Abstract
Virulence-associated type III secretion systems (T3SS) serve the injection of bacterial effector proteins into eukaryotic host cells. They are able to secrete a great diversity of substrate proteins in order to modulate host cell function, and have evolved to sense host cell contact and to inject their substrates through a translocon pore in the host cell membrane. T3SS substrates contain an N-terminal signal sequence and often a chaperone-binding domain for cognate T3SS chaperones. These signals guide the substrates to the machine where substrates are unfolded and handed over to the secretion channel formed by the transmembrane domains of the export apparatus components and by the needle filament. Secretion itself is driven by the proton motive force across the bacterial inner membrane. The needle filament measures 20–150 nm in length and is crowned by a needle tip that mediates host-cell sensing. Secretion through T3SS is a highly regulated process with early, intermediate and late substrates. A strict secretion hierarchy is required to build an injectisome capable of reaching, sensing and penetrating the host cell membrane, before host cell-acting effector proteins are deployed. Here, we review the recent progress on elucidating the assembly, structure and function of T3SS injectisomes.
Keywords: type III secretion systems, microbial pathogenesis, membrane proteins, protein secretion, Salmonella; host-pathogen interaction
Type III secretion systems: complex devices for the delivery of bacterial effector proteins into eukaryotic host cells.
INTRODUCTION
Virulence-associated type III secretion systems (T3SS) that serve the injection of bacterial effector proteins into eukaryotic host cells are among the most complex membrane-localized molecular machines known. These 6 MDa machines, also called injectisomes, consist of almost 20 different proteins with copy numbers ranging from 1 to more than 100 (Zilkenat et al.2016) (Fig. 1). Their principle mechanism of protein secretion has evolved from the T3SS responsible for self-assembly of bacterial flagella (Abby and Rocha 2012), however, injectisomes are able to secrete a much greater diversity of substrate proteins (Galán 2007). In addition, they have evolved to sense host-cell contact and to inject their substrates through a translocon pore in the host cell membrane (Rosqvist et al.1995; Collazo and Galan 1997; Park et al.2018). T3SS injectisomes are utilized by many different Gram-negative pathogens and symbionts to modulate interaction with host cells, among them human pathogens like Chlamydia, enterophathogenic Escherichia coli (EPEC), Pseudomonas aeruginosa, Salmonella, Shigella or Yersinia, plant pathogens like Erwinia, Pseudomonas syringae or Xanthomonas, and plant symbionts like Rhizobium (Büttner 2012). As different as the lifestyles of T3SS-utilizing bacteria are the diverse cellular functions of the effector repertoire secreted by them. Injected effector proteins serve in prevention of phagocytosis (Yersinia), in killing of macrophages (Salmonella), in bacterial entry into nonphagocytic cells (Salmonella), in interference with immune responses (e.g. Salmonella, Yersinia and Xanthomonas), in mediating tight adhesion (EPEC), or in acquisition of nutrients (Chlamydia and Salmonella) (Galán 2009). Although secreted effectors fulfill very different functions and are structurally diverse, they are all secreted in an unfolded conformation by a common ATP and proton-motive force (PMF)-driven one-step secretion mechanism involving an N-terminal secretion signal, and often also cognate targeting chaperones.
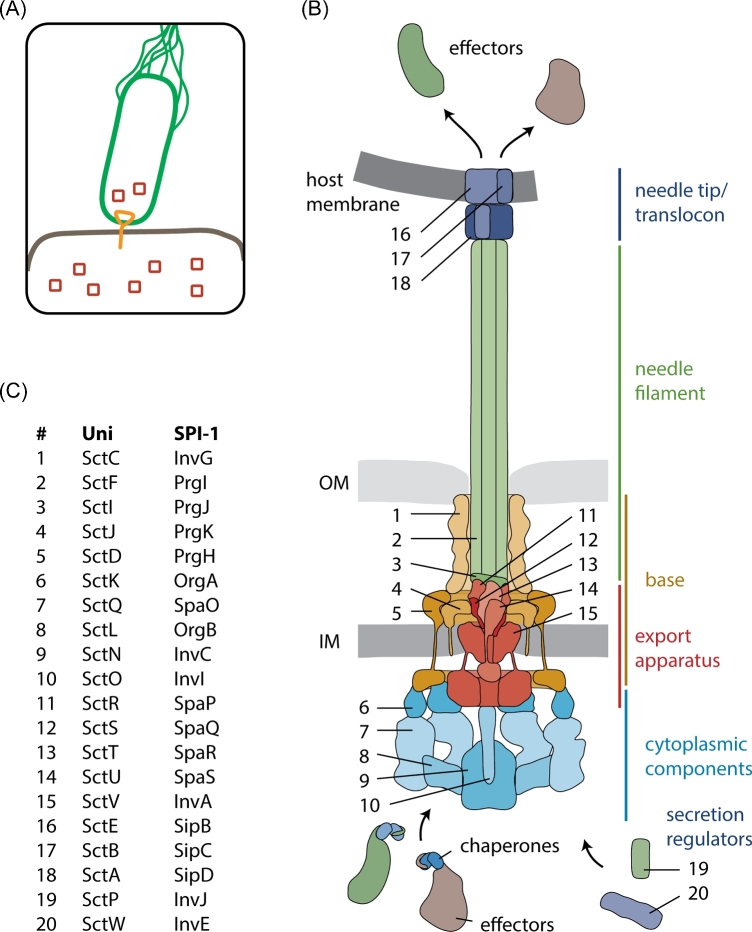
Figure 1.
Structure and function of the T3SS injectisome. A. Bacteria utilize T3SS to inject bacterial effector proteins into eukaryotic host cells. B. Structure of the T3SS indicating the different structural units (color-coded) and the individual components. Protein names are given in C. C. Nomenclature of T3SS components. The unified nomenclature (Uni) according to Hueck (1998) is shown on the left, the nomenclature of the T3SS encoded by Salmonella pathogenicity island 1 (SPI-1) is shown on the right. The nomenclature of the most commonly studied systems in Yersinia and pathogenic Escherichia coli follow the unified nomenclature with Ysc and Esc, respectively, instead of Sct. Abbreviations: IM, inner membrane; OM, outer membrane.
Here, we review the recent progress on elucidating assembly, structure and function of T3SS injectisomes that mediate secretion and injection of these effectors. Throughout this review we will refer to the components of the injectisome according to the unified nomenclature introduced previously (Hueck 1998) (Fig. 1). For a comprehensive table of the nomenclature, we refer to a recent review (Deng et al.2017) and to the STEP database.
STRUCTURE OF THE INJECTISOME
Base
The injectisome is anchored in the bacterial cell envelope by a base composed of the proteins SctC, SctD and SctJ; 12–15 SctC form an outer membrane secretin complex, similar to the secretins found in type II secretion systems (Kowal et al.2013; Worrall et al.2016; Filloux and Voulhoux 2018). T3SS secretins consist of three N-terminal N-domains (N0, N1 and N3) that protrude deeply into the periplasm, a C-domain that forms the outer membrane portal, and an S-domain through which a cognate pilotin binds the secretin and mediates its targeting and assembly (Worrall et al.2016). The N0- and N1-domains form structural motifs that are also found in SctD and SctJ. They were coined ring-building motifs for their ability to mediate ring formation of these base components (Spreter et al.2009). The C-domain forms a unique double-layered β-barrel. Its outer barrel extends towards the outer membrane where it forms a concave membrane association domain. The inner barrel extends into the channel lumen to form a gate separating the periplasmic space from the outside (Worrall et al.2016).
SctC is linked to the inner membrane-localized SctD by its N0-domain (Schraidt et al.2010). SctD contains a single transmembrane segment (TMS), a periplasmic domain composed of two ring-building motifs as well as a small N-terminal cytoplasmic FHA domain (Spreter et al.2009). SctJ contains an N-terminal lipid anchor and often a single C-terminal transmembrane segment. The periplasmic domain of SctJ is composed of two ring-building motifs (Yip et al.2005): 24 SctD and 24 SctJ form two concentric rings with SctD on the outside and SctJ on the inside, hence, SctD is also referred to as outer inner ring protein and SctJ as inner inner ring protein (Schraidt and Marlovits 2011).
