Abstract
Beef cow stayability is a complex, economically important trait often used as an indicator of a cow’s potential lifetime productivity. Stayability is defined as capability of a cow to maintain a perfect record up to 6 yr of age. This age is commonly cited as a financial break-even point, where initial costs of cow development and maintenance are recovered by her cumulative net income from yearly calf receipts. Later-maturing Bos indicus–Bos taurus crossbred cows may experience reproductive difficulty early in life but have a high potential for a long reproductive life span. It was the objective of this study to identify genetic variants associated with measures of beef cow stayability. A population of B. indicus–B. taurus crossbred cows (n = 305) from central Texas was used. Phenotypes for various measures of stayability to 6 yr of age were produced by artificially imposing five different culling criteria on data from the population. Cows were scored either as a 1 (indicating a perfect record through 6 yr) or a 0 (indicating failure at or before 6 yr), under each criterion. Genome-wide association studies (GWAS) were conducted for each criterion using univariate procedures and prefitting the fixed effect of cow contemporary group. SNP associations for two criteria surpassed the false discovery threshold of 0.15, when a cow was scored as 0 upon her first failure to wean a calf, regardless of reason, through 6 yr (criterion 2), and when a cow was scored as 0 upon her first failure to give birth to a calf, through 6 yr (criterion 3). Associated SNP were found on bovine chromosomes (BTA) 1, 2, 5, 9, 18, and 21 for criterion 2 and on BTA 1, 5, 11, 15, and 24 for criterion 3. A critical region on BTA 5: 43–50 Mb was identified for each criterion. Due to the similarities to prior work, the tendency for B. indicus cattle to experience reproductive difficulties early in life, and due to the large proportion of cows that left the herd at an early age under these criteria, these results suggest that the associations are likely driven by an early life trait such as age at puberty or rate of heifer development.
Keywords: beef cow, Bos indicus, GWAS, stayability
INTRODUCTION
Beef cow stayability is a complex trait often used as an indicator of a cow’s potential lifetime productivity. Stayability was first defined as a cow’s probability of surviving to a specific age, given the opportunity to first reach that age (Hudson and Van Vleck, 1981). Now, stayability usually refers to a cow’s ability to maintain a perfect weaning record and produce five calves by 6 yr of age, typically with respect to Bos taurus cattle (Snelling et al., 1995). Heritability estimates for stayability range from 0.1 to 0.22 (Snelling et al., 1995; Van Melis et al., 2007; Cavani et al., 2015). Stayability is an important metric for producers, as longevity in the cow herd is related to lifetime productivity and economic value or profitability of the productive asset (Rogers, 1972).
Bos indicus–Bos taurus crossbred cows are recognized as having high potential for long reproductive life spans, likely due to the combined advantages of heterosis and adaptation to the climates in which they are raised (Riley et al., 2001; Thrift and Thrift, 2003). However, B. indicus–influenced females reach puberty later than B. taurus heifers, are significantly less likely to first calve at 2 yr of age, and if successful, experience difficulty rebreeding during the subsequent breeding period (Chenoweth, 1994), decreasing their likelihood of meeting the stayability benchmark. As a popular choice among producers in tropical and subtropical climates, it would be economically advantageous to understand the underlying genetic contributors of stayability in B. indicus–B. taurus crossbred cows. Therefore, the objective of this study was to identify genetic variants associated with measures of beef cow stayability.
MATERIALS AND METHODS
Population
Cows used in this study were part of the McGregor Genomics Cycle 1 Population, an experimental population housed at the Texas A&M AgriLife Research Center at McGregor, Texas. When this population was developed, the primary objective was to understand cow lifetime productivity traits. These females (n = 305) were born in 2003 through 2007, and they were from either 13 full-sibling F2 families produced through embryo transfer (ET) or four paternal half-sibling families produced through natural service (NS) matings. These cows were all B. indicus–B. taurus crosses, specifically, Nellore–Angus F2 crosses, Nellore–Angus × Brahman–Angus crosses, or Nellore–Angus × Brahman–Hereford crosses. All procedures involving animals were approved by the Texas A&M Institutional Animal Care and Use Committee.
