Abstract
The increasing world population is driving demand for improved efficiency of feed resources of livestock. However, the molecular mechanisms behind various feed efficiency traits and their regulation by nutrition remain poorly understood. Here, we aimed to identify differentially expressed (DE) genes in the liver tissues of fattening Merino lambs and differences in metabolites accumulated in plasma to identify modified metabolic pathways as a consequence of milk restriction during the suckling period. Twenty-four male Merino lambs (4.81 ± 0.256 kg) were divided into 2 groups (n = 12 per dietary treatment). The first group (ad libitum, ADL) was kept permanently with the dams, whereas the other group (restricted, RES) was milk restricted. When they reached 15 kg of live body weight (LBW), all the animals were offered the same complete pelleted diet at the same level (35 g DM/kg LBW per day) to ensure no differences in dry matter intake. All the lambs were harvested when they reached 27 kg of LBW. For transcriptomic analysis, 4 liver samples from each group (8 samples in total) were selected for RNA sequencing (RNA-seq), and plasma samples from all animals (24 samples in total) were used to perform a nontargeted metabolomic analysis on a hybrid quadrupole-time-of-flight mass spectrometer coupled to an ultra-high performance liquid chromatographic system (UHPLC/QTOF-MS). Thirty-eight DE annotated genes were identified by RNA-seq, with 23 DE genes being down-regulated and 15 up-regulated in the liver of RES lambs relative to the ADL group (P < 0.10). Also, the metabolomic assay identified 38 differentially accumulated compounds (P < 0.10). In general, those genes and pathways involved in protein synthesis or protease inhibitors were down-regulated in the RES group, whereas those related to proteolytic degradation were up-regulated, thus suggesting a higher catabolism of proteins in these lambs. RES lambs showed over-expression of xenobiotic metabolism pathways, whereas those genes related to β-oxidation of fatty acids were down-regulated. According to the data obtained, early feed restriction during the suckling period of Merino lambs promoted long-term effects on both the hepatic transcriptomic profile and plasma metabolic profile, which might have modified fatty acids metabolism, catabolism of proteins, and detoxification of xenobiotics, thus reducing feed efficiency during the fattening period.
Keywords: feed efficiency, feed restriction, lamb, metabolomics, transcriptomics
INTRODUCTION
The search for solutions towards a more efficient use of resources is being promoted in the context of an increasing world population. Concerning the livestock sector, environmental factors (such as prenatal nutrition) have been postulated as important under the term “early nutritional programming,” which may affect metabolism, growth, and development, and thereby the feed efficiency and health of the offspring (Roseboom et al., 2011; Ruchat et al., 2014). Although studies addressing nutrition during the neonatal period of lambs are scarce, some have demonstrated early nutritional programming during this phase (Davies and Owen, 1967; Greenwood et al., 2004; Galvani et al., 2014). Moreover, several experiments have described the molecular mechanisms promoted by early feed restriction in children (Ruchat et al., 2014), cattle (Alexandre et al., 2015), and pigs (Vincent et al., 2015), whereas this has not been described for fattening lambs. Consequently, a deeper knowledge in this field is needed in order to design strategies to achieve greater efficiency when fattening lambs, because this will determine the overall profitability and sustainability of this production system.
Modern molecular and -omic tools (i.e., epigenomics, transcriptomics, proteomics, and metabolomics) are further increasing our understanding of the cascade from genes to phenotypes (Tizioto et al., 2015). Therefore, such studies can be used as an initial step to investigate the molecular mechanisms triggered by early feed restriction of lambs. Here, we aim to describe the long-term effects on metabolic pathways promoted by early feed restriction of Merino lambs, attempting to identify both differentially expressed (DE) genes in the liver and differentially accumulated metabolites in the plasma, and reasoning that this approach might highlight potential strategies to improve feed efficiency during the fattening period through either selection or improved management practices.
MATERIALS AND METHODS
Care and Use of Animals
All handling practices involving animals followed the recommendations of the Directive 2010/63/EU of the European Parliament and the Council on the Protection of Animals used for Scientific Purposes and the IGM-CSIC Animal Experimentation Committee (protocol number 2015-04).
Animal Diets and Sampling
Twenty-four male Merino lambs, penned individually with their corresponding ewe during the suckling period, were used in this experiment. The lambs were stratified by live body weight (LBW) at birth (4.81 ± 0.256 kg) and then assigned randomly to one of the two experimental treatments (n = 12 per dietary treatment) during the suckling period. The first group of lambs (ad libitum [ADL]) was kept permanently with the dams, whereas the other group (restricted [RES]) was separated within 9 to 18 h. Dams of the RES group were injected with oxytocin to remove alveolar milk and then milked at 17 h (an hour before the reintroduction of lambs). The milk yield of all the ewes (RES and ADL groups) was recorded on weeks 3, 5, and 7, according to the McCance (1959) method, using oxytocin (intravenous injection of oxytocin before each milking, Oxitón, Laboratorios Ovejero, España) and a milking machine. All lambs were weighed twice per week throughout the suckling period and vaccinated against enterotoxemia and Pasteurella (Heptavac P Plus, MSD Animal Health) when they were 4-weeks-old.
When each lamb reached 13.5 kg of LBW, it was weaned progressively (free access to a complete pelleted diet [CPD] and alfalfa, whereas it was allowed only 2 h with the dam) until it weighed 15 kg. Once the target weaning LBW was reached (15 kg), each lamb was penned individually and offered the same CPD at the same level (35 g/kg LBW each day) to ensure no variations in DM intake during the fattening phase. Fresh drinking water was always available. All of the lambs received the CPD once per day at 09:00 h; the daily amount of feed offered was adjusted twice per week based on LBW. Leftovers (if any) were collected daily, pooled in weekly composites for each animal, and analyzed for DM content. Ingredients and chemical composition of the CPD administered are summarized in Table 1.
Table 1.
Ingredients and chemical composition (g/kg DM unless otherwise stated) of the complete pelleted diet administered during the fattening period of Merino lambs
| Ingredients, g/kg | |
|---|---|
| Barley | 433 |
| Corn | 150 |
| Soybean meal 44 | 237 |
| Barley straw | 150 |
| Vitamin-mineral premix | 30 |
| Chemical composition, g/kg DM | |
| DM, g/kg | 900 |
| NDF | 227 |
| ADF | 121 |
| CP | 174 |
| Fat | 30 |
| Ash | 68 |
| Metabolizable energy, kcal/kg | 2464 |
Blood samples were collected from all of the animals during the fattening period by jugular venepuncture into 10-mL vacutainer tubes containing lithium-heparin, placed in iced water, and then centrifuged at 3,520 × g for 16 min at 5 °C. The obtained plasma was stored at −80 °C until metabolomic profiling. Finally, all the animals were harvested with a target body weight of 27 kg. A piece of liver was excised immediately, stabilized with RNAlater (Ambion, Inc., Austin, TX, USA) overnight at 4 °C, and then stored at −80 °C until transcriptomics analysis (RNA-seq).
Transcriptomics Analysis of Liver Samples Using RNA-seq
RNA extraction.
A comparative liver transcriptome analysis was carried out using next-generation sequencing (RNA-seq). The minimum number of liver samples to cover the complete range of values of average daily gain (feed efficiency) in each group was selected for RNA extraction (4 animals per group and 8 samples in total), which was performed using RNeasy Mini kit and RNase-Free DNase Set kit (Qiagen, Hilden, Germany). Concentration and quality of the extracted RNA were first verified by NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, DE, USA) and further evaluated with an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) after separation using an RNA 6000 Nano Kit. All of the samples had an RNA integrity number (RIN) larger than 8 (8.5 ± 0.04).
