste and smell GPCRs in the lung: evidence for a previously unrecognized widespread chemosensory system. Cell. Signal. 41:82–88. doi:10.1016/j.cellsig.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baly C., Aioun J., Badonnel K., Lacroix M. C., Durieux D., Schlegel C., Salesse R., and Caillol M.. 2007. Leptin and its receptors are present in the rat olfactory mucosa and modulated by the nutritional status. Brain Res. 1129:130–141. doi:10.1016/j.brainres.2006.10.030 [DOI] [PubMed] [Google Scholar]
- Bates L. A., Sayialel K. N., Njiraini N. W., Moss C. J., Poole J. H., and Byrne R. W.. 2007. Elephants classify human ethnic groups by odor and garment color. Curr. Biol. 17:1938–1942. doi:10.1016/j.cub.2007.09.060 [DOI] [PubMed] [Google Scholar]
- Braun T., Voland P., Kunz L., Prinz C., and Gratzl M.. 2007. Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology 132:1890–1901. doi:10.1053/j.gastro.2007.02.036 [DOI] [PubMed] [Google Scholar]
- Buck L. B. 2000. The molecular architecture of odor and pheromone sensing in mammals. Cell 100:611–618. doi:10.1016/S0092-8674(00)80698-4 [DOI] [PubMed] [Google Scholar]
- Buck L. and Axel R.. 1991. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65:175–187. doi:10.1016/0092-8674(91)90418-X [DOI] [PubMed] [Google Scholar]
- Caillol M., Aïoun J., Baly C., Persuy M. A., and Salesse R.. 2003. Localization of orexins and their receptors in the rat olfactory system: possible modulation of olfactory perception by a neuropeptide synthetized centrally or locally. Brain Res. 960:48–61. doi:10.1016/S0006-8993(02)03755-1 [DOI] [PubMed] [Google Scholar]
- Chang A. J., Ortega F. E., Riegler J., Madison D. V., and Krasnow M. A.. 2015. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature 527:240–244. doi:10.1038/nature15721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. W., Wu P. J., and Chiang B. H.. 2012. In vitro neuropeptide Y mRNA expressing model for screening essences that may affect appetite using Rolf B1.T cells. J. Agric. Food Chem. 60:7824–7829. doi:10.1021/jf302320f [DOI] [PubMed] [Google Scholar]
- Chen Z., Zhao H., Fu N., and Chen L.. 2018. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J. Cell. Physiol. 233:2104–2115. doi:10.1002/jcp.25929 [DOI] [PubMed] [Google Scholar]
- Choquette A. C., Bouchard L., Drapeau V., Lemieux S., Tremblay A., Bouchard C., Vohl M. C., and Pérusse L.. 2012. Association between olfactory receptor genes, eating behavior traits and adiposity: results from the Quebec Family Study. Physiol. Behav. 105:772–776. doi:10.1016/j.physbeh.2011.10.015 [DOI] [PubMed] [Google Scholar]
- Dalton R. P. and Lomvardas S.. 2015. Chemosensory receptor specificity and regulation. Annu. Rev. Neurosci. 38:331–349. doi:10.1146/annurev-neuro-071714-034145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do D. N., Strathe A. B., Ostersen T., Pant S. D., and Kadarmideen H. N.. 2014. Genome-wide association and pathway analysis of feed efficiency in pigs reveal candidate genes and pathways for residual feed intake. Front. Genet. 5:307. doi:10.3389/fgene.2014.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson N., Wu S., Do C. B., Kiefer A. K., Tung J. Y., Mountain J. L., Hinds D. A., and Francke U.. 2012. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour 1:22. doi:10.1186/2044-7248-1-22 [Google Scholar]
- Flegel C., Vogel F., Hofreuter A., Schreiner B. S., Osthold S., Veitinger S., Becker C., Brockmeyer N. H., Muschol M., Wennemuth G.,. et al. 2016. Characterization of the olfactory receptors expressed in human spermatozoa. Front. Mol. Biosci. 2:73. doi:10.3389/fmolb.2015.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer J., Breer H., and Strotmann J.. 2009. Mammalian olfactory receptors. Front. Cell. Neurosci. 3:9. doi:10.3389/neuro.03.009.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer J., Bumbalo R., Bautze V., Strotmann J., and Breer H.. 2015. Expression of odorant receptor Olfr78 in enteroendocrine cells of the colon. Cell Tissue Res. 361:697–710. doi:10.1007/s00441-015-2165-0 [DOI] [PubMed] [Google Scholar]