Cows from this population were born either during spring or fall calving seasons. They were vaccinated against clostridial diseases at 2 to 3 mo of age and then again at weaning. On average, these cows were weaned as calves at 214.8 ± 0.93 d of age. After weaning, but before the first breeding season, they were also vaccinated against bovine viral diarrhea virus, bovine respiratory syncytial virus, infectious bovine rhinotracheitis, parainfluenza type 3, leptospirosis, vibriosis and treated for internal parasites. As heifers, they were developed on native, warm-season perennial pastures, were nutritionally managed for a target body condition score of 5 to 6 by the first breeding and calving seasons, and were given a protein supplement as necessary.
On average, heifers were first exposed to Angus bulls at 433.3 ± 0.99 d of age for the opportunity to first calve at approximately 2 yr of age. Spring-born heifers (ET and NS) were all managed to first calve at 2 yr of age. Fall-born heifers (ET only) were exposed to Angus bulls from the first week in December to the second week in February and given the opportunity to first calve at 2 yr of age in the following fall. Those that initially failed to conceive were transitioned to a spring calving schedule and were bred to first calve at 2.5 yr of age, without a failure to calve counted against them. Any fall-born heifers that first calved during the fall were held through the winter without mating opportunity and rebred in the following spring breeding season to be on a spring calving schedule, with their second calf born at 3.5 yr of age. Subsequently, all cows were managed for spring calving only, and they were typically exposed to bulls from the third week in May to the third week in July. Across the study, the average length of the breeding season was 68 d.
Females were allowed to remain in the herd until their second failure to wean a calf. Calves were kept with their dams until weaning at an average of 209.8 ± 0.57 d of age. If a cow was ever deemed unfit to care for her calf, and the calf was removed from her, that was counted as a failure to wean. Records used for this analysis span from the date of this population’s first possible calving season in 2005 through 2014.
Phenotypes
Phenotypes for various measures of stayability to 6 yr of age were produced by artificially imposing culling criteria on data from the population. Any cow that left the herd prior to 6 yr for a reason other than the criterion being considered was omitted from that analysis, so the number of cows included in each analysis differed and ranged from 169 to 300 cows. Cows were scored either as a 1 to indicate a perfect record under each criterion through 6 yr or as a 0 to indicate failure at or before 6 yr under each criterion (Table 1). The first culling criterion corresponded to the actual management of the herd, in which cows were removed from the herd upon their second incidence of failure to wean a calf, regardless of reason (criterion 1).
Table 1.
Description of culling criteria
| Criterion | Description |
|---|---|
| 1 | Cows were removed from the herd upon their second incidence of failure to wean a calf, regardless of reason, through 6 yr |
| 2 | Cows were scored as culled upon their first failure to wean a calf, regardless of reason, through 6 yr |
| 3 | Cows were scored as culled upon their first failure to give birth to a calf, through 6 yr |
| 4 | Cows were scored as culled upon their first incidence of failure to wean a calf, but not considering calving failure as a reason, through 6 yr |
| 5 | Cows were scored as culled upon their first incidence of failure to wean a calf, provided that they had never previously experienced calving failure, through 6 yr |
For the second constructed culling criterion, cows were scored as a 0 (failure) upon their first failure to wean a calf, regardless of reason (criterion 2). This criterion corresponds to the Beef Improvement Federation (BIF, 2016) definition of stayability. For the third culling criterion, a cow was scored as a 0 upon her first failure to give birth to a calf (criterion 3). Criterion 3 was used as an indication of pregnancy, so a full-term, stillborn calf was not considered a failure.
Under the fourth criterion, a cow was scored as a 0 when she gave birth to a calf and then for any reason failed to wean that calf (criterion 4). Note that under this criterion, a prior failure to calve was ignored. For the fifth criterion, a cow was scored as a 0 upon her first instance of failing to wean a calf, provided that she had no prior instances of calving failure (criterion 5). For each of these criteria, lifetime productivity records were analyzed, and scores were manually assigned.
