Abstract
The Sine oculis homeobox 1 (Six1) gene is important for skeletal muscle growth and fiber specification; therefore, it is considered as a promising candidate gene that may influence porcine growth and meat quality traits. Nevertheless, the association of Six1 with these processes and the mechanisms regulating its expression remain unclear. The objectives of this study were to identify variant sites of Six1 in different pig breeds, conduct association analysis to evaluate the relationship between polymorphisms of these variants and porcine production traits in Pietrain × Duroc × Landrace × Yorkshire commercial pigs, and explore the potential regulatory mechanisms of Six1 affecting production traits. A total of 12 variants were identified, including 10 single- nucleotide variations (SNVs), 1 insertion– deletion (Indel), and 1 (AC)n microsatellite. Association analysis demonstrated that the SNV, g.1595A>G, was significantly associated with meat color (redness, a*); individuals with the G allele had greater a* values (P < 0.05). Moreover, our results demonstrated that the (AC)n polymorphism in the Six1 promoter was significantly associated with weaning weight (P < 0.05), carcass weight (P < 0.05), and thoracic and lumbar back fat (P < 0.01).In addition, we found that the (AC)n variant was closely related with Six1 expression levels and demonstrated this polymorphism on promoter activity by in vitro experiments. Overall, this study provides novel evidence for elucidating the effects of Six1 on porcine production traits as promising candidate and describes two variants with these traits, which are potential reference markers for pig molecular breeding. In addition, our data on the relationship between porcine Six1 expression and the polymorphic (AC)n microsatellite in its promoter may facilitate similar studies in other species.
Keywords: microsatellite, pig, production traits, Six1, variation
INTRODUCTION
In the field of porcine molecular breeding, several major genes affecting important economic traits have been identified, including ryanodine receptor (RYR1) gene (Fujii et al., 1991), regulatory γ subunit of adenosine monophosphate-activated protein kinase (PRKAG3) gene (Milan et al., 2000), insulin-like growth factor 2 (IGF2) (Van Laere et al., 2003) and melanocortin-4 receptor (MC4R) (Kim et al., 2000), and two novel major genes, vertnin (VRTN), which is related to vertebral number, and phosphorylase kinase catalytic subunit gamma 1 (PHKG1), which is associated with glycolytic potential (Mikawa et al., 2011; Ma et al., 2014). Nevertheless, the major genes or variants directly applicable to porcine molecular breeding remain limited. Sine oculis homeobox 1 (Six1) belongs to the Six gene family, which is important in regulation of muscle growth (Wu et al., 2014) and has been demonstrated to drive fast-type fiber specification in mice and zebrafish (Grifone et al., 2004; Bessarab et al., 2008; Sakakibara et al., 2016). Our previous results demonstrate that porcine Six1 expression is most abundant in adult skeletal muscles and that the polymorphism, g.1595A>G, in first intron is associated with meat color in a Yorkshire × Meishan F2 pig population (Wu et al., 2011). Therefore, we hypothesized that Six1 can affect porcine growth and meat quality traits. In this study, in an effort to generate more evidence to support our initial findings, we re- sequenced ~2 kb of the region upstream of the Six1 translation start site (ATG) and part of its first intron. The objectives of this study were to identify the variant sites in Six1 and analyze their diversity in seven pig breeds, to conduct association analysis between the variants identified and porcine production traits in a Pietrain × Duroc × Landrace × Yorkshire (P×D×L×Y) commercial pig population, and to evaluate the effect of the identified (AC)n microsatellite promoter polymorphism on Six1 expression and transcriptional activity.
MATERIALS AND METHODS
Animals and Tissues
All procedures involving animals were in compliance with the guidelines for the care and use of experimental animals established by the Ministry of Agriculture of China and this study was approved by the Institutional Animal Care and Use Committee of Nanjing Agricultural University, Nanjing, Jiangsu, P.R. China.
A total of 144 individuals, representing seven pig breeds, were used to identify the SNPs in Six1, they are as follows: two western lean-type breeds (20 Yorkshire and 20 Landrace pigs), four Chinese indigenous breeds (20 Meishan, 24 Mi, 20 Shawutou, and 20 Erhualian pigs), and a cultivated breed (20 Suhuai pigs composed of 75% Yorkshire and 25% Huai). A commercial pig population, comprising 1,310 Pietrain (25%) × Duroc (25%) × Landrace (25%) × Yorkshire (25%) (P×D×L×Y) animals, generated by random crossing of 12 P×D boars with 144 L×Y sows, was used for association analysis with the porcine traits. Detailed information about the experimental pig population is provided in our previous study (Li et al., 2016). In addition, the longissimus dorsi muscles were harvested from the commercial pig population (n = 1,310), immediately frozen in liquid nitrogen, and then stored at −80 °C.
Targeted Sequencing and Data Analysis
The previous study had confirmed that there were no mutations in Six1 coding region, but the potential mutations in the promoter region were not validated completely (Wu et al., 2011). For comprehensive identification of genetic variations in the Six1 promoter, genomic DNA was extracted from ear tissues of seven different pig breeds containing 144 individuals and muscle tissues of 509 P×D×L×Y commercial pigs. Approximately 2 kb of sequence upstream of the Six1 translation start site (ATG) and part of its first intron sequence, which contained an identified single-nucleotide polymorphism site (g.1595A>G) in our previous study (Wu et al., 2011), were included in the subsequent analyses.
FastTarget technology developed by Genesky Biotechnologies (Shanghai, China) was adopted to identify the potential genetic variation sites. Ten pairs of primers were designed (Table 1) according to the reference sequence (NC_010443.5), with the addition of a universal sequence (Illumina, San Diego, CA), and polymerase chain reaction (PCR) conditions were optimized for amplification using each primer pair and for multiplex reactions. The multiplex PCR amplification was evaluated by a specific method based on capillary electrophoresis. According to the results of multiplex PCR optimization, the 10 pairs of primers were divided into 4 groups (4 panels) to conduct multiplex PCR. Then, equal amounts of the multiplex PCR products resulting from amplification using the four panels were pooled for each sample, and sample-specific sequence labels (Illumina) were added to the mixed multiplex PCR products by PCR amplification with index primers. Furthermore, the labeled PCR products from a specific number of samples (n = 80–100 samples) were mixed equally, and then sequencing was performed on the Illumina MiSeq 2000 platform using 2 × 300 bp paired-end sequencing mode.