Library preparation, RNA sequencing, and data processing.
Library preparation for RNA-seq (transcriptome sequencing) was performed on an Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA, USA) to obtain readings of 100 base pairs (bp) in length from each sample; cDNA libraries were obtained according to the manufacturer’s instructions. The quality of the libraries was analyzed in an Agilent 2100 Bioanalyzer and quantified by real-time PCR in a LightCycler 480 (Roche). Finally, libraries were pooled to perform paired-end multiplex sequencing (2 × 100 bp) on the Illumina HiSeq 2500 sequencer. The quality of the obtained data was evaluated with the FastQC program to identify possible artifacts in the sequencing. The readings generated were expurgated through the BBDuk program. The sequence and annotation of the most recent ovine genome assembly were obtained from the National Center for Biotechnology Information (NCBI) database (accession number GCF_000298735.2, assembly Oar_v4.0). The expurgated readings were mapped on the sequence of this genome with the BBMap program (part of the BBTools package).
Annotation, standardization, and statistical analysis of differentially expressed genes.
The identification of new and annotated transcripts that presented differential transcription between the experimental groups was made by mapping with HISAT, the assembly with StringTie, and the statistical analysis of Ballgown, following the workflow of Pertea et al. (2016). A differential expression analysis was performed with the Bioconductor package DESeq2 (version 1.12.4). Statistical analyses of differentially expressed genes were performed in R (version 3.3.1). Also, the Transcripts Per Kilobase Million (TPM) values were calculated according to Wagner et al. (2012; PMID 22872506) using DESeq2-normalized read counts. The data were deposited in the Gene Expression Omnibus database ([GEO] accession number GSE107064). Genes were considered to be differentially expressed if their P-value, adjusted for multiple testing using the Benjamini and Hochberg method (false discovery rate [FDR]), was less than 0.1 (Benjamini and Hochberg, 1995).
Functional enrichment analysis.
Gene ontology (GO) term enrichment analysis was performed through the Database for Annotation, Visualization, and Integrated Discovery (DAVID), version 6.8. The Functional Annotation Tool of DAVID was used to determine the most relevant GO terms within the list of DE genes. Moreover, Kyoto Encyclopedia Genes and Genomes (KEGG) pathways analysis was carried out with the kegga function of the edgeR package. The results of both procedures allowed us to interpret the DE gene lists.
Metabolomic Analysis of Blood Plasma Samples
Extraction and ultra-high performance liquid chromatographic system (UHPLC/QTOF-MS) analysis.
Plasma samples were analyzed in duplicate to obtain the metabolomic profile. Aliquots of each sample (150 μL) were diluted twice in 10 volumes of acetonitrile:methanol:trichloroacetic acid mixture (50:47:3 vol/vol, LCMS grade from VWR, Milan, Italy), vortex-mixed, and then left at −18 °C overnight. Extracts were then centrifuged (8,000 × g) at 4 °C and the supernatant filtered through a 0.2-μm cellulose syringe filter. The metabolomic analysis was then carried out on a hybrid quadrupole-time-of-flight mass spectrometer coupled to an UHPLC/QTOF-MS. An Agilent 1290 liquid chromatography system was used, equipped with binary pump and a Dual Electrospray Jet Stream ionization system, and then coupled to an Agilent G6550 mass spectrometer detector (Agilent Technologies, Santa Clara, CA, USA). The instrument was run in positive mode and acquired MS-only spectra in the range of 100 to 1,600 m/z. Chromatographic separation was achieved using a Knauer BlueOrchid C18 column (100 × 2 mm i.d., 1.8 μm). The LC mobile phase was a mixture of water (proteomic grade from VWR) and methanol (LCMS grade from VWR); formic acid (0.1%, vol/vol) and ammonium formate (5 mM) (both from Sigma Aldrich, St. Louis, MO, USA) were added to both phases. UHPLC/QTOF-MS chromatographic gradient and source parameters were taken from previous experiments (Lucini et al., 2015). Briefly, the gradient started with 5% methanol and was increased to 90% methanol within 34 min, and the solvents temperature was set to 35 °C. The flow rate was 220 μL/min, and the injection volume was 3.5 μL.
QTOF raw data were processed by the Profinder B.06 software (from Agilent Technologies) using the naïve find-by-molecular-feature algorithm (isotopic model: small compounds after chromatography, no halogens). Molecular features were recursively identified after deconvolution followed by mass and retention time alignment. Compounds that were not present in 80% of the replicates within at least 1 treatment were discarded. Compound identification was based on both accurate mass and isotope pattern (accurate spacing and isotopes ratio) and performed using the database Metlin (from Agilent Technologies). The compounds passing alignment and filtering were finally exported in Mass Profiler Professional B.12.05 (Agilent technologies) for statistics.
Statistical analysis.
Compounds were log2 normalized, and their abundances normalized at the 75th percentile and baselined against the median of control (ADL). Unsupervised cluster analysis (hierarchical cluster algorithm; Euclidean similarity measure) and partial least squares discriminant analysis ([PLS-DA], N-fold validation with n = 4) were performed on the baseline dataset. Concerning PLS-DA, the compounds’ weight within the latent vectors in the class prediction model was plotted (loading plot), and the most relevant ones exported and recorded. ANOVA analysis (P < 0.1, Bonferroni multiple testing corrections) and fold-change analysis (cut-off = 3) were then combined into volcano plots to identify differential metabolites.
RESULTS AND DISCUSSION
Understanding the causes of low feed efficiency might be the first step towards selecting efficient animals or finding solutions to express their utmost potential, thus helping to reduce farm feeding costs. Therefore, the aim of the present study was to establish the molecular mechanisms behind the long-term effects on feed efficiency traits of fattening Merino lambs promoted by early feed restriction during the suckling period.
Milk production was similar for all the ewes (ADL and RES groups) under experimental conditions (week 3, 110 ± 8.47 g/h; week 5, 102 ± 6.52 g/h; week 8, 97 ± 6.97 g/h). Hence, as expected, the lambs separated from the dams during 9 h each day (RES) grew more slowly during the suckling period (Supplementary Table 1) probably because after being milked, milk production of the RES ewes was insufficient to satisfy the demand of the RES lambs. However, once the weaning weight was reached, RES lambs also showed lower average daily gain (165 vs. 202 g/d for RES and ADL lambs, respectively; P < 0.01) even though during this period dry matter intake was fixed for both groups (35 g/kg LBW each day) to avoid confounding effects with early feed restriction. Thus, RES animals took more days (62 vs. 74 days for the ADL and RES groups, respectively; P < 0.001) to reach the intended LBW (27 kg LBW) and, consequently, the total DM intake during the fattening period was increased (39.3 vs. 47.5 kg; P < 0.001; Supplementary Table 1). Therefore, as explained before (Santos et al., 2018a,b), RES lambs showed reduced feed efficiency traits, this effect not being associated with age or intake differences during the fattening period. This is extremely important because according to our results, early feed restriction due to health problems of the lambs, milk production of the dam, or pathologies in the udder may result in significant costs in the profitability of the farm during post-weaning phases.