Genome-Wide Association Study
Filtered genotypes for these cows were the same as those described by Hulsman Hanna et al. (2014). Briefly, DNA was extracted from white blood cells and genotyped using the Illumina BovineSNP50v1 chip (Illumina Inc., San Diego, CA, United States). Chromosomal assignments and positions of SNP were based on the UMD3.1 B. taurus sequence assembly. Genotypes from the whole population (males and females) were filtered in PLINK (Purcell et al., 2007) to remove SNP with completion rates <90%, minor allele frequencies <0.05, and those deviating from Hardy–Weinberg equilibrium proportions at P <0.0001. After filtering, average SNP spacing was 75.9 kb with a median of 50.5 kb. Each of the five stayability phenotypes (0 = left herd, 1 = remaining in the herd through 6 yr of age) was preadjusted for the fixed effect of contemporary group (birth year and season of birth) using linear model procedures in R, and the residuals from these models were used in genome-wide association studies (GWAS). GWAS for beef cow reproductive longevity were performed using the univariate procedures in GEMMA (Zhou and Stephens, 2012) that fitted a single, standardized, genomic relationship matrix to account for genetic covariances among animals. The default SNP filters in GEMMA (missingness 0.05, minor allele frequency 0.01, r-squared threshold 0.999) were used, and because a different number of cows were part of each analysis, some SNP that passed the PLINK filters were subsequently excluded by GEMMA. The Benjamini and Hochberg false discovery rate (Benjamini and Hochberg, 1995) was initially constrained to 0.05 to correct for multiple testing. Given the complex nature of stayability, we anticipated that the SNP heritability would be explained by many small SNP effects (Manolio et al., 2009; Boyle et al., 2017). To minimize failure to detect true associations and reduce the type II error in our study, a false discovery threshold of 0.15 was ultimately used.
RESULTS AND DISCUSSION
Stayability typically refers to a cow’s ability to remain productive in the herd and produce five calves by 6 yr of age, provided that she first calved at 2 yr (Snelling et al., 1995). Although alternatives to this definition at earlier ages have been proposed, a cow’s potential to maintain a perfect weaning record through 6 yr is the definition adopted by BIF (2016) and numerous breed associations. Using this definition, EPD for stayability have been developed by the Red Angus Association of America, the American Simmental Association, and the American Gelbvieh Association (Snelling et al., 1995). For these reasons, the analyses were conducted using the most commonly accepted definition of stayability, where a cow must remain in the herd through 6 yr of age.
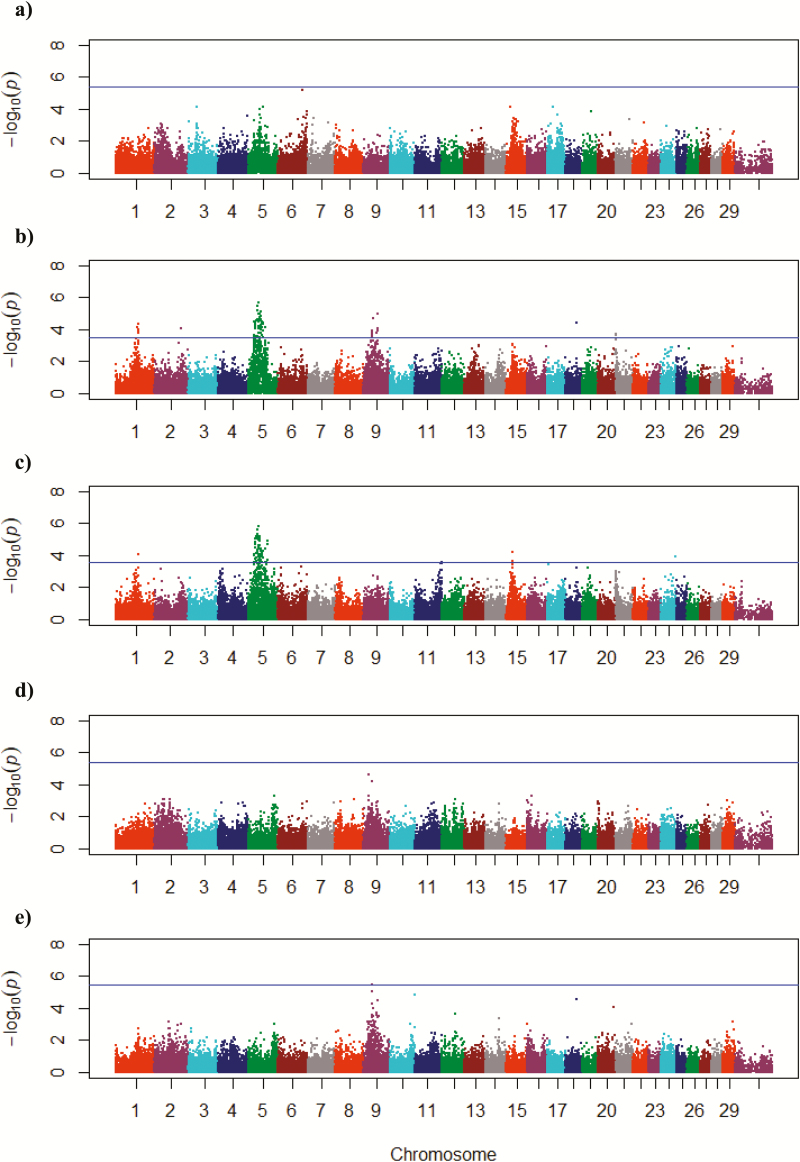