Table 1.
Primers used in this study
| Gene name | Length of fragment | Primers* | Location | Application |
|---|---|---|---|---|
| Six1 | 247 | F1: CTCCCACTCCCACTTATGTCTCA R1: ATGCAAGGAGATAAGGGGAGCCA |
Chr1:189625957-189626204 | Amplification |
| Six1 | 307 | F2: TCAGAGYTAACTAGAGCCATAAAGTCACT R2: TGTGTAGATTGCAATGCTGGCCT |
Chr1:189625716-189626023 | Amplification |
| Six1 | 255 | F3: GGACATCTCTCATTCTCTGGCAGT R3: CCCTGGGAACTTCACACGAAAGG |
Chr1:189625517-189625768 | Amplification |
| Six1 | 326 | F4: CCTCTCAGAACAGGGCAAGGATT R4: AAAGGCCGCTACTATTTGGGACA |
Chr1:189625301-189625627 | Amplification |
| Six1 | 313 | F5: TCAGATGTGGAAACTGAGCCCA R5: ACGCGATAAATAATACAAGAGCAAGCA |
Chr1:189625131-189625444 | Amplification |
| Six1 | 304 | F6: GGCAGGAGGGAAGAGAGGAGAAA R6: CCTCTGCAGTGTCATCTAAGGGC |
Chr1:189624938-189625242 | Amplification |
| Six1 | 316 | F7: AGTCATTGATTTGTGCAGAGATGAGAGCGGCT R7: AGTGTCCCCGGCGCGCTGATTGG |
Chr1:189624677-189624993 | Amplification |
| Six1 | 305 | F8: TCGCGGTCCCACTCCTCCCACCA R8: TGAGTAGGGCGCGCACTGGACAGG |
Chr1:189624532-189624837 | Amplification |
| Six1 | 305 | F9: AACGAGCAGCATCCACCCGGCGG R9: TGTGTGAAGCCGAACGATGGCAGCA |
Chr1:189624324-189624629 | Amplification |
| Six1 | 236 | F10: CTCTGGACACCACTTCCCATGAC R10: ACAAGCACGCATTTAGAAGTCACT |
Chr1:189622076-189622312 | Amplification |
| Six1 | 329 | F11: CCGTCCGTCCTTTAAGTCAG R11: AGCACGCATTTAGAAGTCAC |
Chr1:189622079-189622408 | PCR–RFLP genotyping |
| Six1 | PF: cggGGTACCATTGTTCGGTGGTGCTGAG PR: cggCTCGAGAAAGGCCGCTACTATTTGG |
Chr1:189625301-189626173 | Promoter activity analysis | |
| Six1 | 189 | F: CGTGTTGCGGGAGTGGTA R: TGCTTGTTGGAGGAGGAGTT |
Chr1:189621136-189623371 | Expression analysis |
| HPRT | 134 | F: GGTCAAGCAGCATAATCCAAAG R: CAAGGGCATAGCCTACCACAA |
ChrX:110352282-110356641 | Expression analysis |
*Restrictive enzyme sites are marked by underline and bold type, and protective base are indicated by bold lowercase.
Subsequently, the raw short reads were assembled to produce longer reads (Fastq file) using FLASH software (Magoc and Salzberg, 2011) after quality control. The FastX tool (http://hannonlab.cshl.edu/fastx_toolkit/index.html) was used to obtain the fa format sequences from the Fastq file, and the clean reads were obtained by alignment with target reference sequences (NC_010443.5) using the Blat tool (Kent, 2002). Later, reference genome alignment was performed using BWA software (Li and Durbin, 2010) and results were analyzed by Picard tool (https://github.com/broadinstitute/picard). Finally, the genetic variants, including single-nucleotide variations (SNVs), and Indels were detected using VarScan (Koboldt et al., 2012) and GATK (McKenna et al., 2010) software. The positions of variations were defined according to the transcription start site (TSS) of porcine Six1 which were identified in previous study (Wu et al., 2011).
Validation of Sequencing Data
In our previous study, we established a PCR–restriction fragment length polymorphism (RFLP) method to genotype the SNP g.1595A>G, in the first intron of Six1 (Wu et al., 2011). In the present study, the PCR–RFLP method was improved by optimization of the amplification primer (Table 1). Using this technique, g.1595A>G were genotyped in the 823 P×D×L×Y commercial pigs, which contains 509 P×D×L×Y commercial pigs used in sequencing analysis and the genotyping results were compared with the sequencing data. In addition, the genotyping results of microsatellite variants by sequencing were validated using a typical microsatellite analysis method with randomly selected 50 P×D×L×Y commercial pigs. Briefly, the forward sequencing primers (Table 1, F3) for (AC)n microsatellite variant were fluorescently labeled with 5-carboxyfluorescein (5ʹ FAM), and the PCR products were separated by size using capillary electrophoresis on an ABI 3730XL sequencing platform. Then, the genotypes and sizes of (AC)n microsatellites were analyzed using GeneMapper software (Version 4.0; Applied Biosystems, Foster City, CA).