Cytoplasmic components
SctK, SctQ, SctL, SctN and SctO are the cytoplasmic components of the injectisome. Because of their role in recruiting chaperone-substrate complexes according to the hierarchy state of secretion, SctK, SctQ and SctL are also referred to as a sorting platform (Lara-Tejero et al.2011). The cytoplasmic components exist in an injectisome-bound form and as free cytoplasmic complexes (Lara-Tejero et al.2011; Diepold et al.2015; Diepold et al.2017; Zhang et al.2017). The injectisome-bound SctK and SctQ form six pods that bind through a single SctK to the cytoplasmic domains of four SctD each (Hu et al.2015; 2017). Because of an internal start codon, sctQ is translated into two polypeptides of different lengths (SctQFL and SctQC), and the presence of both polypeptides is essential for injectisome assembly and T3SS function (Bzymek, Hamaoka and Ghosh 2012; Diepold et al.2015; McDowell et al.2016; Song et al.2017). Stoichiometry estimates based on fluorescence microscopy (Diepold et al.2017; Zhang et al.2017) and structural analysis of SctQ complexes (Bzymek, Hamaoka and Ghosh 2012; Notti et al.2015; McDowell et al.2016) suggest that 24 SctQFL and 48 SctQC are associated with each injectisome. Based on the density observed by cryo-electron tomography (Hu et al.2017) it is unlikely, however, that each pod contains four SctQFL and eight SctQC, and thus it is improbable that all SctQ are part of these visible structures. A homodimer of SctL connects the six pods to the hexameric ATPase SctN (Notti et al.2015; Imada et al.2016; Diepold et al.2017; Hu et al.2017). A single SctO is located at the center of the SctN hexamer and protrudes towards the cytoplasmic domain of the major export apparatus protein SctV (Ibuki et al.2011; Hu et al.2017).
Export apparatus
The export apparatus of T3SS consists of five hydrophobic proteins that are highly conserved between injectisome and flagellar T3SS: SctR, SctS, SctT, SctU and SctV (Wagner et al.2010). Nine protomers of the major export apparatus protein SctV are located in the central membrane patch of the base (Abrusci et al.2012). The transmembrane domain (TMD) of SctV is predicted to contain four plus four TMS with a sizable conserved loop in between that may form the entrance of the translocation channel in the inner membrane (Barker et al.2016; Erhardt et al.2017). SctV contains a large cytoplasmic C-terminus composed of four subdomains that forms a nonameric ring with a central pore of 50 Å in diameter (Worrall, Vuckovic and Strynadka 2010; Abrusci et al.2012). SctU is a protein with four predicted TMS and a cytoplasmic C-terminal domain that undergoes autocleavage at a conserved NPTH motif (Allaoui et al.1994; Deane et al.2008; Zarivach et al.2008; Lountos et al.2009; Wiesand et al.2009). Unfortunately, cryo-electron tomography of injectisomes or flagella has not so far resulted in assigning a specific location within the complex to SctU, but crosslinking analysis identified protein-protein interactions between the predicted TMDs of SctU and SctR, also placing SctU in the central membrane patch of the injectisome (Dietsche et al.2016). The minor export apparatus proteins SctR, SctS and SctT were predicted to contain four, two and six TMS, respectively; however, the recently solved cryo-electron microscopy structure of an SctRST complex of the flagellar homologs revealed that these proteins hardly make membrane contact in the assembled injectisome but are clamped within the SctJ ring at a periplasmic position above the membrane (Kuhlen et al.2018). They form an unusual helical assembly with five SctR most distal to the membrane, followed by one SctT and four SctS. The predicted transmembrane hairpins form the building blocks of the helix, of which SctR contributes two, SctS one, and SctT three. SctT is a structural fusion of SctR and SctS. It serves to compensate for the helical rise between the first and fifth SctR and stabilizes the assembled structure (Dietsche et al.2016; Kuhlen et al.2018). The assembled SctRST complex forms a translocation pore with an inner diameter of about 15 Å (Radics, Königsmaier and Marlovits 2014; Dietsche et al.2016; Kuhlen et al.2018). In the pre-secretion state, the complex possesses three gates: highly conserved Gln and Met line the mouth of the pore, a loop provided by SctT closes the pore entrance from the inside, and the N-termini of SctR and SctT constrict the exit of the pore (Kuhlen et al.2018). Upon assembly of the helical filament, this most distal end of the SctRST complex makes intimate contacts with the inner rod (Dietsche et al.2016), which might stabilize the pore in an open conformation.
Filament, needle tip and translocon
The helical needle filament composed of more than 100 copies of SctF (Broz et al.2007) is connected to the distal end of the SctRST complex by about six copies of the inner rod protein SctI (Marlovits et al.2006; Dietsche et al.2016; Zilkenat et al.2016; Kuhlen et al.2018). SctF forms a helical hairpin with a disordered and weakly conserved outward-facing N-terminus and a conserved loop and inward-facing C-terminus (Loquet et al.2012; Demers et al.2013; 2014). The SctRST complex and the needle filament have a very similar helical rise (4.0 and 4.3 Å per subunit, respectively) and number of subunits per turn (5.7 and 5.6, respectively), ensuring a tight anchoring of the filament to the export apparatus and base (Kuhlen et al.2018). However, while the termini of the subunits of the SctRST complex face outwards, the termini of the needle filament protein SctF face inwards, which may explain why the adapter SctI is needed to tightly connect export apparatus and needle filament. The inner diameter of the needle filament is 20 Å, which only allows passage of unfolded substrates (Loquet et al.2012). The highly conserved inner surface of the needle filament is mostly polar.
The needle of most injectisomes ends with a pentameric needle tip complex formed by the hydrophilic translocator protein SctA (Mueller et al.2005; Broz et al.2007; Epler et al.2012). Pathogenic Escherichia coli (EPEC and EHEC) possess an additional filament of up to 1 μm length instead (Sekiya et al.2001). Based on their structure, two classes of tip proteins can be distinguished: those similar to IpaD from Shigella (Erskine et al.2006; Johnson et al.2007; Chatterjee et al.2011; Lunelli et al.2011; Rathinavelan et al.2014) and those similar to LcrV from Yersinia (Derewenda et al.2004). One prominent difference between these two classes of tip proteins is the way they prevent self-aggregation in the bacterial cytoplasm: while IpaD-like proteins possess an N-terminal alpha-helical hairpin that fulfills a self-chaperoning function (Johnson et al.2007), LcrV-like proteins are bound by specific chaperones (e.g. LcrG in Yersinia) (DeBord, Lee and Schneewind 2001; Matson and Nilles 2001). Penetration of the host cell membrane is achieved by formation of a so-called translocon complex, which is composed of multiple copies of two different hydrophobic translocator proteins, SctB and SctE (Montagner, Arquint and Cornelis 2011; Dickenson et al.2013; Park et al.2018). These proteins are predicted to contain 1–2 TMS that insert into the host cell membrane with support from the tip complex (Marenne et al.2003). One copy of SctE may form part of the tip complex in order to fulfill this function (Veenendaal et al.2007; Harmon et al.2013; Cheung et al.2014).
TYPE III SECRETION-INDEPENDENT ASSEMBLY OF THE INJECTISOME
Assembly of the injectisome is a highly coordinated process to ensure the efficient formation of secretion- and membrane translocation-competent machines. The details of the assembly process have been reviewed extensively (Diepold and Wagner 2014), so we will only provide an overview and focus on the most recently gained insights. Two phases of injectisome assembly are distinguished: assembly of the principle secretion-competent machine and type III secretion-dependent assembly of the needle filament, tip and translocon.
Assembly of the principle secretion-competent machine is restricted by two factors: the confinement of the inner and outer membrane-embedded parts in a two-dimensional space and the penetration of the peptidoglycan layer. We previously proposed a bipolar model of assembly stating that (i) dependence of base assembly on nucleation by the export apparatus components ensures secretion competence (Fig. 2A), and (ii) dependence of base assembly on secretin formation ensures outer membrane translocation (Fig. 2B) (Diepold and Wagner 2014).
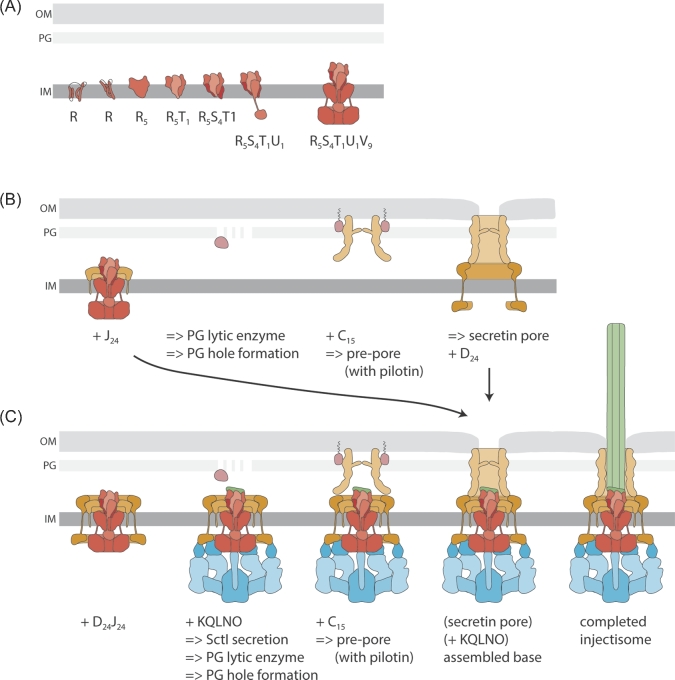
Figure 2.