Under actual management (criterion 1), just 19% of the cows were removed from the herd before 7 yr of age (Table 2). By giving cows two opportunities for failure before removing them from production, a similar number of cows (3% to 6%) were removed from the herd each year. Criterion 1 resulted in limited detectable associations (Table 3). Although some SNP were found to be significant, no SNP survived correction for multiple testing, and no distinct structure was visible in the corresponding Manhattan plot (Figure 1a). This criterion also gave us no insight into the herd’s reproductive performance at 2 yr.
Table 2.
Proportion of cows remaining in the herd that were culled under each criterion by each age
| Criteriona | Culled | Not culled | ||||
|---|---|---|---|---|---|---|
| 2 yr | 3 yr | 4 yr | 5 yr | 6 yr | ||
| 1 | N/A | 0.06 | 0.06 | 0.05 | 0.03 | 0.81 |
| 2 | 0.23 | 0.20 | 0.07 | 0.05 | 0.06 | 0.39 |
| 3 | 0.17 | 0.18 | 0.06 | 0.03 | 0.04 | 0.52 |
| 4 | 0.07 | 0.07 | 0.04 | 0.05 | 0.04 | 0.73 |
| 5 | 0.11 | 0.07 | 0.04 | 0.05 | 0.04 | 0.69 |
a1 = cows were removed from the herd upon their second incidence of failure to wean a calf, regardless of reason, through 6 yr; 2 = cows were scored as culled upon their first failure to wean a calf, regardless of reason, through 6 yr; 3 = cows were scored as culled upon their first failure to give birth to a calf, through 6 yr; 4 = cows were scored as culled upon their first incidence of failure to wean a calf, but not considering calving failure as a reason, through 6 yr; 5 = cows were scored as culled upon their first instance of failing to wean a calf, provided that they had no prior instances of calving failure, through 6 yr.
Table 3.
Number of significant markers for each culling criterion
| Criteriona | n | Counts | Total No. SNP | No. significant SNP | After FDRb | PVEc | ||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | P < 0.01 | P < 0.0001 | |||||
| 1 | 291 | 60 | 231 | 34,640 | 371 | 6 | 0 | 0 |
| 2 | 300 | 182 | 118 | 34,675 | 517 | 46 | 69 | 0.218 |
| 3 | 294 | 141 | 153 | 34,632 | 492 | 44 | 61 | 0.181 |
| 4 | 266 | 70 | 196 | 34,655 | 306 | 2 | 0 | 0 |
| 5 | 169 | 51 | 118 | 34,596 | 238 | 7 | 1 | 0 |
a1 = cows were removed from the herd upon their second incidence of failure to wean a calf, regardless of reason, through 6 yr; 2 = cows were scored as culled upon their first failure to wean a calf, regardless of reason, through 6 yr; 3 = cows were scored as culled upon their first failure to give birth to a calf, through 6 yr; 4 = cows were scored as culled upon their first incidence of failure to wean a calf, but not considering calving failure as a reason, through 6 yr; 5 = cows were scored as culled upon their first instance of failing to wean a calf, provided that they had no prior instances of calving failure, through 6 yr.
bFalse discovery rate = 0.15.
cProportion of variance explained.