Traits and Association Analysis
Finishing pigs were slaughtered, and 13 traits were measured following the international and national standard protocols. Experimental pigs were slaughtered humanely in a standardized commercial processing plant (Jiangsu Sushi Group, Huaian City, China) at an average age of 170 d. Meat color values, including lightness (L*), redness (a*), and yellowness (b*), were determined with longissimus dorsi muscles from the last thoracic vertebrae using a portable Minolta colorimeter (CR-10, Minolta, Osaka, Japan) at 45 min after slaughter, according to the manufacturer’s instructions. Intramuscular fat (IMF) content was measured using the Soxhlet extraction method, with petroleum ether as the solvent. Percentage of IMF was calculated from the ratio of IMF weight to dry meat weight. Water loss rate was measured using pieces of meat (diameter, 2.523 cm; thickness, 1 cm), under 35 kg pressure on 18 layers of thick filter paper; the percentage loss was calculated as follows: water loss rate = ([initial weight − final weight]/initial weight) × 100.
According to the results of sequencing and PCR–RFLP genotyping, the genotype and allele frequencies were calculated in each of the seven pig breeds and the P×D×L×Y commercial pig population. The effects of single genotypes on traits were calculated by the least-squares method and association analysis was performed using the Mixed Linear Model (MLM) procedure in SAS version 8.0 (SAS Institute, Cary, NC), according to the following statistical model:
Model 1 for birth weight:
Model 2 for weaning weight:
Model 3 for meat quality traits:
Model 4 for carcass traits:
where Yijk or Yijkl represents the observed values of traits; μ, the overall mean; Genotypei, the fixed effect of genotype (i = 3 for SNP or 5 for microsatellite); Sexj, the fixed effect of sex (j = 1 for male or 2 for female); Famk, the random effect of family (k = 144); Batchl, the random effect of batch (l = 7); aijkl, the regression coefficient of the weaning days for weaning weight; Qijk, the weaning days; bijkl, the regression coefficient of the carcass weight for meat quality; Xijkl, the carcass weight; cijkl, the regression coefficient of the slaughter days for carcass weight; Zijkl, the slaughter days; and eijk or eijkl, the random residual. Additive and dominant effects were estimated using the REG procedure in SAS 8.0, where the additive effects −1, 0, and 1 represent the AA, AG, and GG genotypes, respectively; and the dominance effects 0, 1, and 0 represent the AA, AG, and GG genotypes, respectively.
Gene Expression Analysis
To investigate the relationship between (AC)n microsatellite variation and gene expression, we quantified the expression levels of Six1 in longissimus dorsi muscles of (AC)19/19, (AC)19/22, and (AC)22/22 genotype individuals derived from the commercial pig population. Briefly, 23, 26, and 25 pigs with the three main types of genotypes, (AC)19/19, (AC)19/22 and (AC)22/22, were selected from the P×D×L×Y commercial pig population. Longissimus dorsi muscles were thawed from −80 °C and ground using a homogenizer (Jingxin, Shanghai, China). Then, the total RNA were extracted using TRIzol reagent (Invitrogen, Life Technologies, Carlsbad, CA), cDNA was synthesized using Prime Script Reverse Transcription (RT) reagent (TaKaRa, Dalian, China), and quantitative RT-PCR was performed on a Step-One Plus Real-Time PCR System (Applied Biosystems), using the AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing City China). All reactions were performed in triplicate for each sample, and porcine Hypoxanthine Guanine Phosphoribosyl Transferase (HPRT) was used as the reference gene. Relative expression levels were calculated using the comparative Ct (△△Ct) value method (Livak and Schmittgen, 2001). Subsequently, the expression levels of Six1 were transformed to log10 using SPSS 20.0 before analysis. Scatter plot was drawn using GraphPad Prism (5.0), and data are presented as means ± SD. Comparisons by one-way analysis of variance were conducted in SPSS 20.0, and the Bonferroni method was used to correct for multiple testing.
Promoter Activity Analysis
To further clarify the impacts of (AC)n microsatellite variations on porcine traits, the promote activities of Six1 with (AC)19/19 and (AC)22/22 homozygous genotypes were compared using the Dual-Luciferase Reporter Assay system (Promega, Madison, WI). The PCR amplification primers (PF and PR) for Six1 promoter fragments were shown in Table 1 and DNA templates were selected from the (AC)19/19 and (AC)22/22 homozygous genotype pigs in P×D×L×Y commercial pig population. Subsequently, promoter activity analysis was evaluated in two cell lines (C2C12 and PK15) in vitro, using previously described methods for cell culture, transfection, and promoter activity evaluation (Wu et al., 2013), except that the transfection reagent was changed to Lipofectamine 3000 (Invitrogen, Life Technologies). Firefly and Renilla luciferase activities were detected using a Glomax 20/20 luminometer (Promega). Six replicate experiments were performed for each group (n = 6), and data are presented as means ± SE. Statistical analyses and graph generation were conducted using GraphPad Prism (5.0), and an unpaired Student’s t-test was used to evaluate the statistical significance of differences between two groups, with P < 0.05 considered significant. The detailed protocol for promoter activity analysis was reported previously (Wu et al., 2013).
RESULTS AND DISCUSSION
Identification of Variants in the Six1 Gene
The porcine Six1 is highly conserved across species, and contains two exons and one intron (GenBank Accession No: GU225953.1). The identities of whole coding region nucleotide and protein sequences between human (NM_005982, NP_005973), mouse (NM_009189, NP_033215), and pig (NM_001199718, NP_001186647) exhibit more than 96.41% and 99.6%, respectively. In our previous study, no mutations were found across the entire Six1-coding region in multiple pig breeds, while 18 potential mutations were detected in other gene regions of Meishan and Yorkshire pigs; however, only two SNPs (g.1595 A<G in the intron and g.-1363C>T in the promoter) were validated in eight pig breeds (Wu et al., 2011). In this study, to identify genuine variants in the Six1 promoter, we performed high-throughput sequencing analysis on the Illumina MiSeq 2000 platform, using the genomic DNA from 144 individuals of two western lean-type breeds (Yorkshire and Landrace), four Chinese indigenous breeds (Meishan pig, Mi pig, Shawutou pig, and Erhualian pig), and a cultivated breed (Suhuai pig), together with a 509 P×D×L×Y commercial pig population. A total of 12 variants were identified, including 10 SNV, 1 Indel, and 1 (AC)n microsatellite (Table 2). Among these 12 variations, 10 were in the Six1 promoter, one of which (g.-1363 C>T) was validated in our previous study, and 2 were in the first intron, of which g.1595 A<G was also validated in our previous study (Wu et al., 2011). To ensure the validity of the sequencing data, we genotyped g.1595 A<G in the first intron by PCR-RFLP, and promoter (AC)n microsatellite variant by capillary electrophoresis, in P×D×L×Y commercial pig population; the genotyping results were identical to those generated by the high-throughput sequencing, indicating that the genotyping data from high-throughput next- generation sequencing were reliable.