- Freeman J. L., Perry G. H., Feuk L., Redon R., McCarroll S. A., Altshuler D. M., Aburatani H., Jones K. W., Tyler-Smith C., Hurles M. E.,. et al. 2006. Copy number variation: new insights in genome diversity. Genome Res. 16:949–961. doi:10.1101/gr.3677206 [DOI] [PubMed] [Google Scholar]
- Gilad Y., Rifkin S. A., Bertone P., Gerstein M., and White K. P.. 2005. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 15:674–680. doi:10.1101/gr.3335705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G., Yanai I., Rubin I., and Lancet D.. 2001. The complete human olfactory subgenome. Genome Res. 11:685–702. doi:10.1101/gr.171001 [DOI] [PubMed] [Google Scholar]
- Hasin-Brumshtein Y., Lancet D., and Olender T.. 2009. Human olfaction: from genomic variation to phenotypic diversity. Trends Genet. 25:178–184. doi:10.1016/j.tig.2009.02.002 [DOI] [PubMed] [Google Scholar]
- Hayden S., Bekaert M., Crider T. A., Mariani S., Murphy W. J., and Teeling E. C.. 2010. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 20:1–9. doi:10.1101/gr.099416.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S. and Teeling E. C.. 2014. The molecular biology of vertebrate olfaction. Anat. Rec. (Hoboken). 297:2216–2226. doi:10.1002/ar.23031 [DOI] [PubMed] [Google Scholar]
- Islam M. A., Fagundo A. B., Arcelus J., Agüera Z., Jiménez-Murcia S., Fernández-Real J. M., Tinahones F. J., de la Torre R., Botella C., Frühbeck G.,. et al. 2015. Olfaction in eating disorders and abnormal eating behavior: a systematic review. Front. Psychol. 6:1431. doi:10.3389/fpsyg.2015.01431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger S. R., McRae J. F., Bava C. M., Beresford M. K., Hunter D., Jia Y., Chheang S. L., Jin D., Peng M., Gamble J. C.,. et al. 2013. a MENDELIAN trait for olfactory sensitivity affects odor experience and food selection. Curr. Biol. 23:1601–1605. doi:10.1016/j.cub.2013.07.030 [DOI] [PubMed] [Google Scholar]
- Julliard A. K., Al Koborssy D., Fadool D. A., and Palouzier-Paulignan B.. 2017. Nutrient sensing: another chemosensitivity of the olfactory system. Front. Physiol. 8:468. doi:10.3389/fphys.2017.00468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang N., Bahk Y. Y., Lee N., Jae Y., Cho Y. H., Ku C. R., Byun Y., Lee E. J., Kim M. S., and Koo J.. 2015. Olfactory receptor Olfr544 responding to azelaic acid regulates glucagon secretion in α-cells of mouse pancreatic islets. Biochem. Biophys. Res. Commun. 460:616–621. doi:10.1016/j.bbrc.2015.03.078 [DOI] [PubMed] [Google Scholar]
- Kang N. and Koo J.. 2012. Olfactory receptors in non-chemosensory tissues. BMB Rep. 45:612–622. doi:10.5483/BMBRep.2012.45.11.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Zhuang H., Chi Q., Vosshall L. B., and Matsunami H.. 2007. Genetic variation in a human odorant receptor alters odour perception. Nature 449:468–472. doi:10.1038/nature06162 [DOI] [PubMed] [Google Scholar]
- Koch R. M., Swiger L. A., Chambers D., and Gregory K. E.. 1963. Efficiency of feed use in beef cattle. J. Anim. Sci. 22:486–494. [Google Scholar]
- Lee K., Nguyen D. T., Choi M., Cha S. Y., Kim J. H., Dadi H., Seo H. G., Seo K., Chun T., and Park C.. 2013. Analysis of cattle olfactory subgenome: the first detail study on the characteristics of the complete olfactory receptor repertoire of a ruminant. BMC Genomics 14:596. doi:10.1186/1471-2164-14-596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles S. D. and Buck L. B.. 2006. a second class of chemosensory receptors in the olfactory epithelium. Nature 442:645–650. doi:10.1038/nature05066 [DOI] [PubMed] [Google Scholar]
- Lindholm-Perry A. K., Kern R. J., Kuehn L. A., Snelling W. M., Miles J. R., Oliver W. T., and Freetly H. C.. 2015. Differences in transcript abundance of genes on BTA15 located within a region associated with gain in beef steers. Gene 572:42–48. doi:10.1016/j.gene.2015.06.076 [DOI] [PubMed] [Google Scholar]