Transcriptomic Profile
The liver is the major metabolic organ in the body and regulates a wide variety of metabolic processes (Ji et al., 2012; Zheng et al., 2016); therefore, global transcript profiling of the liver is a logical approach for identifying the long-term effects on feed efficiency traits promoted by early feed restriction during the suckling period of lambs. Differential gene expression analysis in the liver tissue of Merino lambs with different nutritional levels during the preweaned period identified 38 DE annotated genes. Positive values of log2-fold change (log2FC) indicated that 15 genes were up-regulated in the RES relative to the ADL group, whereas negative log2FC values denoted the down-regulation of 23 genes (Table 2).
Table 2.
Differential expression of genes in the livers of fattening lambs allowed ADL or RES milk intake during the suckling period
| Gene ID and function | Description | TPMa mean ADL | TPMa mean RES | log2FCb | P-value | FDRc |
|---|---|---|---|---|---|---|
| LOC101110202 | Cytochrome P450 3A24-like | 508 | 749 | 0.433 | 8.45E-08 | 0.0005 |
| CYP2C19 | Cytochrome P450 2C19 | 347 | 819 | 0.708 | 3.48E-05 | 0.0218 |
| CYP3A24 | Cytochrome P450 CYP3A24 | 1,150 | 1,956 | 0.548 | 6.48E-05 | 0.0304 |
| CDC37L1 | Cell division cycle 37 like 1 | 15 | 22 | 0.381 | 2.35E-06 | 0.0026 |
| APOH | Apolipoprotein H | 5,423 | 4,484 | −0.359 | 9.67E-06 | 0.0091 |
| F2 | Coagulation factor II, thrombin | 1,270 | 1,038 | −0.366 | 1.00E-04 | 0.0433 |
| TF | Serotransferrin | 9,775 | 7,643 | −0.420 | 1.75E-05 | 0.0123 |
| GUCY2C | Guanylate cyclase 2C | 5 | 12 | 0.619 | 4.30E-04 | 0.0893 |
| ACSM3 | Acyl-CoA synthetase medium-chain family member 3 | 755 | 604 | −0.382 | 3.00E-04 | 0.0877 |
| ACOT13 | Acyl-CoA thioesterase 13 | 166 | 128 | −0.419 | 4.20E-04 | 0.0893 |
| ACOX3 | Acyl-CoA oxidase 3, pristanoyl | 44 | 28 | −0.561 | 4.80E-04 | 0.0893 |
| FADS6 | Fatty acid desaturase 6 | 186 | 136 | −0.459 | 5.20E-04 | 0.0893 |
| ND3 | NADH dehydrogenase subunit 3 | 1,751 | 1,057 | −0.686 | 3.21E-07 | 0.0005 |
| APOM | Apolipoprotein M | 121 | 96 | −0.392 | 2.30E-04 | 0.0843 |
| ABCA10 | ATP binding cassette subfamily A member 10 | 118 | 96 | −0.364 | 6.50E-04 | 0.0972 |
| CST3 | Cystatin C | 314 | 222 | −0.515 | 5.22E-05 | 0.0280 |
| SERPINA11 | Serpin family A member 11 | 235 | 199 | −0.318 | 4.70E-04 | 0.0893 |
| DNAJA1 | DnaJ heat shock protein family member A1 | 14 | 21 | 0.422 | 4.10E-04 | 0.0893 |
| LOC101105132 | DnaJ homolog subfamily A member 1 | 24 | 37 | 0.430 | 4.20E-04 | 0.0893 |
| LOC105605968 d | DnaJ homolog subfamily A member 1 | 36 | 56 | 0.468 | 5.47E-05 | 0.0280 |
| LOC101101922 d | DnaJ homolog subfamily A member 1 | 29 | 46 | 0.474 | 2.90E-04 | 0.0877 |
| LOC101102122 d | 60S acidic ribosomal protein P1 | 596 | 471 | −0.420 | 3.84E-07 | 0.0005 |
| RPLP1 | Ribosomal protein lateral stalk subunit P1 | 871 | 692 | −0.403 | 1.37E-05 | 0.0110 |
| RPL3 | Ribosomal protein L3 | 548 | 494 | −0.245 | 2.90E-04 | 0.0877 |
| LOC101114033 | 60S ribosomal protein L17 | 208 | 179 | −0.297 | 6.50E-04 | 0.0972 |
| DPYS | Dihydropyrimidinase | 358 | 233 | −0.541 | 4.20E-04 | 0.0893 |
| ACP5 | Acid phosphatase 5, tartrate resistant | 82 | 56 | −0.571 | 3.55E-07 | 0.0005 |
| RBMS2 | RNA binding motif single stranded interacting protein 2 | 8 | 6 | −0.436 | 4.50E-04 | 0.0893 |
| SERINC2 | Serine incorporator 2 | 13 | 28 | 0.633 | 2.40E-04 | 0.0843 |
| GCAT | Glycine C-acetyltransferase | 190 | 139 | −0.454 | 6.60E-04 | 0.0972 |
| NUFIP2 | NUFIP2, FMR1 interacting protein | 16 | 25 | 0.454 | 2.40E-04 | 0.0843 |
| PLXNA2 | Plexin A2 | 4 | 2 | −0.555 | 5.70E-04 | 0.0911 |
| LOC101106024 | Secreted and transmembrane protein 1A-like | 47 | 92 | 0.592 | 3.30E-04 | 0.0893 |
| ELK4 | ELK4, ETS transcription factor | 10 | 14 | 0.402 | 5.40E-04 | 0.0893 |
| EPS8 | Epidermal growth factor receptor pathway substrate 8 | 10 | 14 | 0.363 | 5.40E-04 | 0.0893 |
| C1H21orf33 | Chromosome 1 open reading frame, human C21orf33 | 161 | 126 | −0.399 | 5.00E-04 | 0.0893 |
| LOC101121401 d | Ferritin heavy chain | 273 | 227 | −0.344 | 4.80E-04 | 0.0893 |
| CCDC117 | Coiled-coil domain containing 117 | 16 | 22 | 0.344 | 5.30E-04 | 0.0893 |
aTPM = Transcripts Per Kilobase Million.
bFold change estimates are relative to ADL group, so positive values indicate greater expression in RES lambs.
cFDR = false discovery rate.
dNonexpressed pseudogenes.
With respect to fatty acid metabolism, several enzymes (e.g., ACSM3 and ACOT13) involved in fatty acid activation before they can be carried into the mitochondria (where fatty acid oxidation takes place) were down-regulated in the RES group. Together with the lower expression of genes involved in the oxidative phosphorylation and mitochondrial electron transport chain (e.g., ND3), this might indicate that β-oxidation of fatty acids (and hence ATP production through this pathway) was down-regulated in RES lambs, which, in accordance with those in the works of Alexandre et al. (2015), Tizioto et al. (2015), and Weber et al. (2016), might have promoted differences in feed efficiency. Also, two genes involved in the biosynthesis of polyunsaturated fatty acids (e.g., ACOX3 and FADS6) were down-regulated in the RES lambs. In fact, these circumstances might be related to the higher storage of carcass, and noncarcass fat depots and the more saturated fatty acid profile observed in the meat of the RES lambs (Santos et al., 2018a).