Figure 1.
Manhattan plots of genome-wide associations for each culling criterion. Horizontal line represents the false discovery rate threshold of 0.15 for each criterion: (a) criterion 1, where cows were removed from the herd upon their second incidence of failure to wean a calf, regardless of reason, through 6 yr; (b) criterion 2, where cows were scored as culled upon their first failure to wean a calf, regardless of reason, through 6 yr; (c) criterion 3, where cows were scored as culled upon their first failure to give birth to a calf, through 6 yr; (d) criterion 4, where cows were scored as culled upon their first incidence of failure to wean a calf, but not considering calving failure as a reason, through 6 yr; (e) criterion 5, where cows were scored as culled upon their first instance of failing to wean a calf, provided that they had no prior instances of calving failure, through 6 yr.
The BIF (2016) definition of stayability was a benchmark of interest for this study and represents the most severe culling strategy. Criterion 2 corresponds to a common culling policy used by U.S. beef producers, where a cow is removed from production upon her first instance of failing to raise a calf. Under criterion 2, significant attrition was observed, such that only 39% of the females would have remained in the herd after 6 yr (Table 2). Most of the failures were observed in the first 2 yr. Although this is unsurprising in a population produced by the inter se mating of B. indicus–B. taurus cattle, if this culling criterion were to actually have been applied, only 57% of the herd would have remained past 3 yr, diminishing the long-term research potential of these cows. Therefore, implementation of such strict culling policy on a B. indicus–influenced research herd may be impractical.
In the GWAS for criterion 2, the null model explained 21.8% of the variance observed, and after correction for multiple testing, resulted in 69 significantly associated SNP (Table 3). These SNP fell on BTA 1, 2, 5, 9, 18, and 21, with defined peaks due to multiple-linked SNP associations on BTA 1, 5, and 9 (Figure 1b). The most highly significant SNP from this analysis fall between 40 and 50 Mb on BTA 5. Independent association between chromosomal peaks was verified by extracting the lead SNP from BTA 5 and modeling it as a covariate in a replication of the criterion 2 GWAS (Supplementary Figure 1). Although significant associations were found, criterion 2 was not specific enough to determine why a cow left the herd. For example, it did not distinguish between different factors such as preweaning calf death loss, failure to maintain pregnancy, or failure to conceive. The purpose of the subsequent analyses was to increase the power of detection by more specifically defining the reasons for reproductive failure and reducing the sources of phenotypic variation.
Criterion 3 was used to identify associations corresponding to failure to give birth to a calf vs. failure to raise and wean a calf. Criterion 3 allowed trends in pregnancy rates to be observed by focusing on yearly calving records. Only 83% of the cows in the herd calved during their first calving season at 2 yr (Table 2). After removing the records of cows that failed to calve at 2 yr, 18% of remaining cows failed to successfully rebreed after the birth of their first calf and experienced calving failure during their second calving season at 3 yr. The strongest associations were observed on BTA 5, and other SNP survived multiple testing correction on BTA 1, 11, 15, and 24 (Figure 1c). These associations were the most significant from 43 to 50 Mb on BTA 5, and this region corresponds to the critical region reported by Hawken et al. (2012) that was associated with measures of age at puberty. These findings are also similar to reports of SNP from 20 to 55 Mb on BTA 5 associated with reproductive efficiency in American B. indicus–B. taurus composite cattle (McDaneld et al., 2014).