- Magalhães A. F. B., de Camargo G. M. F., Junior G. A. F., Gordo D. G. M., Tonussi R. L., Costa R. B., Espigolan R., Silva R. M.de. O., Bresolin T., de Andrade W. B. F.,. et al. 2016. Genome-wide association study of meat quality traits in Nellore cattle. PLoS One 11:e0157845. doi:10.1371/journal.pone.0157845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainland J. D., Keller A., Li Y. R., Zhou T., Trimmer C., Snyder L. L., Moberly A. H., Adipietro K. A., Liu W. L., Zhuang H.,. et al. 2014. The missense of smell: functional variability in the human odorant receptor repertoire. Nat. Neurosci. 17:114–120. doi:10.1038/nn.3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B., Godfrey P. A., and Buck L. B.. 2004. The human olfactory receptor gene family. Proc. Natl. Acad. Sci. USA 101:2584–2589. doi:10.1073/pnas.0307882100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B., Hirono J., Sato T., and Buck L. B.. 1999. Combinatorial receptor codes for odors. Cell 96:713–723. doi: 10.1016/S0092-8674(00)80581-4 [DOI] [PubMed] [Google Scholar]
- McRae J. F., Mainland J. D., Jaeger S. R., Adipietro K. A., Matsunami H., and Newcomb R. D.. 2012. Genetic variation in the odorant receptor OR2J3 is associated with the ability to detect the “grassy” smelling odor, cis-3-hexen-1-ol. Chem. Senses 37:585–593. doi:10.1093/chemse/bjs049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I., Abaffy T., Hasin Y., Goshen S., Yahalom V., Luetje C. W., and Lancet D.. 2007. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 5:e284. doi:10.1371/journal.pbio.0050284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan N. and Pluznick J. L.. 2016. Olfaction in the kidney: ‘smelling’ gut microbial metabolites. Exp. Physiol. 101:478–481. doi:10.1113/EP085285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y., Matsui A., and Touhara K.. 2014. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 24:1485–1496. doi:10.1101/gr.169532.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender T., Waszak S. M., Viavant M., Khen M., Ben-Asher E., Reyes A., Nativ N., Wysocki C. J., Ge D., and Lancet D.. 2012. Personal receptor repertoires: olfaction as a model. BMC Genomics 13:414. doi:10.1186/1471-2164-13-414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri B. F., Mercadante M. E., Cyrillo J. N., Branco R. H., Bonilha S. F., de Albuquerque L. G., Silva R. M., and Baldi F.. 2016. Genomic regions associated with feed efficiency indicator traits in an experimental Nellore cattle population. PLoS One 11:e0164390. doi:10.1371/journal.pone.0164390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palouzier-Paulignan B., Lacroix M. C., Aimé P., Baly C., Caillol M., Congar P., Julliard A. K., Tucker K., and Fadool D. A.. 2012. Olfaction under metabolic influences. Chem. Senses 37:769–797. doi:10.1093/chemse/bjs059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel Y., Madsen O., Megens H. J., Frantz L. A., Bosse M., Crooijmans R. P., and Groenen M. A.. 2015. Copy number variation in the speciation of pigs: a possible prominent role for olfactory receptors. BMC Genomics 16:330. doi:10.1186/s12864-015-1449-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlath G. K. 2010. a new function for odorant receptors: mor23 is necessary for normal tissue repair in skeletal muscle. Cell Adh. Migr. 4:502–506. doi:10.4161/cam.4.4.12291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priori D., Colombo M., Clavenzani P., Jansman A. J., Lallès J. P., Trevisi P., and Bosi P.. 2015. The olfactory receptor OR51E1 is present along the gastrointestinal tract of pigs, co-localizes with enteroendocrine cells and is modulated by intestinal microbiota. PLoS One 10:e0129501. doi:10.1371/journal.pone.0129501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme M. J., Lacroix M. C., Badonnel K., Gougis S., Baly C., Salesse R., and Caillol M.. 2009. Nutritional status modulates behavioural and olfactory bulb Fos responses to isoamyl acetate or food odour in rats: roles of orexins and leptin. Neuroscience 162:1287–1298. doi:10.1016/j.neuroscience.2009.05.043 [DOI] [PubMed] [Google Scholar]