Table 2.
Variations identified in the porcine Six1 gene
| Gene localization | Position (bp) | Polymorphism | Variant ID |
|---|---|---|---|
| Promoter | −1480 | AGA/− | — |
| −1449 | T/C | rs323621547 | |
| −1434 | G/A | rs344565956 | |
| −1363 | T/C | rs328947567 | |
| −1030 | (AC)n | — | |
| −639 | G/A | rs340799460 | |
| −379 | G/A | rs331557076 | |
| −157 | T/C | rs321465095 | |
| −136 | T/C | rs336933580 | |
| Exon 1 | +294 | G/A | rs327199393 |
| Intron 1 | 1595 | G/A | rs319868732 |
| 1648 | G/A | rs337137823 |
Genetic Diversity of Six1 Gene Variations
To understand the genetic diversity of 12 identified variants, we performed genotype and allele frequencies analysis in eight pig breeds. The genotype and allele frequencies of the SNVs and (AC)n microsatellite variations were presented in Supplementary Table S1 and Table 3, respectively. Overall, the diversities of the SNVs and (AC)n microsatellite were greater in Chinese indigenous breeds. Notably, no diversity of any SNVs was observed in the western lean-type breeds (Yorkshire and Landrace); however, in Suhuai pigs, which comprise 75% Yorkshire and 25% Huai pig lineages, the diversity was identified. For the (AC)n microsatellite, the alleles (AC)19 and (AC)22, corresponding to genotypes (AC)19/19, (AC)19/22, and (AC)22/22, dominated in the western lean-type breeds, while (AC)20 and (AC)22 alleles, corresponding to (AC)20/20, (AC)20/22, and (AC)22/22, were most common in Chinese indigenous pigs. Moreover, only the g.1595A>G and (AC)n microsatellite sites were polymorphic in the P×D×L×Y commercial pig population. A total of 823 individuals and 509 P×D×L×Y individuals were genotyped successfully for the g.1595A>G and (AC)n microsatellite sites, respectively. The genotype frequency results for the g.1595A>G site and (AC)n microsatellite sites in the P×D×L×Y commercial pig population are presented in Tables 4 and 5, respectively.
Table 3.
Genotype distribution of (AC)n microsatellite in different pig breeds
| Genotypes | Breeds | ||||||
|---|---|---|---|---|---|---|---|
| Yorkshire | Landrace | Suhuai | Meishan | Mi | Shawutou | Erhualian | |
| 18/22 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 19/19 | 7 | 0 | 7 | 0 | 0 | 3 | 0 |
| 19/20 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| 19/22 | 6 | 5 | 6 | 0 | 1 | 0 | 0 |
| 19/24 | 4 | 0 | 2 | 0 | 0 | 1 | 0 |
| 20/20 | 0 | 0 | 0 | 5 | 1 | 3 | 8 |
| 20/22 | 0 | 0 | 1 | 10 | 7 | 7 | 7 |
| 20/25 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 20/26 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| 21/22 | 0 | 2 | 0 | 1 | 1 | 2 | 0 |
| 21/26 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| 22/22 | 2 | 11 | 4 | 3 | 7 | 1 | 0 |
| 22/24 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22/26 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| 23/26 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 26/26 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| Number | 20 | 19* | 20 | 20 | 24 | 20 | 20 |
*One of Landrace pigs genotypes failed.
Table 4.
Association of g. 1595 A>G of Six1 polymorphisms with porcine traits
| Genotype traits | Least squares means (LSM) ± SE | Genetic effect (mean ± SE ) | |||
|---|---|---|---|---|---|
| AA(n = 33) | AG(n = 263 ) | GG (n = 527) | Additive effect | Dominant effect | |
| Birth weight (kg) | 1.553 ± 0.103 | 1.597 ± 0.036 | 1.570 ± 0.026 | 0.007 ± 0.0537 | −0.036 ± 0.064 |
| Weaning weight (kg) | 5.895 ± 0.157 | 5.917 ± 0.052 | 5.999 ± 0.037 | 0.0548 ± 0.076 | 0.036 ± 0.091 |
| Carcass weight (kg) | 82.674 ± 1.499 | 81.997 ± 0.530 | 82.176 ± 0.375 | 0.123 ± 0.818 | −0.315 ± 0.993 |
| Color a * | 4.081 b ± 0.339 | 4.799 a ± 0.119 | 4.509 b ± 0.085 | 0.237 ± 0.179 | −0.541† ± 0.217 |
| Leaf fat (mm) | 0.985 ± 0.048 | 0.938 ± 0.017 | 0.901 ± 0.012 | −0.0356 ± 0.030 | −0.007 ± 0.037 |
| Shoulders back fat (mm) | 31.595 ± 0.931 | 30.774 ± 0.326 | 30.401 ± 0.232 | −0.453 ± 0.565 | −0.007 ± 0.686 |
| Thoracic and lumbar back fat (mm) | 19.439 ± 0.793 | 19.116 ± 0.278 | 18.855 ± 0.197 | −0.060 ± 0.502 | −0.331 ± 0.610 |
| Lumbar sacral back fat (mm) | 15.407 ± 0.778 | 15.208 ± 0.273 | 14.609 ± 0.194 | −0.001 ± 0.467 | −0.680 ± 0.567 |
| Average back fat (mm) | 22.147 ± 0.718 | 21.699 ± 0.252 | 21.288 ± 0.179 | −0.174 ± 0.462 | −0.345 ± 0.562 |
| IMF (%) | 10.102 ± 1.013 | 9.933 ± 0.362 | 9.695 ± 0.255 | −0.122 ± 0.516 | −0.138 ± 0.630 |
| Color L* | 41.005 ± 0.618 | 40.314 ± 0.216 | 40.294 ± 0.154 | −0.342 ± 0.311 | 0.354 ± 0.377 |
| Color b* | 5.583 ± 0.236 | 5.656 ± 0.083 | 5.586 ± 0.059 | 0.015 ± 0.122 | −0.078 ± 0.149 |
| Water loss rate (%) | 0.191 ± 0.015 | 0.196 ± 0.005 | 0.198 ± 0.004 | 0.001 ± 0.008 | −0.116 ± 0.860 |
Bold values indicate significant association events and values in each row with different lowercase superscript letters are significantly different at P < 0.05.