Assembly of the T3SS injectisome. Protein names are indicated by their last letter, omitting ‘Sct’. The assembly pathways are shown from left to right. A. Assembly of the inner membrane export apparatus starts with membrane-integrated SctR. SctR (and also SctS and SctT, not shown) folds into a conformation that permits helical assembly. Then five SctR, followed by one SctT and four SctS, assemble into a helical assembly, winding up from the membrane. Recruitment of one SctU and then nine SctV finish the export apparatus assembly. B. Outside-in assembly model assuming independent secretin assembly. The export apparatus (the last step in A.) recruits 24 subunits of the inner inner ring protein SctJ. In an independent step, 15 secretin subunits assemble, supported by pilotins, into a pre-pore. PG penetration of the secretin is facilitated by hole-forming PG lytic enzymes. The secretin forms a pore in the outer membrane and recruits the outer inner ring protein SctD. Closure of the outer inner ring is not permitted until the SctJ-export apparatus assembly has been incorporated. C. Inside-out assembly model. The export apparatus (the last step in A.) recruits 24 SctJ and 24 SctD, followed by recruitment of the cytoplasmic components. Type III-dependent secretion of early substrates starts, which locally activates PG lytic enzymes in the periplasm to form a hole for secretin assembly. Just above the inner membrane complex, 15 secretin subunits assemble, supported by pilotins, into a pre-pore. Secretin pore formation and subsequent assembly of the needle filament complete the assembly of the injectisome. Abbreviations: IM, inner membrane; OM, outer membrane; PG, peptidoglycan.
Assembly of the base is nucleated by prior assembly of the core export apparatus components SctR, SctS and SctT, as shown for the T3SS encoded by Salmonella pathogenicity island 1 (SPI-1) (Wagner et al.2010). We showed that SctR assembles independently into multimers that are stabilized by recruitment of SctT (Dietsche et al.2016). SctS-SctT interactions were only detected in the presence of SctR (Dietsche et al.2016), further strengthening the notion that export apparatus assembly begins with SctR and proceeds through recruitment of SctT and SctS. Also, the helical structure of the complex of the flagellar SctRST homologs FliPQR suggests that assembly starts with five SctR and proceeds through subsequent recruitment of one SctT and four SctS, even though it cannot be ruled out at this point that the SctRST complex assembles from SctRS heterodimers followed by association of SctT. SctR assembly may be mediated by formation of intramolecular salt bridges in the predicted TMS that stabilize the step-like conformation of the protein (Kuhlen et al.2018). Likewise, intermolecular salt bridges seem to facilitate assembly of SctS onto the SctRT subcomplex. Strong hydrophobic interactions between the predicted TMS of SctR, SctS and SctT are believed to support stepwise winding of the assembling SctRST complex out of the membrane towards the periplasm (Kuhlen et al.2018); however, the details of this unusual process remain to be elucidated. In vivo photocrosslinking data suggest that the switch protein SctU assembles onto the SctRST complex before recruitment of SctV (Dietsche et al.2016). In some T3SS, SctV is completely dispensable for assembly of the base (Wagner et al.2010) while it is necessary in others (Diepold et al.2010). Formation of the SctV ring around the assembled SctRSTU complex may further drive the lifting of the helical SctRST complex from the membrane and support interaction with SctJ and formation of the 24-mer SctJ ring (Fig. 2AB).
Outer membrane targeting and assembly of the secretin has been studied mainly for secretins of type II secretion systems but it is likely that the main conclusions can be inferred. Secretin targeting and assembly requires no other T3SS factors except in most cases a so-called pilotin (Sukhan et al.2001; Burghout et al.2004; Diepold et al.2010). The pilotin is a small lipoprotein that is targeted to the outer membrane by the Lol machinery (Collin et al.2011). It binds the C-terminal S-domain of cognate secretins (Okon et al.2008) and facilitates outer membrane targeting of secretin subunits and assembly of the secretin complex in the outer membrane (Burghout et al.2004; Collin et al.2011; Yin, Yan and Li 2018). Multimerization of secretin subunits leading to pre-pore formation is probably driven by interactions between adjacent N3-domains as well as by interactions of the S-domain with the C-domain of an adjacent subunit (Worrall et al.2016). In particular, this latter interaction may be supported by binding of the pilotin to the S-domain (Yin, Yan and Li 2018). Both mutations localizing to the N3-domain interaction site as well as to the S-domain–C-domain interaction site were shown to compromise multimerization of the secretin (Guilvout et al.2008; 2011; Worrall et al.2016). Outer membrane insertion of the pre-pore is a Bam independent process (Hoang et al.2011) and believed to occur through membrane association of an amphipathic helical loop followed by subsequent insertion of the distal β-barrel lip region of the C-domain (Worrall et al.2016) (Fig. 2BC).
The secretin protrudes about 18 nm into the lumen of the periplasm and penetrates the peptidoglycan layer at the N3-domain (Hu et al.2015) (Fig. 2BC). The assembled secretin complex is about 150 Å in diameter. Since the hole size of peptidoglycan is only about 25 Å (Vollmer, Blanot and de Pedro 2008), peptidoglycan reconstruction is required in order to allow penetration of the assembled secretin complex. Some pathogenicity islands encoding for T3SS also contain genes of peptidoglycan lytic enzymes to locally clear the peptidoglycan (Büttner 2012) (Fig. 2BC). These enzymes often seem to be regulated by type III secreted substrates to limit peptidoglycan degradation to the site of complex assembly (Oh et al.2007; Burkinshaw et al.2015) (Fig. 2C). Such a regulation necessitates ongoing type III secretion before interaction of the inner membrane assembly with the secretin, which is incompatible with a strict outside-in assembly model that starts with the secretin. So, two models for integration of the inner and outer membrane assemblies are currently discussed: (i) independent secretin assembly followed by interaction with the outer inner ring protein SctD and subsequent integration of the SctJ-export apparatus assembly (Diepold et al.2010) (Fig. 2B); and (ii) secretin assembly facilitated by peptidoglycan clearing above the already active inner membrane assembly (Fig. 2C) (Burkinshaw et al.2015). The extensive interaction of SctD with SctJ compared with the limited interaction of SctD with SctC also speak in favor of an assembly pathway where the secretin is recruited to a completely finished inner membrane subcomplex; however, different assembly pathways may occur in different bacterial species.
The cytoplasmic components are recruited to the injectisome by interactions of SctK with the cytoplasmic domain of SctD (Hu et al.2017). Interactions of SctO with the cytoplasmic domain of SctV do not seem to contribute to recruitment of the cytoplasmic components as judged by evidence from cryo-electron tomography and fluorescence microscopy (Diepold et al.2010; 2012; Hu et al.2017; Zhang et al.2017). Recruitment of the cytoplasmic components was shown to depend on SctK, SctQ, SctL and SctN in Yersinia while SctN was less critical for injectisome recruitment of SctK, SctQ and SctL in Salmonella (Lara-Tejero et al.2011; Diepold et al.2017; Zhang et al.2017). The cytoplasmic components already assemble into complexes before recruitment to the injectisome (Lara-Tejero et al.2011; Diepold et al.2017; Zhang et al.2017), however, there does not seem to be one clearly defined injectisome-free complex of cytoplasmic components but a dynamic and adaptive interaction network where new components can be incorporated at any point (Diepold et al.2017). Thus, while incomplete injectisome-free complexes of cytoplasmic components can be observed, injectisome recruitment requires the presence of all components except SctO, at least in Yersinia. In Salmonella, recruitment of the pod components SctK, SctQ and SctL to the injectisome may precede the association of SctN and SctO instead (Zhang et al.2017).
SUBSTRATE TARGETING AND SECRETION
Once assembly of the cytoplasmic components is completed, the machine is secretion competent. An N-terminal T3SS signal and often a chaperone-binding domain (CBD) guide export through the injectisome (Wattiau et al.1994; Sory et al.1995). The T3SS secretion signal comprises the first 10–25 residues of T3SS substrates (Sory et al.1995; Rüssmann et al.2002). Its sequence is highly variable and enriched in polar but depleted in charged and hydrophobic amino acids, which is reflected in a lack of secondary structure (Samudrala, Heffron and McDermott 2009). The role of the secretion signal in the targeting mechanism is still unclear. The 20–50 amino acids-long CBD is located downstream of the signal sequence (Cheng, Anderson and Schneewind 1997; Stebbins and Galán 2001). Cognate T3SS chaperones that are often encoded adjacent to their T3SS substrate bind to the CBD and guide the substrate to the cytoplasmic components (Stebbins and Galán 2001; Lara-Tejero et al.2011).