Additionally, it has been suggested that the efficiency of energy production for maintenance and growth can be altered by manipulating either mitochondrial number or function through epigenetic modifications (Meyer et al., 2012). Moreover, mitochondrial density or the number of mitochondria per unit of tissue is highly correlated with capillary density because the oxygen source (the capillary bed) should be closely matched with the oxygen-utilizing machinery (the mitochondrion). Consequently, the down regulation of various genes encoding proteins related to vascularization, such as serotransferrin (TF, an iron binding transport protein responsible for the transport of iron in the plasma from sites of absorption to sites of storage and utilization, such as the heme prosthetic group of haemoglobin, myoglobin, and cytochromes), erythropoiesis (TF), and coagulation factor II (F2, which plays a role in maintaining vascular integrity during development and postnatal life) in the RES lambs, might also be related to the lower feed efficiency of this group of animals.
On the other hand, genes involved in protein synthesis, such as those encoding ribosomal proteins (LOC101102122, RPLP1, RPL3, and LOC101114033) or protease inhibitors (CST3 and SERPINA11), were down-regulated in the RES group, whereas those related to proteolytic degradation (DNAJA1, LOC101105132, LOC105605968, and LOC101101922) were up-regulated, suggesting a higher catabolism of proteins. These results differ from those obtained by Keogh et al. (2016) in cattle, who observed an increased capacity for protein synthesis after a feed restriction period. However, the animals studied by Keogh et al. (2016) were fed ad libitum during the refeeding period and underwent compensatory growth, whereas this was avoided in our study to prevent confounding effects with early feed restriction.
Additionally, in RES lambs, there was a lower expression of genes related to cellular growth and proliferation (ACP5, TF, RBMS2, F2, and DPYS). Especially remarkable was the lower expression of DPYS in the liver of RES lambs, since this condition has been related in humans to villous atrophy in the small intestine, thus leading to malabsorption problems and failure to gain weight at the expected rate (Kuilenburg et al., 2010).
Also, this study revealed that early feed RES animals showed over-expression of several cytochrome P450 forms, such as CYP3A24 and CYP2C19. The cytochrome P450 constitutes the mayor enzyme family capable of oxidizing a variety of structural compounds, including steroids, fatty acids, and xenobiotics (Zanger and Schwab, 2013). Therefore, early feed restriction might provoke differences in reproductive traits (Rhind et al., 1998; Robinson et al., 2006; Swelum et al., 2017), but also on detoxification of xenobiotics in the RES animals when compared with the ADL group. This theory seems to be supported by the presence of lower levels of alkaloids in the plasma or RES lambs when compared with those in the ADL group, as will be described below.
Also, our results are similar to previous studies in humans describing that retarded early growth is strongly related to high plasma concentrations of the hemostatic factors fibrinogen and factor VII (Barker et al., 1992), and thus to a more severe inflammatory response (Spurlock, 1997). Additionally, the lower expression of TF in RES lambs might be related to increased inflammation in these animals (Ritchie et al., 1999), which seems to be corroborated by the higher expression of the GUCY2C gene (Wang et al., 2017). These results suggest that, compared with the ADL lambs, the RES lambs might have responded in a different way to hepatic proinflammatory stimulus, potentially expending more energy towards combating systemic inflammation and redirecting less nutrients towards growth and protein accretion (Paradis et al., 2015).
A lower expression of C1H21orf33, which encodes a potential mitochondrial protein with protective effects against oxidative stress, was also noticed in RES lambs. This might be related to a lower oxidative stress in the RES lambs, probably due to a reduced activity at the mitochondrial level, as will be explained below. Finally, and according to the results obtained, several other functions might have been affected by long-term effects caused by feed restriction during the suckling period; thus, genes related to cholesterol metabolism (APOM, ABCA10) or alterations in the nervous system development (NUFIP2 and PLXNA2) were differentially expressed when comparing the lamb groups. However, there is a tissue-specific modulation of messenger RNAs (mRNA), so the existence of differences at the tissue level does not guarantee the same effect at other locations (Tizioto et al., 2015).
When analyzed using DAVID software, the identified functional gene clusters were related to the cytochrome P450, ribosome and ribosomal proteins, and several components of the membrane/transmembrane cell, among others (Table 3). Moreover, KEGG annotation and enrichment analysis with the kegga function of the edgeR package allowed us to identify those enriched pathways in which DE genes were involved (Table 4).
Table 3.
Annotation clusters identified by the Functional Annotation Tool of DAVID from the list of 38 differentially expressed genes [DESeq2 (P. BH < 0.10)]
| Term | No.a | P-value | Genes symbolsb |
|---|---|---|---|
|
Annotation cluster 1
Enrichment score: 1.900 | |||
| Cytochrome P450, conserved site | 3 | 0.002 | CYP2C19, CYP3A24, LOC101110202 |
| Cytochrome P450 | 3 | 0.003 | CYP2C19, CYP3A24, LOC101110202 |
| Monooxygenase | 3 | 0.004 | CYP2C19, CYP3A24, LOC101110202 |
| Oxidoreductase | 4 | 0.005 | CYP2C19 , ND3, CYP3A24, LOC101110202 |
| Heme | 3 | 0.009 | CYP2C19, CYP3A24, LOC101110202 |
| Iron | 3 | 0.020 | CYP2C19, CYP3A24, LOC101110202 |
| Heme binding | 3 | 0.026 | CYP2C19, CYP3A24, LOC101110202 |
| Iron ion binding | 3 | 0.034 | CYP2C19, CYP3A24, LOC101110202 |
| Metal-binding | 3 | 0.560 | CYP2C19, CYP3A24, LOC101110202 |
|
Annotation cluster 2
Enrichment score: 0.440 | |||
| Ribosomal protein | 3 | 0.014 | RPLP1, LOC101114033, RPL3 |
| Ribonucleoprotein | 3 | 0.017 | RPLP1, LOC101114033, RPL3 |
| Structural constituent of ribosome | 3 | 0.074 | RPLP1, LOC101114033, RPL3 |
| Ribosome | 3 | 0.100 | RPLP1, LOC101114033, RPL3 |
|
Annotation cluster 3
Enrichment score: 0.090 | |||
| Membrane | 8 | 0.700 | FADS6, ABCA10, GUCY2C , ND3, CYP3A24, LOC101110202, SERINC2, PLXNA2 |
| Transmembrane helix | 7 | 0.810 | FADS6, ABCA10, GUCY2C , ND3, LOC101110202, SERINC2, PLXNA2 |
| Transmembrane | 7 | 0.820 | FADS6, ABCA10, GUCY2C , ND3, LOC101110202, SERINC2, PLXNA2 |
| Integral component of membrane | 6 | 0.910 | FADS6, ABCA10, GUCY2C , ND3, LOC101110202, SERINC2 |
aNumber of genes involved.
bDifferentially expressed genes up-regulated in the early feed restricted group are marked in bold.
Table 4.