†The significant effect at P < 0.01.
Table 5.
Effects of (AC)n microsatellite in Six1 promoter on porcine traits
| Genotype traits | Least squares means ± SE | |||||
|---|---|---|---|---|---|---|
| 19/19 (n = 50) | 19/20 (n = 34) | 19/22 (n = 278) | 20/22(n = 13) | 22/22 (n = 131) | 19/24(n = 3) | |
| Birth weight, kg | 1.555 ± 0.100 | 1.592 ± 0.122 | 1.615 ± 0.042 | 1.690 ± 0.195 | 1.536 ± 0.062 | __ |
| Weaning weight, kg | 5.740 b ± 0.123 | 5.974 ab ± 0.151 | 6.036 a ± 0.050 | 5.776 ab ± 0.274 | 5.898 ab ± 0.077 | __ |
| Carcass weight, kg | 81.948 ab ± 1.218 | 85.096 a ± 1.480 | 83.196ab ± 0.517 | 83.463 ab ± 2.381 | 81.793 b ± 0.756 | __ |
| Color a* | 4.590 ± 0.293 | 4.222 ± 0.356 | 4.749 ± 0.123 | 5.058 ± 0.571 | 4.614 ± 0.181 | __ |
| Leaf fat, mm | 0.945 ± 0.041 | 0.958 ± 0.050 | 0.928 ± 0.017 | 0.832 ± 0.080 | 0.919 ± 0.025 | __ |
| Shoulders back fat, mm | 30.787 ± 0.778 | 29.908 ± 0.945 | 30.698 ± 0.328 | 30.202 ± 1.517 | 30.623 ± 0.481 | __ |
| Thoracic and lumbar backfat, mm | 19.006 ABab ± 0.680 | 17.384 Bb ± 0.826 | 19.365 ABa ± 0.287 | 21.284 Aa ± 1.327 | 19.580 Aa ± 0.421 | __ |
| Lumbar sacral back fat, mm | 15.060 ± 0.649 | 14.216 ± 0.788 | 15.375 ± 0.273 | 16.557 ± 1.269 | 15.607 ± 0.401 | __ |
| Average back fat, mm | 21.618 ± 0.608 | 20.512 ± 0.738 | 21.817 ± 0.256 | 22.681 ± 1.185 | 21.937 ± 0.378 | __ |
| IMF, % | 10.193 ± 0.414 | 9.584 ± 0.482 | 9.412 ± 0.169 | 8.837 ± 0.740 | 9.448 ± 0.245 | __ |
| Color L* | 39.877 ± 0.489 | 40.940 ± 0.594 | 40.469 ± 0.206 | 39.468 ± 0.955 | 40.731 ± 0.302 | __ |
| Color b* | 5.688 ± 0.183 | 5.367 ± 0.221 | 5.596 ± 0.077 | 5.612 ± 0.355 | 5.538 ± 0.113 | __ |
| Water loss rate, % | 17.659 ± 1.119 | 19.180 ± 1.33 | 19.712 ± 0.471 | 20.412 ± 2.322 | 19.277 ± 0.694 | __ |
Bold values indicate significant association events, and values in each row with different lower case superscripts are significantly different at P <0.05 and with different capital superscript letters are significantly different at P <0.01.
Association of Six1 Polymorphisms With Traits
The detailed mean values, SD, and coefficient of variation (CV) of trait phenotypes are presented in Table 6. The results demonstrate that the CV of measured traits were all >11.00%. In particular, the CV for the meat color a*, IMF, and water loss rate were 67.77%, 55.45%, and 40.24%, respectively, indicating that the large variation of trait phenotypic values is existed in P×D×L×Y commercial pig population, which provides a good basis for trait association analysis. Interestingly, the porcine Six1 was mapped to several production traits QTLs region by alignment analysis with Pig QTL Database (https://www.animalgenome.org/cgi-bin/QTLdb/SS/search), e.g., back fat at last rib, weaning body weight, carcass weight, and meat color CIE-a*. To assess the association of the g.1595A>G and (AC)n microsatellite polymorphisms in Six1 with porcine traits, we conducted association analysis using multiple single marker MLM for different traits. The results demonstrated that g.1595A>G was significantly associated with meat color a*(P < 0.05); individuals carrying the G allele had greater meat color a* values (Table 4). In this study, the meat color values, lightness (L*), redness (a*), and yellowness (b*), were determined using a portable Minolta colorimeter (CR-10, Minolta). The a* value indicates redness, with greater values indicating redder meat color. The G allele was mainly found in western lean-type pigs, suggesting the G allele is associated with superior meat color in western lean-type breeds. Notably, the A allele of this SNV was dominated in Chinese indigenous pigs (Supplementary Table S1), which suggests that the other alleles might coordinately control the redness in Chinese indigenous pigs.
Table 6.