The role of cognate T3SS chaperones often goes beyond the mere targeting of substrates to the injectisome. Many secreted effectors contain localization signals for targeting inside the host cell, e.g. membrane localization domains for peripheral membrane targeting or transmembrane domains for insertion into host membranes. Inside the bacterial cell, chaperones turned out to serve also in preventing the erroneous targeting of membrane-localized effector proteins to the bacterial inner membrane by binding to membrane localization or even transmembrane domains (Letzelter et al.2006; Nguyen et al.2015; Krampen et al.2018). Furthermore, binding of the Salmonella chaperone SicA was found to prevent degradation of both SipB and SipC by cytosolic proteases (Tucker and Galan 2000).
The path substrates take through the secretion system is still unclear. It has been shown that chaperone-substrate complexes interact with sorting platform components (Spaeth, Chen and Valdivia 2009; Lara-Tejero et al.2011), with the ATPase (Akeda and Galán 2005), and with the cytoplasmic domain of the major export apparatus protein SctV (Portaliou et al.2017; Xing et al.2018). In light of the assembled injectisome, it is not clear, however, how chaperone-substrate complexes are handed over from one of these components to the other. In fact, there is no direct evidence that chaperone-substrate complexes bind the injectisome-assembled sorting platform or ATPase, and super-resolution microscopy data indicate a disperse distribution of substrates in the cytoplasm that is independent of injectisome components (Zhang et al.2017). In contrast, it was proposed that chaperone-substrate complexes already bind to the injectisome-free sorting platform as well as to the ATPase (Lara-Tejero et al.2011; Diepold et al.2015) (Fig. 3). The dynamic exchange of these cytoplasmic components may reflect the targeting and handing over of chaperone-substrate complexes from the cytoplasm to the injectisome, even though the kinetics of exchange were regarded as too slow to directly reflect secretion (Diepold et al.2015). For the related flagella T3SS, a model was proposed in which a free SctL2N complex (FliH2I in flagella) binds chaperones and serves as a dynamic carrier, while a flagellum-associated SctN6O (FliI6J) complex functions as a static substrate loader towards SctV (FlhA) (Bai et al.2014). Similar to flagellar T3SS, SctL was also shown to regulate the ATPase activity of the Shigella SctN in an oligomeric state-dependent manner, thus potentially reflecting the free and injectisome-bound functions of this protein complex (Case and Dickenson 2018).
Figure 3.
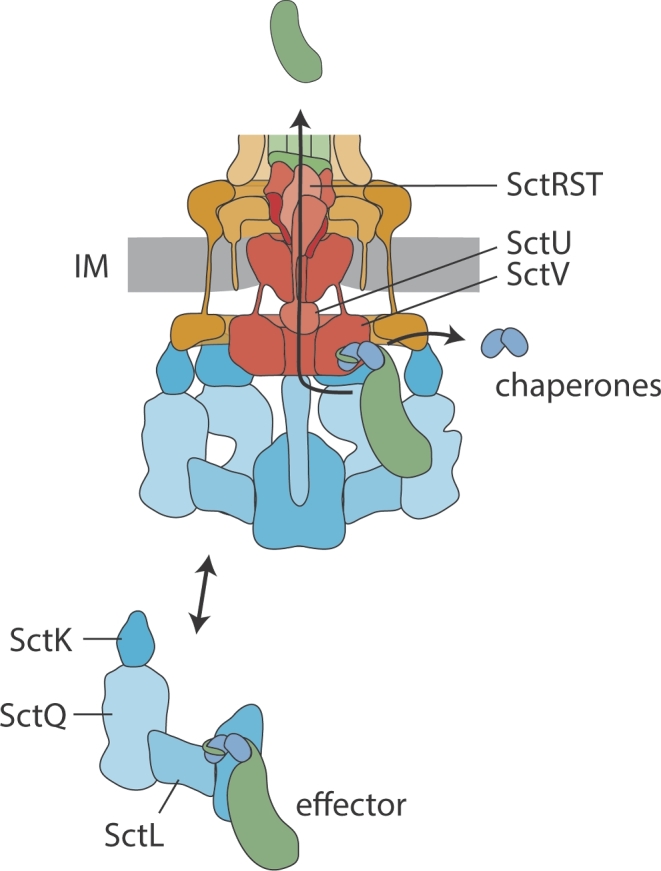
The secretion pathway through the T3SS injectisome. Chaperone-substrate complexes are loaded onto free complexes of cytoplasmic components. These complexes are believed to serve as dynamic substrate carriers and deliver chaperone-substrate complexes to the injectisome. Here, chaperone-substrates complexes bind to the cytoplasmic domain of the major export apparatus protein SctV. Supported by the action of the ATPase, substrates are dechaperoned, unfolded and inserted into the secretion channel formed by the export apparatus components and the needle filament. Abbreviation: IM, inner membrane.
Chaperones are stripped from the substrates prior to secretion and substrates are unfolded to accommodate transport through the narrow secretion channel. The ATPase SctN was reported to independently achieve dechaperoning and unfolding of substrates in vitro (Akeda and Galán 2005), however, the reports of folded flagellar chaperone-substrate complexes bound to the downstream of the ATPase-located cytoplasmic domain of the major export apparatus protein SctV suggest that the mechanism is more complex in vivo (Bange et al.2010; Xing et al.2018). It was even shown that the function of the ATPase is neither required for principal secretion in flagella nor in injectisomes (Minamino and Namba 2008; Paul et al.2008; Erhardt et al.2014) and that only very infrequent ATP hydrolysis is needed for efficient assembly of flagella (Minamino et al.2014). Thus, dechaperoning and substrate unfolding can be accomplished by SctV alone, but the function of the ATPase seems to enhance the efficiency of this process. Interestingly, the ATPase alone is sufficient to fuel secretion in flagella in vitro, even in the absence of the PMF (Terashima et al.2018). It is conceivable that the ATPase and SctV dechaperone and unfold substrates in a concerted action linked by SctO.
Secreted substrates are thought to translocate through the pore of SctV towards the entrance of the secretion channel in the inner membrane, which is most likely formed by the TMDs of the nine SctV (Erhardt et al.2017) (Fig. 3). Access to the entrance may be gated by the cytoplasmic domain of SctU, which was proposed to serve as a docking platform for substrates before entering the secretion channel (Evans et al.2013). Substrate translocation itself is mainly fueled by the PMF across the bacterial inner membrane (Lee et al.2014; Shen and Blocker 2016). Several conserved charged residues reside within the predicted TMD of SctV that seem to be implicated in utilization of the PMF (Hara, Namba and Minamino 2011; Minamino et al.2011; Erhardt et al.2017). After translocation across the membrane, substrates travel through the SctRST complex, the inner rod, the needle filament and the needle tip towards the host cell (Dohlich et al.2014; Radics, Königsmaier and Marlovits 2014).
FILAMENT ASSEMBLY, NEEDLE-LENGTH CONTROL AND SUBSTRATE SPECIFICITY SWITCHING
Secretion through T3SS begins with the export of early substrates. In most T3SS, only three different early substrates are secreted: the inner rod protein SctI, the needle filament protein SctF, and the needle-length control protein SctP (Kimbrough and Miller 2000; Kubori et al.2000). While these early substrates seem to be secreted without the help of cognate chaperones in most T3SS, SctF homologs of Yersinia Ysc-like T3SS require chaperoning by a heterodimeric complex of class III chaperones (YscG and YscE in Yersinia) (Quinaud et al.2007; Sun et al.2008). More detailed studies on the YscG and YscE homologs of Pseudomonas (PscG and PscE) revealed that PscG is the main SctF chaperone while PscE serves as a PscG-stabilizing cochaperone (Ple et al.2010). Interestingly, besides specialized SctF chaperones, secretion of early substrates requires the action of the additional components YscX and YscY in Ysc-like T3SS that do not seem to be present in other systems. YscX and YscY bind to the major export apparatus component YscV (SctV), most likely in a 1:1:1 stoichiometry (Diepold et al.2012; Gurung et al.2018). YscY is regarded as a dedicated chaperone of YscX, which is secreted after substrate specificity switching (Day and Plano 2000; Gurung et al.2018). It is tempting to speculate that YscX and YscY serve in recruiting YscF-YscG-YscE complexes to YscV in a manner analogous to the role of the gatekeeper protein in secretion of translocon-type substrates (see below).