- Purves D., Augustine G. J., Fitzpatrick D., Katz L. C., LaMantia A.-S., McNamara J. O., and Williams S. M.. 2001. Neuroscience, 2nd ed Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Rolls E. T. 2015. Taste, olfactory, and food reward value processing in the brain. Prog. Neurobiol. 127–128:64–90. doi:10.1016/j.pneurobio.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Ruijschop R. M., Boelrijk A. E., de Graaf C., and Westerterp-Plantenga M. S.. 2009. Retronasal aroma release and satiation: a review. J. Agric. Food Chem. 57:9888–9894. doi:10.1021/jf901445z [DOI] [PubMed] [Google Scholar]
- Seabury C. M., Oldeschulte D. L., Saatchi M., Beever J. E., Decker J. E., Halley Y. A., Bhattarai E. K., Molaei M., Freetly H. C., Hansen S. L.,. et al. 2017. Genome-wide association study for feed efficiency and growth traits in U.S. beef cattle. BMC Genomics 18:386. doi:10.1186/s12864-017-3754-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S., Miyamichi K., and Sakano H.. 2004. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 20:648–653. doi:10.1016/j.tig.2004.09.006 [DOI] [PubMed] [Google Scholar]
- Shen J., Niijima A., Tanida M., Horii Y., Maeda K., and Nagai K.. 2005a. Olfactory stimulation with scent of grapefruit oil affects autonomic nerves, lipolysis and appetite in rats. Neurosci. Lett. 380:289–294. doi:10.1016/j.neulet.2005.01.058 [DOI] [PubMed] [Google Scholar]
- Shen J., Niijima A., Tanida M., Horii Y., Maeda K., and Nagai K.. 2005b. Olfactory stimulation with scent of lavender oil affects autonomic nerves, lipolysis and appetite in rats. Neurosci. Lett. 383:188–193. doi:10.1016/j.neulet.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Spehr M. and Munger S. D.. 2009. Olfactory receptors: G protein-coupled receptors and beyond. J. Neurochem. 109:1570–1583. doi:10.1111/j.1471-4159.2009.06085.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreckley E. and Murphy K. G.. 2015. The L-cell in nutritional sensing and the regulation of appetite. Front. Nutr. 2:23. doi:10.3389/fnut.2015.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida M., Niijima A., Shen J., Nakamura T., and Nagai K.. 2006. Olfactory stimulation with scent of lavender oil affects autonomic neurotransmission and blood pressure in rats. Neurosci. Lett. 398:155–160. doi:10.1016/j.neulet.2005.12.076 [DOI] [PubMed] [Google Scholar]
- Vandewege M. W., Mangum S. F., Gabaldón T., Castoe T. A., Ray D. A., and Hoffmann F. G.. 2016. Contrasting patterns of evolutionary diversification in the olfactory repertoires of reptile and bird genomes. Genome Biol. Evol. 8:470–480. doi:10.1093/gbe/evw013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerkamp R. F., Coffey M., Berry D., de Haas Y., Strandberg E., Bovenhuis H., Calus M., and Wall E.. 2012. Genome-wide associations for feed utilisation complex in primiparous Holstein–Friesian dairy cows from experimental research herds in four European countries. Animal 6:1738–1749. doi:10.1017/S1751731112001152 [DOI] [PubMed] [Google Scholar]
- Voigt J. P. and Fink H.. 2015. Serotonin controlling feeding and satiety. Behav. Brain Res. 277:14–31. doi:10.1016/j.bbr.2014.08.065 [DOI] [PubMed] [Google Scholar]
- Wu C., Jia Y., Lee J. H., Kim Y., Sekharan S., Batista V. S., and Lee S. J.. 2015. Activation of OR1A1 suppresses PPAR-γ expression by inducing HES-1 in cultured hepatocytes. Int. J. Biochem. Cell Biol. 64:75–80. doi:10.1016/j.biocel.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Zhuang H. and Matsunami H.. 2008. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat. Protoc. 3:1402–1413. doi:10.1038/nprot.2008.120 [DOI] [PMC free article] [PubMed] [Google Scholar]