Characteristics of traits measured in this study
| Traits (n = 1,310) | Mean ± SD | CV (%) |
|---|---|---|
| Birth weight, kg | 1.57 ± 0.51 | 32.68 |
| Weaning weight, kg | 5.97 ± 0.77 | 12.78 |
| Carcass weight, kg | 82.66 ± 9.31 | 11.26 |
| Color a* | 4.64 ± 3.14 | 67.77 |
| Leaf fat, kg | 0.90 ± 0.33 | 36.74 |
| Shoulders back fat, mm | 30.28 ± 6.38 | 21.07 |
| Thoracic and lumbar back fat, mm | 18.77 ± 5.55 | 29.60 |
| Lumbar sacral back fat, mm | 14.66 ± 5.15 | 35.11 |
| Average back fat, mm | 21.22 ± 5.15 | 24.25 |
| IMF, % | 9.66 ± 5.36 | 55.45 |
| Color L* | 39.75 ± 5.88 | 14.78 |
| Color b* | 5.54 ± 1.47 | 26.59 |
| Water loss rate, % | 20.38 ± 8.20 | 40.24 |
Moreover, significant associations were also observed between the (AC)n microsatellite polymorphism and porcine traits (Table 5). In this study, six (AC)n microsatellite genotypes were identified in P×D×L×Y commercial pig population, including (AC)19/19, (AC)19/20, (AC)19/22, (AC)20/22, (AC)22/22, and (AC)19/24. Three pigs with the (AC)19/24 genotype were removed from the association analysis. The results indicated that the (AC)n polymorphism was significantly associated with weaning weight (P < 0.05), carcass weight (P < 0.05), and thoracic and lumbar back fat (P < 0.01). Pigs with (AC)19/20 heterozygous genotype had greater weaning and carcass weights, and lower thoracic and lumbar back fat. Intriguingly, the (AC)20/22 genotype was mainly observed in Chinese indigenous pigs, and was absent in the western lean-type breeds, Yorkshire and Landrace (Table 3), which is consistent with the observation that pigs with (AC)20/22 genotype in the P×D×L×Y commercial pig population had greater thoracic and lumbar back fat. Pigs with (AC)20/22 genotype also had greater meat color a* value, lumbar sacral back fat, and average back fat than pigs with the other genotypes, although the differences were not statistically significant (Table 5).
In addition, we considered the multiple testing for association analysis with Bonferroni method according to the number of variants, and defined the raw P-value < 0.05/N is significant. In this study, two variants were analyzed, thus the raw P-value < 0.025 was considered significant. The corrected results showed that the (AC)n polymorphism was still significantly associated with weaning weight and thoracic and lumbar back fat, although the significant associations were not observed between the (AC)n polymorphism with carcass weight and the g.1595A>G polymorphism with meat color a*. Weaning weight of the individuals with (AC)19/22 genotype was significantly greater than the individuals with (AC)19/19 genotype, while thoracic and lumbar back fat of the pigs with (AC)19/20 genotype was significantly lower than the pigs with (AC)19/22 and (AC)22/22 genotypes.
Effect of Six1 Promoter (AC)n Microsatellite Variation on Its Expression and Promoter Activity
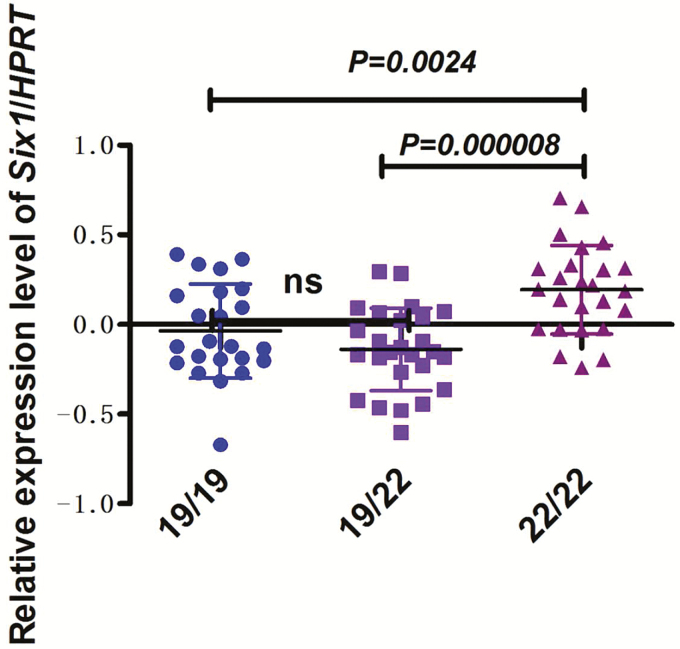
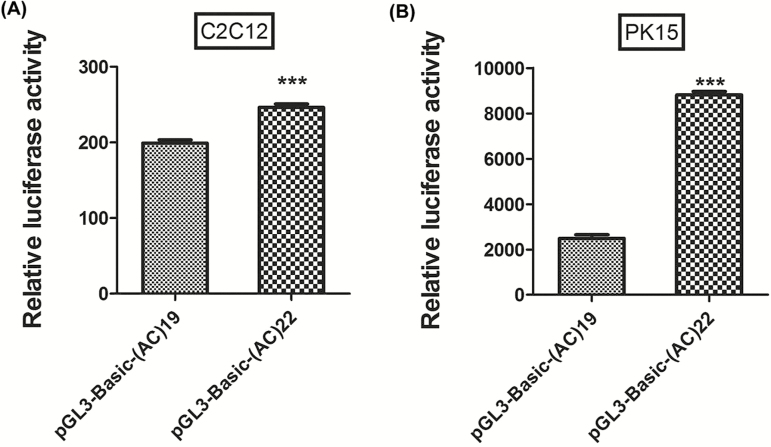
In the P×D×L×Y commercial pig population, three main genotypes, (AC)19/19, (AC)19/22, and (AC)22/22, were detected at the polymorphic microsatellite in the Six1 promoter. To explore the possible mechanisms underlying the influence of the (AC)n microsatellite variants on phenotypic traits, we detected the differences in the expression levels of Six1 among pigs with the (AC)19/19, (AC)19/22, and (AC)22/22 genotypes. The results demonstrate that the expression level of Six1 in pigs with the (AC)22/22 genotype was significantly greater than in those with (AC)19/19 (P < 0.01) and (AC)19/22(P < 0.00001) genotypes (Figure 1), suggesting that (AC)22 allele may promote gene expression of Six1. Previous studies demonstrated that Six1 is critical factor related to skeletal muscle growth and development (Wu et al., 2014) and found that Six1−/− homozygous mice show smaller body weight due to extensive muscle hypoplasia (Laclef et al., 2003), which is consistent with the pigs with (AC)22 allele that have greater weaning weight and carcass weight. To further confirm the effect of Six1 promoter (AC)n microsatellite variation on expression of this gene, we constructed two dual-luciferase reporter plasmids containing (AC)19/19 and (AC)22/22 homozygous genotype sequences and evaluated their promoter activities in two cell lines (C2C12 and PK15) in vitro. The results demonstrate that (AC)22/22 homozygous genotype promoter activity was significantly greater than that of the (AC)19/19 homozygous genotype promoter (P < 0.001) (Figure 2), consistent with the results of gene expression analysis (Figure 1).