The inner rod assembles at the distal end of the SctRST complex, intimately interacting with the N-termini of SctR and SctT (Dietsche et al.2016; Kuhlen et al.2018). A growing body of evidence suggests that the inner rod does not constitute a rod but merely a thin adapter between the export apparatus and the needle filament, hence we suggest the term needle adapter rather than inner rod (Zilkenat et al.2016). The needle is believed to subsequently assemble onto the needle adapter, although this has not been formally proven. However, it has been shown that the needle grows at its distal end (Poyraz et al.2010), implying that needle subunits travel all the way through the needle conduit to the growing tip, where they insert into and elongate the filament. Needle filament elongation is promoted by a special early substrate in some T3SS (e.g. OrgC of SPI-1) that is structurally similar to flagellar capping proteins (Kato et al.2018). Needle elongation is expected to induce the opening of the secretin gate in the outer membrane (Worrall et al.2016).
Based on studies of the flagellar SctP homolog FliK, it is believed that intermittent secretion of the needle-length control protein SctP measures the length of the growing needle (Erhardt et al.2011). Different hypotheses for how this results in the stopping of further needle elongation have been put forward, but no conclusive answer has emerged yet (Galán et al.2014). Folding of the secreted N-terminus of SctP may result in pulling the remainder of the protein out of the growing needle. At a discrete needle length, SctPN folding may no longer be possible, as the relevant part of SctP does not emerge at the needle tip. The deficiency of SctPN folding would result in a lack of pulling, which then may allow the folded C-terminal domain of SctP to interact with the cytoplasmic domain of SctU (Kinoshita et al.2017). The interaction of SctPC with SctUC is believed to bring needle elongation to a halt and to induce a substrate specificity switch, leading to secretion of intermediate substrates. In vitro, the interaction of SctU and SctP has been shown (Botteaux et al.2008; Monjarás Feria et al.2012; Ho et al.2016); however, their in vivo interaction remains to be proven. According to this model of needle-length control, the length of SctP, and more specifically, the length of its intrinsically disordered internal ruler domain, dictates the length of the needle, which in fact has been shown in a number of T3SS of flagella and injectisomes (Journet et al.2003; Wagner et al.2009; Erhardt et al.2010; Wee and Hughes 2015).
Due to its role in specificity switching from the secretion of early to intermediate substrates, SctU is also called switch protein. Switching relies on autocleavage of the cytoplasmic domain into two polypeptides (Minamino and Macnab 2000; Lavander et al.2002; Ferris et al.2005). While needle-length control and substrate specificity switching are often seen as one mechanism, they can in fact be separated into two independent processes by preventing autocleavage of SctU (Sorg et al.2007; Shen et al.2012; Monjarás Feria et al.2015). These mutants are able to control needle length but are not capable of switching to the secretion of intermediate substrates. This observation suggests that needle-length control (or rather, stopping of needle elongation) is neither executed by assembly of the needle tip at the distal end of the needle, thereby preventing further needle elongation, nor by the mere switching on of secretion of intermediate substrates, which would concomitantly lead to a reduced secretion of early substrates. Consequently, the stopping of further needle elongation upon needle-length control needs to be executed by a hitherto unidentified mechanism that might involve the active stopping of further secretion of early substrates, or a conformational rearrangement of the needle filament disallowing further assembly at its distal end.
As unclear as the mechanism of the stopping of further needle elongation is the mechanism of substrate specificity switching. The central role of SctU in this process was deduced from mutations within the cytoplasmic domain of this protein that could partly suppress the substrate specificity switching-deficient phenotype of a ΔsctP mutant (Hirano et al.1994; Kutsukake, Minamino and Yokoseki 1994; Williams et al.1996; Edqvist et al.2003). Originally, it was proposed that the autocleavage event of SctU might be the switch, however, we could show that SctU autocleavage occurs directly after folding of the cytoplasmic domain of SctU and even before incorporation into the base of the T3SS (Monjarás Feria et al.2015). We could also show that it is not the altered surface properties of the cleaved SctU that are critical for substrate specificity switching but the conformational flexibility gained by cleavage (Monjarás Feria et al.2015). The effect of the SctU ΔsctP suppressor mutations on the conformational flexibility of SctUC support this notion (Frost et al.2012) and suggest that suppression acts through an increased probability of autonomous switching caused by a greater conformational flexibility. It has been shown that the prevention of autocleavage of SctU simply disallows secretion of intermediate substrates because of differences in their signal sequence. Grafting of a late effector signal sequence onto a needle tip protein fully restored its secretion and its assembly at the needle tip (Sorg et al.2007).
Substrate specificity switching leads to secretion of needle tip proteins and their subsequent assembly at the distal end of the needle filament (Sorg et al.2007). It also leads to a charging of the sorting platform of the injectisome with hydrophobic translocators and to some premature secretion of these proteins (Lara-Tejero et al.2011). In this state, assembly of the machine is completed. It is ready for host-cell contact, translocon pore formation and subsequent injection of effector proteins.
HOST-CELL SENSING AND INJECTION
Full secretion through the injectisome is only induced upon host-cell sensing by the needle tip in vivo but can be mimicked by chemical signals (e.g. congo red and calcium chelation) in vitro (Bahrani, Sansonetti and Parsot 1997; Lee, Mazmanian and Schneewind 2001; Blocker et al.2008). The sensing signal is transmitted to the cytoplasmic side of the injectisome to switch to the secretion of late substrates. A T3SS component critical for induction of late substrate secretion is the so-called gatekeeper protein, SctW. Deletion of the gatekeeper typically results in abrogation of the secretion of translocators and oversecretion of late substrates (Forsberg et al.1991; Kubori and Galán 2002; Botteaux et al.2009). SctW was shown to bind to subdomain 2 of SctVC (Lee et al.2014; Shen and Blocker 2016; Portaliou et al.2017; Gaytán et al.2018). The SctV–SctW interaction participates in secretion regulation by providing a dedicated binding site for translocator-chaperone pairs and by occluding a binding site for late substrates (Portaliou et al.2017). Upon sensing host cells, SctW may be released from SctV to permit late substrate secretion. Free SctW is removed from the injectisome through secretion or degradation (Cheng, Kay and Schneewind 2001; Botteaux et al.2009; Yu et al.2010). It was also reported that the autocleaved C-terminal fragment of SctU is dislocated from the remainder of the protein and subsequently secreted upon host-cell sensing of Yersinia (Frost et al.2012); however, the functional implications of this release have not been elucidated.
Two different kinds of trigger for induction of late substrate secretion have been proposed: chemical or mechanical sensing of host cells. The needle tip and translocon components may be involved in both of these mechanisms (Veenendaal et al.2007; Armentrout and Rietsch 2016). In the T3SS encoded on Salmonella pathogenicity island 2 (SPI-2), transmission of the signal involves a change of pH through the lumen of the needle down to the cytoplasmic components to dislodge a gatekeeper complex (Yu et al.2010). T3SS of other species respond to changes in the calcium concentration by upregulating secretion (Forsberg et al.1987; Deng et al.2005). Whether calcium is sensed on the outside by the tip and the needle or inside by the export machinery is a matter of debate (Deng et al.2005; Shaulov et al.2017; Gaytán et al.2018). For other T3SS, transmission of the host-sensing signal may be of a mechanical nature by propagating a conformational change of protein subunits from the needle tip through the needle and inner rod towards the export apparatus and the cytoplasmic components. A number of mutants in the needle filament subunit SctF and in the inner rod protein SctI support this hypothesis (Kenjale et al.2005; Torruellas et al.2005; Davis and Mecsas 2007; Lefebre et al.2014).
In some T3SS of enteropathogens, effector protein injection is modulated by interaction of the tip complex with environmental small molecules like deoxycholate, which is present at high concentrations in the small intestine. Binding of deoxycholate induces substantial conformational changes of the tip proteins in Shigella (Dickenson et al.2011; Bernard et al.2017), leading to an injection-enhancing effect by supporting the recruitment of hydrophobic translocator proteins to the maturing tip complex in preparation of host membrane contact. Even though the tip proteins of the T3SS of Shigella and Salmonella SPI-1 are structurally very similar, deoxycholate binding does not have an enhancing effect on secretion in Salmonella (Wang et al.2010; Lunelli et al.2011), rather, invasion of Salmonella is inhibited in the presence of high concentrations of bile salts (Prouty and Gunn 2000).
Upon host-cell sensing, the hydrophobic translocators are inserted into the host membrane, forming multimeric pores for substrate deployment directly into the host cell cytosol (Montagner, Arquint and Cornelis 2011). Formation of the translocon pores completes the pathway for injection of effector proteins from the bacterial cytoplasm directly into the host cell (Park et al.2018). Whether the secretion path is always continuous, or whether effector proteins can also be deployed from the surface of the bacterium through a translocon that is not directly injectisome-associated (Akopyan et al.2011; Edgren et al.2012), remains to be further elucidated.