Enriched KEGG terms from the list of 38 differentially expressed genes [DESeq2 (P. BH < 0.10)]
| Enriched KEGG pathways | No.a | P-value | Genes symbolb |
|---|---|---|---|
| Linoleic acid metabolism | 3 | <0.001 | CYP2C19, CYP3A24, LOC101110202 |
| Retinol metabolism | 3 | 0.001 | CYP2C19, CYP3A24, LOC101110202 |
| Steroid hormone biosynthesis | 3 | 0.001 | CYP2C19, CYP3A24, LOC101110202 |
| Chemical carcinogenesis | 3 | 0.001 | CYP2C19, CYP3A24, LOC101110202 |
| Ribosome | 3 | 0.025 | LOC101114033, RPL3, RPLP1 |
| Metabolic pathways | 7 | 0.045 | ACOX3, ACP5, ACSM3, CYP2C19, CYP3A24, DPYS,LOC101110202 |
| Pantothenate and CoA biosynthesis | 1 | 0.046 | DPYS |
| Biosynthesis of unsaturated fatty acids | 1 | 0.068 | ACOX3 |
| Protein processing in endopl. reticulum | 2 | 0.072 | DNAJA1, LOC101105132 |
| α-Linolenic acid metabolism | 1 | 0.073 | ACOX3 |
| Butanoate metabolism | 1 | 0.073 | ACSM3 |
| β-Alanine metabolism | 1 | 0.092 | DPYS |
| Fatty acid degradation | 1 | 0.106 | ACOX3 |
| Drug metabolism—other enzymes | 1 | 0.108 | DPYS |
| Glycine, serine, threonine metabolism | 1 | 0.118 | GCAT |
| Mineral absorption | 1 | 0.120 | TF |
| Fatty acid metabolism | 1 | 0.122 | ACOX3 |
| ABC transporters | 1 | 0.158 | ABCA10 |
| PPAR signaling pathway | 1 | 0.176 | ACOX3 |
| Peroxisome | 1 | 0.197 | ACOX3 |
aNumber of genes involved in the KEGG term.
bDifferentially expressed genes up-regulated in the early feed restricted group are marked in bold.
Metabolomic Profiling
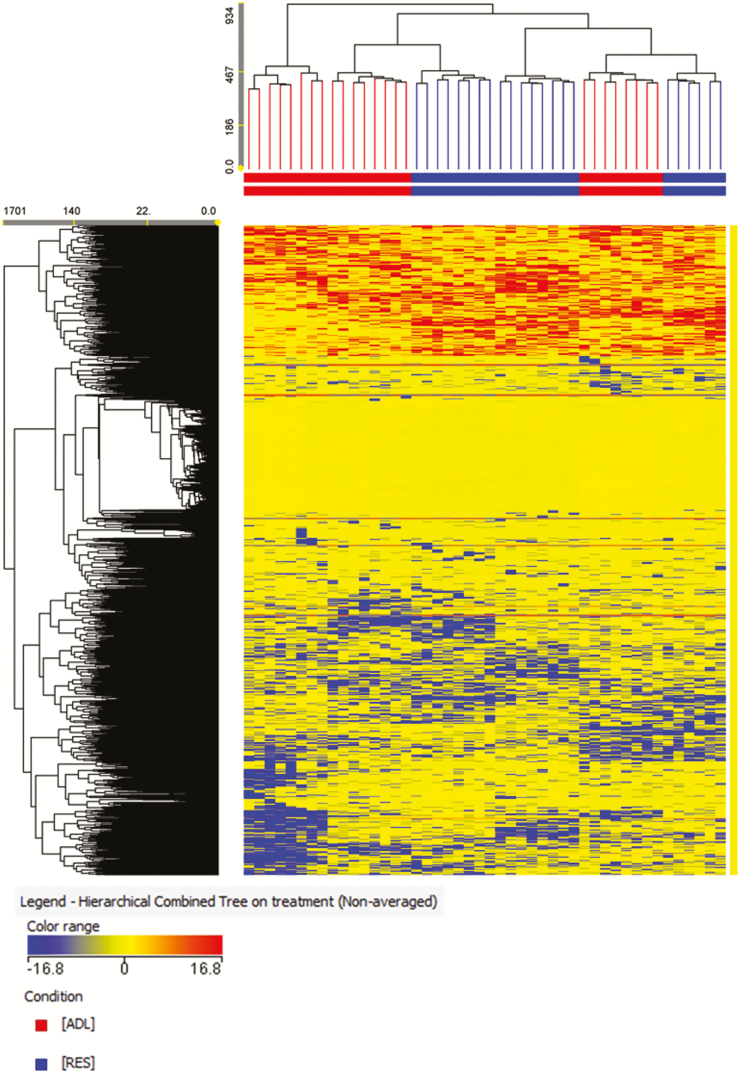
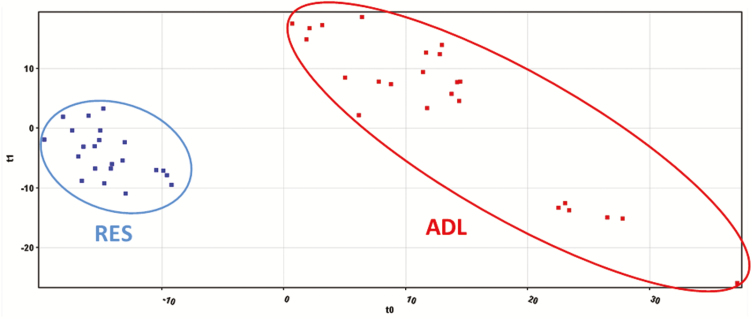
Also, a nontargeted metabolomic profile was assessed as a key tool to identify metabolic pathways modified by early feed restriction, because this procedure aims to detect, identify, and quantify a total population of low molecular weight compounds that might have a significant impact on metabolic, immunological, and physiological processes (Krastanov, 2010). The metabolomic assay enabled the deconvoluting of 4,026 metabolites in the plasma of the animals studied. The unsupervised hierarchical cluster analysis, produced on the basis of the fold-change-based heatmap, showed 3 main clusters (Figure 1). A tendency to differentiate the treatments according to metabolic profile could be observed from this unsupervised approach (no mixed clusters could be identified, and an intracluster homogeneity was observed for each of the 3 clusters). Nonetheless, a subcluster of ADL animals clustered together with the RES group, likely suggesting that the diet restriction was not the only clustering factor. However, clustering became evident when a supervised PLS-DA multivariate approach was adopted (Figure 2), as confirmed by a 100% accuracy after model N-fold validation. As expected, this latter statistical approach was more efficient in discriminating ADL vs. RES animals based on long-term changes of the plasma metabolome profile promoted by early feed restriction. In any case, the ANOVA analysis revealed that only 198 metabolites showed significant differences between groups, 38 of them being identified with the database used. From this set of compounds, 25 metabolites were relatively more accumulated and 13 relatively less accumulated in the RES when compared with the ADL lambs (Table 5).
Figure 1.
Not averaged unsupervised cluster analysis of the plasma metabolomic profile of fattening lambs fed ADL or RES during the suckling period. Clusters were generated on the basis of fold-change heat maps using an Euclidean similarity (linkage rule: Ward).
Figure 2.
PLS-DA of plasma samples using the metabolomic profile of fattening lambs fed ADL or RES during the suckling period.
Table 5.