Figure 1.
Effect of Six1 promoter (AC)n microsatellite variations on its expression. The pigs with three main types of genotypes (AC)19/19 (n = 23), (AC)19/22 (n = 26), and (AC)22/22 (n = 25) were selected from the P×D×L×Y commercial pig population and real-time PCR was conducted to detect the expression of Six1. The relative expression levels were calculated using comparative Ct (△△Ct) value method and HPRT was used as reference. Data were shown as the mean ± SD. One-way analysis of variance was used in SPSS 20.0 and Bonferroni method was used for multiple testing. P-values were shown in scatter plot. ns, no significant difference.
Figure 2.
Effect of Six1 promoter (AC)n microsatellite variations on promoter activity. Promoter activity analyses were performed with Dual-Luciferase Reporter Assay system (Promega) in C2C12 and PK15 in vitro. PGL3-Basic-(AC)19 and PGL3-Basic-(AC)22 represent the promoters derived from (AC)19/19 and (AC)22/22 homozygous genotype pigs, respectively. Each group was performed in six repetitions (n = 6) and data were presented as mean ± SE. Statistical analysis and graph drawing were conducted using GraphPad Prism (5.0), and an unpaired Student’s t-test was used to evaluate the statistical significance between the two groups. ***The significant effect at P < 0.001.
Microsatellites, also referred to as simple sequence repeats, are tandem repeats of DNA composed of 1–6 bp long units, which is a ubiquitous feature of prokaryote and eukaryote genomes, and have important roles in numerous biological processes, including evolution, development, phenotypic variation, and disease (Toth et al., 2000; Hannan, 2010). For example, Fondon and Garner (2004) found that the length variations of microsatellite in the coding regions of the aristaless-like 4 (Alx-4) and runt-related transcription factor 2 (Runx-2) genes were significantly associated with dog limb and skull morphology (Fondon and Garner, 2004). In the present study, we found that the length variation of the (AC)n microsatellite in the Six1 promoter was significantly associated with weaning and carcass weights and lower thoracic and lumbar back fat traits, suggesting that this microsatellite variation may be an important factor affecting pig phenotypic variation. Mononucleotide (A) and dinucleotide (AC) repeats are most abundant in eukaryotic genomes, and primarily distributed in promoters (Toth et al., 2000; Sawaya et al., 2013). The variation of dinucleotide (AC) repeats can modulate gene expression in multiple different species (Sawaya et al., 2012). In this study, a relationship between length variation of (AC)n the microsatellite in the porcine Six1 promoter and gene expression was observed by in vitro and in vivo experiments (Figures 1 and 2). A total of 9 alleles of the (AC)n microsatellite, corresponding to 15 genotypes, were identified in different pig breeds (Table 3). Unfortunately, due to lack of RNA samples from individuals corresponding to each genotype, definitive determination of which genotype has the strongest promoter activity could not be confirmed.
Six1 is a critical transcription factor involved in many diseases, particularly for tumorigenesis (Wu et al., 2015), and early research found that human SIX1 was absent or expressed at low levels in normal mammary tissue, but overexpressed in primary breast cancer and metastatic lesions (Ford et al., 1998). Notably, we found that the (AC)n microsatellite is factually existed in the pig, human, and mouse Six1 promoter region by sequence alignment. However, whether variation of the (AC)n microsatellite exists in the human SIX1 promoter and the potential links of this feature with tumorigenesis warrant further investigation.
CONCLUSIONS
In summary, 12 variants at the porcine Six1 locus were identified in this study, of which the intronic polymorphism, g.1595A>G, was significantly associated with the meat color trait a*, while the (AC)n microsatellite variant in the promoter was significantly associated with weaning and carcass weights and thoracic and lumbar back fat. These data provide potential reference markers for molecular breeding of pigs. In addition, the identification of a polymorphic (AC)n microsatellite in the Six1 promoter and its relationship with expression of the gene in this study may provide a helpful reference for similar studies in other species.
Footnotes
This work was supported by the National Natural Science Foundation of China (31501920), the Fundamental Research Funds of the Central Universities (KJQN201605), and the National Major Project of Breeding for Transgenic Pig (2016ZX08006001-003).