CONCLUSIONS AND OUTLOOK
Impressive progress has been made towards elucidating the assembly, structure and function of bacterial type III secretion systems in recent years, not least because of the cryo-electron microscopy revolution. Unfortunately, however, our understanding of the molecular mechanisms of substrate targeting and secretion, of needle-length control and substrate specificity switching, of host-cell sensing and translocon assembly, is still rudimentary and awaits detailed biochemical investigation. Advances in our understanding of T3SS will foster the exploitation of this intricate system for therapeutic (Bai et al.2018; González-Prieto and Lesser 2018) and biotechnological (Widmaier et al.2009; Singer et al.2012; Guo et al.2014; Azam and Tullman-Ercek 2016) purposes, as well as the development of anti-virulence drugs targeting these machines essential for the pathogenicity of many bacteria (Duncan, Linington and Auerbuch 2012; McShan and De Guzman 2015; Mühlen and Dersch 2016).
Acknowledgements
Work in the laboratory of Samuel Wagner related to this article is supported by the Deutsche Forschungsgemeinschaft (DFG) as part of the Collaborative Research Center (SFB) 766 Bacterial Cell Envelope, project B14, and by the German Center for Infection Research (DZIF), project TTU 06.801 WP1a.
Conflict of interest. None declared.
REFERENCES
- Abby SS, Rocha EPC. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet 2012;8:e1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrusci P, Vergara-Irigaray M, Johnson S et al. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 2013;20:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akeda Y, Galán JE. Chaperone release and unfolding of substrates in type III secretion. Nature 2005;437:911–5. [DOI] [PubMed] [Google Scholar]
- Akopyan K, Edgren T, Wang-Edgren H et al. Translocation of surface-localized effectors in type III secretion. Proc Natl Acad Sci 2011;108:1639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaoui A, Woestyn S, Sluiters C et al. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol 1994;176:4534–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armentrout EI, Rietsch A. The type III secretion translocation pore senses host cell contact. PLoS Pathog 2016;12:e1005530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam A, Tullman-Ercek D. Type-III secretion filaments as scaffolds for inorganic nanostructures. J R Soc Interface 2016;13:20150938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrani FK, Sansonetti PJ, Parsot C. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect Immun 1997;65:4005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Li Z, Umezawa A et al. Bacterial type III secretion system as a protein delivery tool for a broad range of biomedical applications. Biotechnol Adv 2018;36:482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Morimoto YV, Yoshimura SDJ et al. Assembly dynamics and the roles of FliI ATPase of the bacterial flagellar export apparatus. Sci Rep 2015;4:6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange G, Kümmerer N, Engel C et al. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci 2010;107:11295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CS, Inoue T, Meshcheryakova IV et al. Function of the conserved FHIPEP domain of the flagellar type III export apparatus, protein FlhA. Mol Microbiol 2016;100:278–88. [DOI] [PubMed] [Google Scholar]
- Bernard AR, Jessop TC, Kumar P et al. Deoxycholate-enhanced Shigella virulence is regulated by a rare π-helix in the type three secretion system tip protein IpaD. Biochemistry 2017;56:6503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker AJ, Deane JE, Veenendaal AKJ et al. What's the point of the type III secretion system needle? Proc Natl Acad Sci 2008;105:6507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botteaux A, Sani M, Kayath CA et al. Spa32 interaction with the inner-membrane Spa40 component of the type III secretion system of Shigella flexneri is required for the control of the needle length by a molecular tape measure mechanism. Mol Microbiol 2008;70:1515–28. [DOI] [PubMed] [Google Scholar]
- Botteaux A, Sory MP, Biskri L et al. MxiC is secreted by and controls the substrate specificity of the Shigella flexneri type III secretion apparatus. Mol Microbiol 2009;71:449–60. [DOI] [PubMed] [Google Scholar]
- Broz P, Mueller CA, Müller SA et al. Function and molecular architecture of the Yersinia injectisome tip complex. Mol Microbiol 2007;65:1311–20. [DOI] [PubMed] [Google Scholar]
- Burghout P, Beckers F, de Wit E et al. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J Bacteriol 2004;186:5366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkinshaw BJ, Deng W, Lameignere E et al. Structural analysis of a specialized type III secretion system peptidoglycan-cleaving enzyme. J Biol Chem 2015;290:10406–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev 2012;76:262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek KP, Hamaoka BY, Ghosh P. Two translation products of Yersinia yscQ assemble to form a complex essential to type III secretion. Biochemistry 2012;51:1669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case HB, Dickenson NE. MxiN differentially regulates monomeric and oligomeric species of the Shigella type three secretion system ATPase Spa47. Biochemistry 2018;57:2266–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Zhong D, Nordhues BA et al. The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci 2011;20:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LW, Anderson DM, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol 1997;24:757–65. [DOI] [PubMed] [Google Scholar]
- Cheng LW, Kay O, Schneewind O. Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J Bacteriol 2001;183:5293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M, Shen D-K, Makino F et al. Three-dimensional electron microscopy reconstruction and cysteine-mediated crosslinking provide a model of the type III secretion system needle tip complex. Mol Microbiol 2015;95:31–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo CM, Galan JE. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol 1997;24:747–56. [DOI] [PubMed] [Google Scholar]
- Collin S, Guilvout I, Nickerson NN et al. Sorting of an integral outer membrane protein via the lipoprotein-specific Lol pathway and a dedicated lipoprotein pilotin. Mol Microbiol 2011;80:655–65. [DOI] [PubMed] [Google Scholar]
- Davis AJ, Mecsas J. Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, YscF, that specifically abrogate effector translocation into host cells. J Bacteriol 2007;189:83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JB, Plano GV. The Yersinia pestis YscY protein directly binds YscX, a secreted component of the type III secretion machinery. J Bacteriol 2000;182:1834–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JE, Graham SC, Mitchell EP et al. Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol Microbiol 2008;69:267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBord KL, Lee VT, Schneewind O. Roles of LcrG and LcrV during type III targeting of effector Yops by Yersinia enterocolitica. J Bacteriol 2001;183:4588–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers J-P, Habenstein B, Loquet A et al. High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy. Nat Commun 2014;5:4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers J-P, Sgourakis NG, Gupta R et al. The common structural architecture of Shigella flexneri and Salmonella typhimurium type three secretion needles. PLoS Pathog 2013;9:e1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Li Y, Hardwidge PR et al. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun 2005;73:2135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Marshall NC, Rowland JL et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Micro 2017;15:323–37. [DOI] [PubMed] [Google Scholar]
- Derewenda U, Mateja A, Devedjiev Y et al. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure 2004;12:301–6. [DOI] [PubMed] [Google Scholar]
- Dickenson NE, Choudhari SP, Adam PR et al. Oligomeric states of the Shigella translocator protein IpaB provide structural insights into formation of the type III secretion translocon. Protein Sci 2013;22:614–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickenson NE, Zhang L, Epler CR et al. Conformational changes in IpaD from Shigella flexneri upon binding bile salts provide insight into the second step of type III secretion. Biochemistry 2011;50:172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A, Amstutz M, Abel S et al. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J 2010;29:1928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A, Kudryashev M, Delalez NJ et al. Composition, formation, and regulation of the cytosolic C-ring, a dynamic component of the type III secretion injectisome. Stock AM. (ed.). PLoS Biol 2015;13:e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A, Sezgin E, Huseyin M et al. A dynamic and adaptive network of cytosolic interactions governs protein export by the T3SS injectisome. Nat Comms 2017;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A, Wagner S. Assembly of the bacterial type III secretion machinery. FEMS Microbiol Rev 2014;38:802–22. [DOI] [PubMed] [Google Scholar]
- Diepold A, Wiesand U, Amstutz M et al. Assembly of the Yersinia injectisome: the missing pieces. Mol Microbiol 2012;85:878–92. [DOI] [PubMed] [Google Scholar]
- Dietsche T, Tesfazgi Mebrhatu M, Brunner MJ et al. Structural and functional characterization of the bacterial type III secretion export apparatus. PLoS Pathog 2016;12:e1006071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlich K, Zumsteg AB, Goosmann C et al. A substrate-fusion protein is trapped inside the type III secretion system channel in Shigella flexneri. PLoS Pathog 2014;10:e1003881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MC, Linington RG, Auerbuch V. Chemical inhibitors of the type three secretion system: Disarming bacterial pathogens. Antimicrob Agents Chemother 2012;56:5433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgren T, Forsberg A, Rosqvist R et al. Type III secretion in Yersinia: injectisome or not? PLoS Pathog 2012;8:e1002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist PJ, Olsson J, Lavander M et al. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol 2003;185:2259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epler CR, Dickenson NE, Bullitt E et al. Ultrastructural analysis of IpaD at the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri. J Mol Biol 2012;420:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Hirano T, Su Y et al. The role of the FliK molecular ruler in hook‐length control in Salmonella enterica. Mol Microbiol 2010;75:1272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Mertens ME, Fabiani FD et al. ATPase-Independent Type-III Protein Secretion in Salmonella enterica. PLoS Genet 2014;10:e1004800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Singer HM, Wee DH et al. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J 2011;30:2948–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Wheatley P, Kim EA et al. Mechanism of type-III protein secretion: Regulation of FlhA conformation by a functionally critical charged-residue cluster. Mol Microbiol 2017;104:234–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erskine PT, Knight MJ, Ruaux A et al. High resolution structure of BipD: an invasion protein associated with the type III secretion system of Burkholderia pseudomallei. J Mol Biol 2006;363:125–36. [DOI] [PubMed] [Google Scholar]