List of identified metabolites up- or down-accumulated in plasma samples from early feed RES fattening lambs when compared with those fed ADL during the suckling period
| Metabolite | P (corr)a | FCb | Regulationc |
|---|---|---|---|
| Lipid metabolites | |||
| 3-Oxohexacosanoic acid | 0.096 | 23 | down |
| 1-Linoleoyl Glycerol | 0.025 | 435 | down |
| MG(0:0/14:0/0:0) | 0.002 | 529 | up |
| 1-Monopalmitin | 8.91E-09 | 16,522 | up |
| DG(16:0/16:0/0:0) | 0.013 | 1,699 | up |
| PI(22:1(11Z)/0:0) | 9.68E-04 | 1,005 | up |
| PS(14:1(9Z)/14:1(9Z)) | 4.43E-06 | 8,888 | up |
| PS(20:1(11Z)/0:0) | 0.020 | 72 | up |
| PS(22:0/18:1(9Z)) | 0.001 | 234 | up |
| 3-Hydroxysuberic acid | 0.045 | 39 | up |
| Methyl 4-[2-(2-formyl-vinyl)-3-hydroxy-5-oxo-cyclopentyl]-butanoate | 0.099 | 125 | up |
| Steroid ester | |||
| 1,3,5(10)-Estratrien-2,3-dial-17-one2-methyl ether | 0.027 | 178 | down |
| 11β,17β-Dihydroxy-9α-fluoro-17α-methyl-5β-androstan-3- one | 2.28E-07 | 16,074 | up |
| Sphingolipids | |||
| C17 Sphinganine | 3.26E-04 | 1,545 | down |
| Cer(t18:0/16:0) | 0.002 | 452 | up |
| Vitamin D metabolism | |||
| 1α,25-dihydroxy-23,24-dinorvitamin D3 | 0.020 | 82 | up |
| Aminoacid metabolism | |||
| Ser Gly His | 0.085 | 39 | down |
| Calpeptin | 0.025 | 98 | down |
| NAc-FnorLRF-amide | 0.021 | 45 | up |
| Phe His | 0.037 | 163 | up |
| Others | |||
| Melilotocarpan E | 0.004 | 742 | down |
| Prosopinine | 0.011 | 204 | down |
| Manoalide | 4.62E-05 | 3,227 | down |
| Pinacidil | 0.045 | 147 | down |
| Pymetrozine | 0.011 | 240 | down |
| CAY10550 | 0.005 | 713 | down |
| CAY10583 | 0.058 | 103 | down |
| Panaxytriol | 0.001 | 295 | up |
| Oxaprozin | 1.15E-04 | 1,409 | up |
| p-Hydroxytiaprofenic acid | 8.27E-09 | 14,511 | up |
| BILA 2185BS | 7.01E-10 | 12,745 | up |
| Idebenone metabolite (benzenehexanoic acid, 2,5-dihydroxy-3,4-dimethoxy-6-methyl-) | 7.29E-04 | 2,235 | up |
| 2-Thio-Acetyl MAGE | 1.81E-09 | 7,094 | up |
| Hemibrevetoxin B | 2.51E-07 | 4,557 | up |
| β-L-Fucose 1-phosphate | 3.47E-05 | 2,374 | up |
| 7-Hydroxy-5-heptynoic acid | 0.012 | 324 | up |
| 7-Methyl-2-hydroxy-6-oxoocta-2,4-dienoate | 1.89E-04 | 1,777 | up |
| Diethyl oxalpropionate | 0.075 | 140 | up |
aBonferroni multiple testing correction.
bFC = fold change.
cAnalysis is done considering RES vs. ADL; hence, “up” means that the compound is over-accumulated in RES, compared with ADL.
Table 5 lists the known compounds detected, grouped into several categories. The reduced calpeptin (inhibitor of calpain protease) content in the RES lambs confirms the higher catabolism of proteins in these animals. Additionally, the over-accumulation of some lipid metabolites [such as MG(0:0/14:0/0:0), DG(16:0/16:0/0:0)] and 3-hydroxysuberic acid (a metabolite derived from the ω-oxidation, which is considered a rescue pathway for fatty acid oxidation disorders) in RES lambs seems to corroborate that β-oxidation might have been negatively affected by early feed restriction. Under conditions interfering with β-oxidation, such as carnitine deficiency or a deficiency in an enzyme of β-oxidation (Brass and Beyerinck, 1988), fatty acids might have been accumulated in the RES animals if not entering the mitochondria, thus impairing energy production and feed efficiency. In fact, a recent study has demonstrated that growth rates of growing lambs can be improved by including carnitine in the diet (Foroozandeh et al., 2014). Another research has demonstrated diacylglycerol accumulation in skeletal muscle and liver when defects in mitochondrial fatty acid oxidation take place, which is associated with muscle and hepatic insulin resistance (Erion and Shulman, 2010). Moreover, modifications in satellite cell proliferation occur under calorie restriction (Halevy et al., 2000), which might inhibit muscle cell formation and subsequent muscle cell growth in neonates, thus lowering growth rates and feed efficiency due to the reduction in carcass muscle content relative to fat (Bark et al., 1992). These findings are also pertinent to our restricted lambs, which presented a trend towards increased intramuscular fat accretion (Santos et al., 2018a); this circumstance suggests that improving the activity of certain respiratory chain complexes might increase both lipid catabolism and feed efficiency traits (Ojano-Dirain et al., 2005).
However, the levels of a very-long-chain fatty acid (C26:0), 3-oxo-hexacosanoic acid, were reduced in the RES group. Very-long-chain fatty acids are oxidized by peroxisomal β- and α-oxidation pathways, which are essentially chain shortening pathways; when a medium chain length of approximately 8 carbons is reached, the fatty acid is transferred to the mitochondria as a carnitine derivative, and β-oxidation is completed (Carrillo et al., 2016). Consequently, the reduced 3-oxo-hexacosanoic acid levels in the RES group might indicate a higher enzymatic activity at a peroxisomal level in these animals.
Peroxisomes are also involved in oxidative stress (H2O2 production), inflammation (including platelet aggregation), and other catabolic processes, such as amino acids or xenobiotics metabolism (Terlecky, 2012), processes that were identified as being modified by early feed restriction with the transcriptome profile (Table 2). These effects were corroborated by some of the metabolites summarized in Table 5. For example, the inflammatory profile of RES lambs was supported by the over-accumulation of ceramide [Cer(t18:0/16:0)], and the lower levels of a precursor of ceramide (C17 Sphinganine) when compared with the ADL animals. This is because the sphingolipid ceramide takes part in arachidonic acid (AA) release, which is used for prostaglandin E2 (PGE2) production (Nixon, 2009).
In addition, the over-accumulation of phosphatidylserine [e.g., PS(14:1(9Z)/14:1(9Z)), PS(20:1(11Z)/0:0), and PS(22:0/18:1(9Z)), which are complex lipids facing the inner side of the membrane cell] in the plasma of RES lambs is consistent with the increased expression of SERINC2, a carrier protein that incorporates serine into membranes and facilitates the synthesis of 2 serine-derived lipids, phosphatidylserine, and sphingolipids (Inuzuka et al., 2005). However, phosphatidylserine plays a role in blood coagulation and apoptosis; thus, considering the lower expression of the gene encoding coagulation factor II (F2) and other coagulation-related proteins (e.g., APOH and TF) in RES lambs, we cannot discard the accumulation of phosphatidylserine metabolites in RES lambs as a mechanism to compensate for another disorder promoted by the long-term effects of early fed restriction. The reduced levels of calpeptin in RES lambs, which inhibits both collagen- and thrombin-induced aggregation of platelets, also seem to support these results.
Moreover, CYP constitutes the major enzyme family capable of oxidizing a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics (Zanger and Schwab, 2013), so the higher expression of some of these genes in RES lambs (e.g., CYP2C19 and CYP3A24, Table 2) might also explain the reduced accumulation of alkaloids (e.g., prosopinine), flavonoids (e.g., melilotocarpan E), and terpenes (e.g., manoalide) in the RES group. However, it is also possible that the higher hepatic catabolism of alkaloids in the RES group might have promoted the activation and appearance of more toxic metabolites, such as pyrroles (Cheeke, 1994), thus also impairing feed efficiency.