LITERATURE CITED
- Bessarab D. A., Chong S. W., Srinivas B. P., and Korzh V.. 2008. Six1a is required for the onset of fast muscle differentiation in zebrafish. Dev. Biol. 323:216–228. doi:10.1016/j.ydbio.2008.08.015 [DOI] [PubMed] [Google Scholar]
- Fondon J. W. III, and Garner H. R.. 2004. Molecular origins of rapid and continuous morphological evolution. Proc. Natl. Acad. Sci. USA. 101:18058–18063. doi:10.1073/pans.0408118101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford H. L., Kabingu E. N., Bump E. A., Mutter G. L., and Pardee A. B.. 1998. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc. Natl. Acad. Sci. US A. 95:12608–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii J., Otsu K., Zorzato F., Leon S. D., Khanna V. K., Weiler J. E., O'Brien P. J., and MacLennan D. H.. 1991. Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science. 253:448–451. doi:10.1126/science.1862346 [DOI] [PubMed] [Google Scholar]
- Grifone R., Laclef C., Spitz F., Lopez S., Demignon J., Guidotti J. E., Kawakami K., Xu P. X., Kelly R., Petrof B. J., Daegelen D., Concordet J. P., and Maire P.. 2004. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol. Cell. Biol. 24:6253–6267. doi:10.1128/mcb.24.14.6253-6267.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan A. J. 2010. Tandem repeat polymorphisms: modulators of disease susceptibility and candidates for ‘missing heritability’. Trends Genet. 26:59–65. doi:10.1016/j.tig.2009.11.008 [DOI] [PubMed] [Google Scholar]
- Kent W. J. 2002. BLAT—the BLAST-like alignment tool. Genome. Res. 12:656–664. doi:10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Larsen N., Short T., Plastow G., and Rothschild M. F.. 2000. A missense variant of the porcine melanocortin-4 receptor (MC4R) gene is associated with fatness, growth, and feed intake traits. Mamm. Genome. 11:131–135. [DOI] [PubMed] [Google Scholar]
- Koboldt D. C., Zhang Q., Larson D. E., Shen D., McLellan M. D., Lin L., Miller C. A., Mardis E. R., Ding L., and Wilson R. K.. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22:568–576. doi:10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laclef C., Hamard G., Demignon J., Souil E., Houbron C., and Maire P.. 2003. Altered myogenesis in Six1-deficient mice. Development. 130:2239–2252. doi:10.1242/dev.00440 [DOI] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26:589–595. doi:10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Liu K., Weng Q., Li P., Wei W., Li Q., Chen J., Huang R., Wu W., and Liu H.. 2016. RNA-seq analysis reveals new candidate genes for drip loss in a Pietrain × Duroc × Landrace × Yorkshire population. Anim. Genet. 47:192–199. doi:10.1111/age.12401 [DOI] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 25:402–408. doi:10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma J., Yang J., Zhou L., Ren J., Liu X., Zhang H., Yang B., Zhang Z., Ma H., Xie X., Xing Y., Guo Y., and Huang L.. 2014. A splice mutation in the PHKG1 gene causes high glycogen content and low meat quality in pig skeletal muscle. PloS Genet. 10:e1004710. doi:10.1371/journal.pgen.1004710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., and Salzberg S. L.. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27:2957–2963. doi:10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. doi:10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa S., Sato S., Nii M., Morozumi T., Yoshioka G., Imaeda N., Yamaguchi T., Hayashi T., and Awata T.. 2011. Identification of a second gene associated with variation in vertebral number in domestic pigs. BMC Genet. 12. doi:10.1186/1471-2156-12-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan D., Jeon J. T., Looft C., Amarger V., Robic A., Thelander M., Rogel-Gaillard C., Paul S., Iannuccelli N., Rask L., Ronne H., Lundström K., Reinsch N., Gellin J., Kalm E., Roy P. L., Chardon P., and Andersson L.. 2000. A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science. 288:1248–1251. doi:10.1126/science.288.5469.1248 [DOI] [PubMed] [Google Scholar]
- Sakakibara I., Wurmser M., M Dos Santos M. Santolini S. Ducommun R. Davaze A. Guernec K. Sakamoto, and Maire P.. 2016. Six1 homeoprotein drives myofiber type IIA specialization in soleus muscle. Skelet Muscle. doi:10.1186/s13395-016-0102-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya S. M., Bagshaw A. T., Buschiazzo E., and Gemmell N. J.. 2012. Promoter microsatellites as modulators of human gene expression. Adv. Exp. Med. Biol. 769:41–54. [DOI] [PubMed] [Google Scholar]
- Sawaya S., Bagshaw A., Buschiazzo E., Kumar P., Chowdhury S., Black M. A., and Gemmell N.. 2013. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PloS One. 8:e54710. doi:10.1371/journal.pone.0054710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth G., Gaspari Z., and Jurka J.. 2000. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10:967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laere A. S., Nguyen M., Braunschweig M., Nezer C., Collette C., Moreau L., Archibald A. L., Haley C. S., Buys N., Tally M., Andersson G., Georges M., and Andersson L.. 2003. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 425:832–836. doi:10.1038/nature02064 [DOI] [PubMed] [Google Scholar]
- Wu W., Ren Z., Wang Y., Chao Z., Xu D., and Xiong Y.. 2011. Molecular characterization, expression patterns and polymorphism analysis of porcine Six1 gene. Mol. Biol. Rep. 38:2619–2632. doi:10.1007/s11033-010-0403-9 [DOI] [PubMed] [Google Scholar]
- Wu W., Ren Z., Liu H., Wang L., Huang R., Chen J., Zhang L., Li P., and Xiong Y.. 2013. Core promoter analysis of porcine Six1 gene and its regulation of the promoter activity by CpG methylation. Gene. 529:238–244. doi:10.1016/j.gene.2013.07.102 [DOI] [PubMed] [Google Scholar]
- Wu W., Huang R., Wu Q., Li P., Chen J., Li B., and Liu H.. 2014. The role of Six1 in the genesis of muscle cell and skeletal muscle development. Int. J. Biol. Sci. 10:10:983–989. doi:10.7150/ijbs.9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Ren Z., Li P., Yu D., Chen J., Huang R., and Liu H.. 2015. Six1: a critical transcription factor in tumorigenesis. Int. J. Cancer. 136:1245–1253. doi:10.1002/ijc.28755 [DOI] [PubMed] [Google Scholar]