- Evans LDB, Poulter S, Terentjev EM et al. A chain mechanism for flagellum growth. Nature 2013;504:287–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris HU, Furukawa Y, Minamino T et al. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem 2005;280:41236–42. [DOI] [PubMed] [Google Scholar]
- Filloux A, Voulhoux R. Multiple structures disclose the secretins' secrets. J Bacteriol 2018;200:914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg A, Bölin I, Norlander L et al. Molecular cloning and expression of calcium-regulated, plasmid-coded proteins of Y. pseudotuberculosis. Microb Pathog 1987;2:123–37. [DOI] [PubMed] [Google Scholar]
- Forsberg A, Viitanen AM, Skurnik M et al. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol 1991;5:977–86. [DOI] [PubMed] [Google Scholar]
- Frost S, Ho O, Login FH et al. Autoproteolysis and Intramolecular Dissociation of Yersinia YscU Precedes Secretion of Its C-Terminal Polypeptide YscUCC. PLoS One 2012;7:e49349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE, Lara-Tejero M, Marlovits TC et al. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 2014;68:415–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE. SnapShot: effector proteins of type III secretion systems. Cell 2007;130:192.e1–2. [DOI] [PubMed] [Google Scholar]
- Galán JE. Common themes in the design and function of bacterial effectors. Cell Host & Microbe 2009;5:571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaytán MO, Monjarás Feria J, Soto E et al. Novel insights into the mechanism of SepL-mediated control of effector secretion in enteropathogenic Escherichia coli. MicrobiologyOpen 2018;7:e00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Prieto C, Lesser CF. Rationale redesign of type III secretion systems: toward the development of non-pathogenic E. coli for in vivo delivery of therapeutic payloads. Curr Opin Microbiol 2018;41:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilvout I, Chami M, Berrier C et al. In vitro multimerization and membrane insertion of bacterial outer membrane secretin PulD. J Mol Biol 2008;382:13–23. [DOI] [PubMed] [Google Scholar]
- Guilvout I, Nickerson NN, Chami M et al. Multimerization-defective variants of dodecameric secretin PulD. Res Microbiol 2011;162:180–90. [DOI] [PubMed] [Google Scholar]
- Guo S, Alshamy I, Hughes KT et al. Analysis of factors that affect FlgM-dependent type III secretion for protein purification with Salmonella Typhimurium. J Bacteriol 2014;196:2333–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung JM, Amer AAA, Francis MK et al. Heterologous complementation studies with the YscX and YscY protein families reveals a specificity for Yersinia pseudotuberculosis type III secretion. Front Cell Infect Microbiol 2018;8:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara N, Namba K, Minamino T. Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLoS One 2011;6:e22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon DE, Murphy JL, Davis AJ et al. A mutant with aberrant extracellular LcrV-YscF interactions fails to form pores and translocate Yop effector proteins but retains the ability to trigger Yop secretion in response to host cell contact. J Bacteriol 2013;195:2244–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi S, Oosawa K et al. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol 1994;176:5439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho O, Rogne P, Edgren T et al. Characterization of the ruler protein interaction interface on the substrate specificity switch protein in the Yersinia type III secretion system. J Biol Chem 2016;jbc.M116.770255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang HH, Nickerson NN, Lee VT et al. Outer membrane targeting of Pseudomonas aeruginosa proteins shows variable dependence on the components of Bam and Lol machineries. mBio 2011;2:e00246–11–e00246–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Lara-Tejero M, Kong Q et al. In situ molecular architecture of the Salmonella type III secretion machine. Cell 2017;168:1065–1074.e10.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Morado DR, Margolin W et al. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci USA 2015;112:1047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 1998;62:379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibuki T, Imada K, Minamino T et al. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat Struct Mol Biol 2011; 18:277–82. [DOI] [PubMed] [Google Scholar]
- Imada K, Minamino T, Uchida Y et al. Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator. Proc Natl Acad Sci USA 2016;113:3633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S, Roversi P, Espina M et al. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem 2007;282:4035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet L, Agrain C, Broz P et al. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 2003;302:1757–60. [DOI] [PubMed] [Google Scholar]
- Kato J, Dey S, Soto JE et al. A protein secreted by the Salmonella type III secretion system controls needle filament assembly. Elife 2018;7:6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenjale R, Wilson J, Zenk SF et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem 2005;280:42929–37. [DOI] [PubMed] [Google Scholar]
- Kimbrough TG, Miller SI. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci 2000;97:11008–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Aizawa S-I, Inoue Y et al. The role of intrinsically disordered C-terminal region of FliK in substrate specificity switching of the bacterial flagellar type III export apparatus. Mol Microbiol 2017;105:572–88. [DOI] [PubMed] [Google Scholar]
- Kowal J, Chami M, Ringler P et al. Structure of the dodecameric Yersinia enterocolitica secretin YscC and its trypsin-resistant core. Structure 2013;21:2152–61. [DOI] [PubMed] [Google Scholar]
- Krampen L, Malmsheimer S, Grin I et al. Revealing the mechanisms of membrane protein export by virulence-associated bacterial secretion systems. Nat Commun 2018;9:3467. 10.1038/s41467-018-05969-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Galán JE. Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J Bacteriol 2002;184:4699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Sukhan A, Aizawa SI et al. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci 2000;97:10225–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlen L, Abrusci P, Johnson S et al. Structure of the core of the type III secretion system export apparatus. Nat Struct Mol Biol 2018;25:583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K, Minamino T, Yokoseki T. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol 1994;176:7625–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M, Kato J, Wagner S et al. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 2011;331:1188–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavander M, Sundberg L, Edqvist PJ et al. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol 2002;184:4500–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P-C, Zmina SE, Stopford CM et al. Control of type III secretion activity and substrate specificity by the cytoplasmic regulator PcrG. Proc Natl Acad Sci 2014;111:E2027–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Mazmanian SK, Schneewind O. A program of Yersinia enterocolitica type III secretion reactions is activated by specific signals. J Bacteriol 2001;183:4970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebre MD, Lefebre MD, Galan JE et al. The inner rod protein controls substrate switching and needle length in a Salmonella type III secretion system. Proc Natl Acad Sci 2014;111:817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzelter M, Sorg I, Mota LJ et al. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J 2006;25:3223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loquet A, Sgourakis NG, Gupta R et al. Atomic model of the type III secretion system needle. Nature 2012;486:276–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lountos GT, Austin BP, Nallamsetty S et al. Atomic resolution structure of the cytoplasmic domain of Yersinia pestis YscU, a regulatory switch involved in type III secretion. Protein Sci 2009;18:467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunelli M, Hurwitz R, Lambers J et al. Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog 2011;7:e1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenne M-N, Journet L, Mota LJ et al. Genetic analysis of the formation of the Ysc–Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, YscF and YopN. Microb Pathog 2003;35:243–58. [DOI] [PubMed] [Google Scholar]
- Marlovits TC, Kubori T, Unger VM et al. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 2006;441:637–40. [DOI] [PubMed] [Google Scholar]
- Matson JS, Nilles ML. LcrG-LcrV interaction is required for control of Yops secretion in Yersinia pestis. J Bacteriol 2001;183:5082–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell MA, Marcoux J, McVicker G et al. Characterisation of Shigella ;Spa33 and Thermotoga FliM/N reveals a new model for C-ring assembly in T3SS. Mol Microbiol 2016;99:749–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShan AC, De Guzman RN. The bacterial type III secretion system as a target for developing new antibiotics. Chem Biol Drug Des 2015;85:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Macnab RM. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J Bacteriol 2000;182:4906–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Morimoto YV, Hara N et al. An energy transduction mechanism used in bacterial flagellar type III protein export. Nat Commun 2011;2:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Morimoto YV, Kinoshita M et al. The bacterial flagellar protein export apparatus processively transports flagellar proteins even with extremely infrequent ATP hydrolysis. Sci Rep 2015;4:7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 2008;451:485–8. [DOI] [PubMed] [Google Scholar]