Finally, as described in Table 5, other metabolites with different abundances in the experimental groups were related to steroids, such as estradiol or testosterone, whose metabolism (mainly mediated by CYP1, CYP2, and CYP19) might have been affected by the restricted feed intake during the suckling period. These changes might involve long-term effects on reproductive traits (Rhind et al., 1998; Robinson et al., 2006; Swelum et al., 2017), which will need to be clarified in further experiments. Vitamin D metabolism (CYP24) and xenobiotic detoxification (CYP1, CYP2, and CYP3) might also have been affected following feed restriction, as suggested by other metabolites described in Table 5. These results were corroborated by DE genes, as previously described for the transcriptomic profile (Table 2). Thus, results obtained with these different “-omics” approaches, when considered together, support that early postnatal nutrition promotes molecular mechanisms with functional effects and consequences on feed efficiency. It must be stressed, however, that the causes behind lower feed efficiency traits might be multifactorial, so the effect of early feed restriction on the gastrointestinal microbiome (which was beyond the scope of the present study) might also have a role (Myer et al., 2016).
CONCLUSIONS
In conclusion, according to the results of the present study, early feed restriction during the postnatal period seems to be involved in metabolic programming, thus impairing feed efficiency traits during the fattening period of Merino lambs. This effect is mainly related to long-term effects on the hepatic transcriptome profile, which impairs β-oxidation of fatty acids and increases catabolism of proteins, thus altering the sources used for obtaining the energy required for maintenance and fattening. The feed restriction during the suckling period also seems to promote long-term effects on the innate immune response (e.g., inflammation), which might account for the different efficiency traits of lambs during the fattening period. Additional studies are now needed to confirm whether these factors will have an impact on the long-term health or reproductive traits since this will determine the overall sustainability and profitability of lamb production.
SUPPLEMENTARY DATA
Supplementary data are available at Animal Frontiers online.
LITERATURE CITED
- Alexandre P. A., L. J., Kogelman M. H., Santana D., Passarelli L. H., Pulz P., Fantinato-Neto P. L., Silva P. R., Leme R. F., Strefezzi L. L., Coutinho et al. 2015. Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle. BMC Genomics. 16:1073. doi:10.1186/s12864-015-2292-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bark L. J., T. S. Stahly G. L. Cromwell, and Miyat J.. 1992. Influence of genetic capacity for lean tissue growth on rate and efficiency of tissue accretion in pigs fed ractopamine. J. Anim. Sci. 70:3391–3400. doi:10.2527/1992.70113391x [DOI] [PubMed] [Google Scholar]
- Barker D. J., Meade T. W., Fall C. H., Lee A., Osmond C., Phipps K., and Stirling Y.. 1992. Relation of fetal and infant growth to plasma fibrinogen and factor VII concentrations in adult life. BMJ 304:148–152. doi:10.1136/bmj.304.6820.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benajmini Y., and Hochberg Y.. 1995. Controlling the false discovery rate : a practical and powerful approach to multiple testing. J. R. Stat. Soc., B. 57:289–300. doi: 10.2307/2346101 [Google Scholar]
- Brass E. P. and Beyerinck R. A.. 1988. Effects of propionate and carnitine on the hepatic oxidation of short- and medium-chain-length fatty acids. Biochem. J. 250:819–825. doi:10.1042/bj2500819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo J. A., Y. He Y. Li J. Liu R. A. Erdman T. S. Sonstegard, and Song J.. 2016. Integrated metabolomic and transcriptome analyses reveal finishing forage affects metabolic pathways related to beef quality and animal welfare. Sci. Rep. 6:25948. doi:10.1038/srep25948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeke P. R. 1994. a review of the functional and evolutionary roles of the liver in the detoxification of poisonous plants, with special reference to pyrrolizidine alkaloids. Vet. Hum. Toxicol. 36:240–247. [PubMed] [Google Scholar]
- Davies D. A. R., and Owen J. B.. 1967. The intensive rearing of lambs 1. Some factors affecting performance in the liquid feeding period. Anim. Prod. 9:501–508. doi:10.1017/S0003356100042070 [Google Scholar]
- Erion D. M. and Shulman G. I.. 2010. Diacylglycerol-mediated insulin resistance. Nat. Med. 16:400–402. doi:10.1038/nm0410-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroozandeh A. D., Amini H. R., Ghalamkari G. R., Shahzeydi M., and Nasrollahi S. M.. 2014. The effect of fat type and l-carnitine administration on growth, feed digestibility and blood metabolites of growing Afshari lambs. Livest. Sci. 164:67–71. doi:10.1016/j.livsci.2014.03.019 [Google Scholar]
- Galvani D. B., Pires C. C., Hübner C. H., Carbalho S., and Wommer T. P.. 2014. Growth performance and cacass traits of early-weaned lambs as affected by the nutritional regimen of lactating ewes. Small Rumin. Res. 120:1–5. doi: 10.2016/j.smallrumres.2014.03.0080921-4488 [Google Scholar]
- Greenwood P. L., A. S. Hunt, and Bell A. W.. 2004. Effects of birth weight and postnatal nutrition on neonatal sheep: IV. Organ growth. J. Anim. Sci. 82:422–428. doi:10.2527/2004.822422x [DOI] [PubMed] [Google Scholar]
- Halevy O., A. Geyra M. Barak Z. Uni, and Sklan D.. 2000. Early posthatch starvation decreases satellite cell proliferation and skeletal muscle growth in chicks. J. Nutr. 130:858–864. doi:10.1093/jn/130.4.858 [DOI] [PubMed] [Google Scholar]
- Inuzuka M., M. Hayakawa, and Ingi T.. 2005. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J. Biol. Chem. 280:35776–35783. doi:10.1074/jbc.M505712200 [DOI] [PubMed] [Google Scholar]
- Ji B., B Ernest, J. R Gooding, S Das, A. M Saxton, J Simon, J Dupont,S Métayer-Coustard, S. R Campagnaand, and B. H Voy. 2012. Transcriptomic and metabolomic profiling of chicken adipose tissue in response to insulin neutralization and fasting. BMC Genomics 13:441. doi:10.1186/1471-2164-13-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh K., D. A. Kenny P. Cormican A. K. Kelly, and Waters S. M.. 2016. Effect of dietary restriction and subsequent re-alimentation on the transcriptional profile of hepatic tissue in cattle. BMC Genomics. 17:244. doi:10.1186/s12864-016-2578-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krastanov A. 2010. Metabolomics - The state of art. Biotechnol. Biotechnol. Equip. 24:1537–1543. doi:10.2478/V10133-010-0001-y [Google Scholar]