- Monjarás Feria J, García-Gómez E, Espinosa N et al. Role of EscP (Orf16) in injectisome biogenesis and regulation of type III protein secretion in enteropathogenic Escherichia coli. J Bacteriol 2012;194:6029–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monjarás Feria JV, Lefebre MD, Stierhof Y-D et al. Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium. mBio 2015;6:e01459–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner C, Arquint C, Cornelis GR. Translocators YopB and YopD from Yersinia enterocolitica Form a Multimeric Integral Membrane Complex in Eukaryotic Cell Membranes. J Bacteriol 2011, DOI: 10.1128/JB.05555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CA, Broz P, Müller SA et al. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 2005;310:674–6. [DOI] [PubMed] [Google Scholar]
- Mühlen S, Dersch P. Anti-virulence strategies to target bacterial infections. Curr Top Microbiol Immunol 2016;398:147–83. [DOI] [PubMed] [Google Scholar]
- Nguyen VS, Jobichen C, Tan KW et al. Structure of AcrH–AopB Chaperone-Translocator Complex Reveals a Role for Membrane Hairpins in Type III Secretion System Translocon Assembly. Structure 2015;23:2022–31. [DOI] [PubMed] [Google Scholar]
- Notti RQ, Bhattacharya S, Lilic M et al. A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat Commun 2015;6:7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H-S, Kvitko BH, Morello JE et al. Pseudomonas syringae lytic transglycosylases coregulated with the type III secretion system contribute to the translocation of effector proteins into plant cells. J Bacteriol 2007;189:8277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okon M, Moraes TF, Lario PI et al. Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure 2008;16:1544–54. [DOI] [PubMed] [Google Scholar]
- Park D, Lara-Tejero M, Waxham MN et al. Visualization of the type III secretion mediated Salmonella-host cell interface using cryo-electron tomography. bioRxiv 2018:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Erhardt M, Hirano T et al. Energy source of flagellar type III secretion. Nature 2008;451:489–92. [DOI] [PubMed] [Google Scholar]
- Ple S, Job V, Dessen A et al. Cochaperone interactions in export of the type III needle component PscF of Pseudomonas aeruginosa. J Bacteriol 2010;192:3801–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portaliou AG, Tsolis KC, Loos MS et al. Hierarchical protein targeting and secretion is controlled by an affinity switch in the type III secretion system of enteropathogenic Escherichia coli. EMBO J 2017;36:3517–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyraz O, Schmidt H, Seidel K et al. Protein refolding is required for assembly of the type three secretion needle. Nat Struct Mol Biol 2010;17:788–92. [DOI] [PubMed] [Google Scholar]
- Prouty AM, Gunn JS. Salmonella enterica serovar typhimurium invasion is repressed in the presence of bile. Infect Immun 2000;68:6763–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinaud M, Plé S, Job V et al. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc Natl Acad Sci 2007;104:7803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radics J, Königsmaier L, Marlovits TC. Structure of a pathogenic type 3 secretion system in action. Nat Struct Mol Biol 2014;21:82–87. [DOI] [PubMed] [Google Scholar]
- Rathinavelan T, Lara-Tejero M, Lefebre M et al. NMR model of PrgI-SipD interaction and its implications in the needle-tip assembly of the Salmonella type III secretion system. J Mol Biol 2014;426:2958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosqvist R, Persson C, Håkansson S et al. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contrib Microbiol Immunol 1995;13:230–4. [PubMed] [Google Scholar]
- Rüssmann H, Kubori T, Sauer J et al. Molecular and functional analysis of the type III secretion signal of the Salmonella enterica InvJ protein. Mol Microbiol 2002;46:769–79. [DOI] [PubMed] [Google Scholar]
- Samudrala R, Heffron F, McDermott JE. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog 2009;5:e1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraidt O, Lefebre MD, Brunner MJ et al. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog 2010;6:e1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraidt O, Marlovits TC. Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science 2011;331:1192–5. [DOI] [PubMed] [Google Scholar]
- Sekiya K, Ohishi M, Ogino T et al. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc Natl Acad Sci 2001;98:11638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulov L, Gershberg J, Deng W et al. The ruler protein EscP of the enteropathogenic Escherichia coli type III secretion system is involved in calcium sensing and secretion hierarchy regulation by interacting with the gatekeeper protein SepL. mBio 2017;8:e01733–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D-K, Blocker AJ. MxiA, MxiC and IpaD regulate substrate selection and secretion mode in the T3SS of Shigella flexneri. PLoS One 2016;11:e0155141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D-K, Moriya N, Martinez-Argudo I et al. Needle length control and the secretion substrate specificity switch are only loosely coupled in the type III secretion apparatus of Shigella. Microbiology 2012;158:1884–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer HM, Erhardt M, Steiner AM et al. Selective purification of recombinant neuroactive peptides using the flagellar type III secretion system. mBio 2012;3:e00115–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Sukovich DJ, Ciccarelli L et al. Control of type III protein secretion using a minimal genetic system. Nat Comms 2017;8:14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg I, Wagner S, Amstutz M et al. YscU recognizes translocators as export substrates of the Yersinia injectisome. EMBO J 2007;26:3015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sory MP, Boland A, Lambermont I et al. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci 1995;92:11998–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeth KE, Chen Y-S, Valdivia RH. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog 2009;5:e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreter T, Yip CK, Sanowar S et al. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat Struct Mol Biol 2009;16:468–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galán JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 2001;414:77–81. [DOI] [PubMed] [Google Scholar]
- Sukhan A, Sukhan A, Kubori T et al. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J Bacteriol 2001;183:1159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Tropea JE, Austin BP et al. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J Mol Biol 2008;377:819–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Kawamoto A, Tatsumi C et al. In Vitro reconstitution of functional type III protein export and insights into flagellar assembly. mBio 2018;9:e00988–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torruellas J, Jackson MW, Pennock JW et al. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol Microbiol 2005;57:1719–33. [DOI] [PubMed] [Google Scholar]
- Tucker SC, Galan JE. Complex function for SicA, a Salmonella enterica serovar typhimurium type III secretion-associated chaperone. J Bacteriol 2000;182:2262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenendaal AKJ, Hodgkinson JL, Schwarzer L et al. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol 2007;63:1719–30. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev 2008;32:149–67. [DOI] [PubMed] [Google Scholar]
- Wagner S, Königsmaier L, Lara-Tejero M et al. Organization and coordinated assembly of the type III secretion export apparatus. Proc Natl Acad Sci USA 2010;107:17745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Sorg I, Degiacomi M et al. The helical content of the YscP molecular ruler determines the length of the Yersinia injectisome. Mol Microbiol 2009;71:692–701. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nordhues BA, Zhong D et al. NMR characterization of the interaction of the Salmonella type III secretion system protein SipD and bile salts. Biochemistry 2010;49:4220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattiau P, Bernier B, Deslée P et al. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci 1994;91:10493–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee DH, Hughes KT. Molecular ruler determines needle length for the Salmonella Spi-1 injectisome. Proc Natl Acad Sci USA 2015;112:4098–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmaier DM, Tullman-Ercek D, Mirsky EA et al. Engineering the Salmonella type III secretion system to export spider silk monomers. Mol Syst Biol 2009;5:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesand U, Sorg I, Amstutz M et al. Structure of the type III secretion recognition protein YscU from Yersinia enterocolitica. J Mol Biol 2009;385:854–66. [DOI] [PubMed] [Google Scholar]
- Williams AW, Yamaguchi S, Togashi F et al. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol 1996;178:2960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall LJ, Hong C, Vuckovic M et al. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature 2016;540:597–601. [DOI] [PubMed] [Google Scholar]
- Worrall LJ, Vuckovic M, Strynadka NCJ. Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci 2010;19:1091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Q, Shi K, Portaliou A et al. Structures of chaperone-substrate complexes docked onto the export gate in a type III secretion system. Nat Commun 2018;9:1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, Yan Z, Li X. Structural insight into the assembly of the type II secretion system pilotin–secretin complex from enterotoxigenic Escherichia coli. Nat Microbiol 2018;3:581–7. [DOI] [PubMed] [Google Scholar]
- Yip CK, Kimbrough TG, Felise HB et al. Structural characterization of the molecular platform for type III secretion system assembly. Nature 2005;435:702–7. [DOI] [PubMed] [Google Scholar]
- Yu X-J, McGourty K, Liu M et al. pH sensing by intracellular Salmonella induces effector translocation. Science 2010;328:1040–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarivach R, Deng W, Vuckovic M et al. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 2008;453:124–7. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lara-Tejero M, Bewersdorf J et al. Visualization and characterization of individual type III protein secretion machines in live bacteria. Proc Natl Acad Sci USA 2017;114:6098–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilkenat S, Franz-Wachtel M, Stierhof Y-D et al. Determination of the stoichiometry of the complete bacterial type III secretion needle complex using a combined quantitative proteomic approach. Mol Cell Proteomics 2016;15:1598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]