- Kuilenburg A. B. P. Van, Dobritzsch D., Meijer J., Meinsma R., Benoist J., Assmann B., Schubert S., Hoffmann G. F., Duran M., De Vries M. C.,. et al. 2010. Dihydropyrimidinase deficiency : phenotype, genotype and structural consequences in 17 patients. Biochim. Biophys. Acta. 1802:639–648. doi:10.1016/j.bbadis.2010.03.013 [DOI] [PubMed] [Google Scholar]
- Lucini L., M. Pellizzoni R. Pellegrino G. P. Molinari, and Colla G.. 2015. Phytochemical constituents and in vitro radical scavenging activity of different aloe species. Food Chem. 170:501–507. doi:10.1016/j.foodchem.2014.08.034 [DOI] [PubMed] [Google Scholar]
- McCance I. 1959. The determination of milk yield in the Merino ewe. Aust. J. Agric. Res. 10:839–853. doi:10.1071/AR9590839 [Google Scholar]
- Meyer A. M., Caton J. S., Hess B. W., Ford S. P., and Reynolds L. P.. 2012. Epigenetics and effects on the neonate that may impact feed efficiency. In: R. A., Hill, editor, Feed Efficiency in the Beef Industry. Wiley-Blackwell, Ames, Iowa: p. 199–223. [Google Scholar]
- Myer P. R., J. E. Wells T. P. Smith L. A. Kuehn, and Freetly H. C.. 2016. Microbial community profiles of the jejunum from steers differing in feed efficiency. J. Anim. Sci. 94:327–338. doi:10.2527/jas.2015-9839 [DOI] [PubMed] [Google Scholar]
- Nixon G. F. 2009. Sphingolipids in inflammation: pathological implications and potential therapeutic targets. Br. J. Pharmacol. 158:982–993. doi:10.1111/j.1476-5381.2009.00281.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojano-Dirain C., Iqbal M., Wing T., Cooper M., and Bottje W.. 2005. Glutathione and respiratory chain complex activity in duodenal mitochondria of broilers with low and high feed efficiency. Poult. Sci. 84:782–788. doi: 10.1093/ps/84.5.782 [DOI] [PubMed] [Google Scholar]
- Paradis F., S. Yue J. R. Grant P. Stothard J. A. Basarab, and Fitzsimmons C.. 2015. Transcriptomic analysis by rna sequencing reveals that hepatic interferon-induced genes may be associated with feed efficiency in beef heifers. J. Anim. Sci. 93:3331–3341. doi:10.2527/jas.2015-8975 [DOI] [PubMed] [Google Scholar]
- Pertea M., D. Kim G. M. Pertea J. T. Leek, and Salzberg S. L.. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11:1650–1667. doi:10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind S. M., Elston D. A., Jones J. R., Rees M. E., McMillen S. R., and Gunn R. G.. 1998. Effects of restriction of growth and development of Brecon Cheviot ewe lambs on subsequent lifetime reproductive performance. Small Rumin. Res. 30:121–126. doi:10.1016/S0921-4488(98)00103-5 [Google Scholar]
- Ritchie R. F., G. E. Palomaki L. M. Neveux O. Navolotskaia T. B. Ledue, and Craig W. Y.. 1999. Reference distributions for the negative acute-phase serum proteins, albumin, transferrin and transthyretin: a practical, simple and clinically relevant approach in a large cohort. J. Clin. Lab. Anal. 13:273–279. doi:10.1002/(SICI)1098–2825(1999)13:6<273::AID-JCLA4>3.0.CO;2-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. J., Ashworth C. J., Rooke J. A., Mitchell L. M., and McEvoy T. G.. 2006. Nutrition and fertility in ruminant livestock. Anim. Feed Sci. Technol. 126:259–276. doi:10.1016/j.anifeedsci.2005.08.006 [Google Scholar]
- Roseboom T. J., R. C. Painter A. F. van Abeelen M. V. Veenendaal, and de Rooij S. R.. 2011. Hungry in the womb: what are the consequences? Lessons from the Dutch famine. Maturitas. 70:141–145. doi:10.1016/j.maturitas.2011.06.017 [DOI] [PubMed] [Google Scholar]
- Ruchat S. M., Bouchard L., and Hivert M. F.. 2014. Early infant nutrition and metabolic programming: what are the potential molecular mechanisms?Curr. Nutr. Rep. 3:281–288. doi:10.1007/s13668-014-0088-0 [Google Scholar]
- Santos A., F. J. Giráldez J. Mateo J. Frutos, and Andrés S.. 2018a. Programming Merino lambs by early feed restriction reduces growth rates and increases fat accretion during the fattening period with no effect on meat quality traits. Meat Sci. 135:20–26. doi:10.1016/j.meatsci.2017.08.007 [DOI] [PubMed] [Google Scholar]
- Santos A., Valdés C., Giráldez F.J., López S., France J., Frutos J., Fernández M., and Andrés S.. 2018b. Feed efficiency and the liver proteome of fattening lambs are modified by feed restriction during the suckling period. Animal . doi:10.1017/S1751731118000046 [DOI] [PubMed] [Google Scholar]
- Spurlock M. E. 1997. Regulation of metabolism and growth during immune challenge: an overview of cytokine function. J. Anim. Sci. 75:1773–1783. doi:10.2527/1997.7571773x [DOI] [PubMed] [Google Scholar]
- Swelum A. A., M. Ayadi I. Alhidary A. Alowaimer, and Abouheif M.. 2017. The relationships between body fatness, leptin, testosterone, and reproductive performance in ram lambs as affected by level and frequency of feeding. Theriogenology. 89:79–85. doi:10.1016/j.theriogenology.2016.10.013 [DOI] [PubMed] [Google Scholar]
- Terlecky S. R., L. J. Terlecky, and Giordano C. R.. 2012. Peroxisomes, oxidative stress, and inflammation. World J. Biol. Chem. 3:93–97. doi:10.4331/wjbc.v3.i5.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizioto P. C., L. L., Coutinho J. E., Decker R. D., Schnabel K. O., Rosa P. S., Oliveira M. M., Souza G. B., Mourão R. R., Tullio A. S., Chaves et al. 2015. Global liver gene expression differences in nelore steers with divergent residual feed intake phenotypes. BMC Genomics. 16:242. doi:10.1186/s12864-015-1464-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A., I. Louveau F. Gondret C. Tréfeu H. Gilbert, and Lefaucheur L.. 2015. Divergent selection for residual feed intake affects the transcriptomic and proteomic profiles of pig skeletal muscle. J. Anim. Sci. 93:2745–2758. doi:10.2527/jas.2015-8928 [DOI] [PubMed] [Google Scholar]
- Wagner G. P., K. Kin, and Lynch V. J.. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131:281–285. doi:10.1007/s12064-012-0162-3 [DOI] [PubMed] [Google Scholar]
- Wang G., Zhao C., Chen M., Chen S., Chang C., and Xu C.. 2017. Relationship analysis between gene expression profiles and rat liver cirrhosis occurrence. Afr. J. Biotechnol. 16:147–162. doi:10.5897/AJB2016.15718 [Google Scholar]
- Weber K. L., B. T. Welly A. L. Van Eenennaam A. E. Young L. R. Porto-Neto A. Reverter, and Rincon G.. 2016. Identification of gene networks for residual feed intake in Angus cattle using genomic prediction and rna-seq. Plos One. 11:e0152274. doi:10.1371/journal.pone.0152274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger U. M. and Schwab M.. 2013. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138:103–141. doi:10.1016/j.pharmthera.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Zheng A., J Luo, K Meng, J Li, W. L Bryden, W Chang, S Zhang,L. X Wang, G Liu, and B Yao. 2016. Probiotic (Enterococcus faecium) induced responses of the hepatic proteome improves metabolic efficiency of broiler chickens (Gallus gallus). BMC Genomics 17:89. doi:10.1186/s12864-016-2371-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.