In contrast, criterion 4 and 5, which both focus on weaning as the reason for failure, had greater proportions of cows remain in the herd through 6 yr than did the previous 2 criteria, indicating that failure to calve is the primary reason for cows to leave the herd. The purpose of criterion 4 was to identify associations with culling due to weaning failure that was not associated with calving failure. It was anticipated that genetic associations coinciding with, for example, such traits as mothering ability, maternally inherited health traits, or milking ability would be observed. Once the false discovery rate threshold was applied, no SNP were found to be significant (Table 3), and no clear patterns were observed in the corresponding Manhattan plot (Figure 1d). The variance explained by the null model for this analysis was zero, indicating that the model did not adequately capture any genetic variation using this trait.
Criterion 5 focused exclusively on cows that either never failed to raise a calf or those that only ever failed to raise a calf from birth to weaning but never experienced calving failure. This strict criterion restricted the tested population to only 169 cows. This resulted in an underpowered model, and unsurprisingly, only one SNP on BTA 9 survived correction for multiple testing (Table 3). Despite the decreased power of detection, a clear peak of linked SNP on BTA 9 was observed, with no peak on BTA 5 (Figure 1e). Comparing this output with results of the GWAS for criterion 2 and 3, we speculate that suggested associations on BTA 5 may be driven by physiological influencers of pregnancy, whereas associations on BTA 9 may be associated with traits related to calf survivability from birth to weaning.
Bos indicus and Bos indicus crossbred females are known to be slower maturing and older at the onset of puberty than B. taurus heifers (Gregory et al., 1979; Chenoweth, 1994; Hearnshaw et al., 1994; Thallman et al., 1999). Slower maturation rate in B. indicus–influenced cattle, especially straight and high percentage B. indicus heifers, often has a negative impact on the proportion of heifers that calve at 2 yr and the subsequent proportion returning to estrus during the following breeding season (Chenoweth, 1994). Age at puberty in Brahman-crossbred heifers has been found to be highly variable depending on proportion of Brahman in the cross, season, and regional weather conditions (Chenoweth, 1994). Using age at first observed estrus as a physiological indicator, average age at puberty in Brahman-cross heifers has been reported at 510 and 398 d by Plasse et al. (1968) and Gregory et al. (1979), respectively, and when considering age at first conception as the physiological indicator of puberty, Riley et al. (2010) reported an average age of 461 d. In a population of Nellore–Angus F1 heifers, average age at puberty was 405 d using first observed estrus as the determinant (Thallman et al., 1999), and in a separate population of B. indicus–B. taurus crossbred heifers, average age at first corpus luteum as a measure of puberty was 656 d (Hawken et al., 2012).
These last two examples resemble the breed composition of the cycle 1 herd and support the theory that substantial variation in the rate of maturity, with late onset of puberty in part of this population, may be the difference between those females that remain productive through 6 yr and those females that skip early in life. Detection of associated SNP is indicative that some of the biological factors influencing stayability are beyond just managerial influences, and there is genetic variation within the cycle 1 cow herd.
Hawken et al. (2012) found that BTA 5: 44 to 50 Mb was significantly associated with age at puberty in B. indicus–B. taurus composites, as defined by age at first corpus luteum and supported by postpartum anestrous interval and detection of preweaning estrus. This critical interval corresponds to the region on BTA 5 identified for criteria 2 and 3. Due to the strong parallels between these findings and those associated with puberty, it is hypothesized that the later-maturing cows, as indicated by inability to successfully calve at 2 yr or to rebreed after the first calving season (Chenoweth, 1994), drove the strong associations to BTA 5.
In a study characterizing reproductive efficiency, as defined by two consecutive years of reproductive success, BTA 5 was the most significantly associated chromosome in B. indicus–B. taurus composites (McDaneld et al., 2014). Looking more critically at the most highly significant SNP between 26.3 and 48.1 Mb on BTA 5, reported by McDaneld et al. (2014), Psaros et al. (2015) found a large B. indicus–derived haplotype in this region in Brahman-influenced cattle. They determined that the influence on reproduction was most likely due to additive gene action by SNP within this region on BTA 5 and concluded that greater B. indicus influence on this region was negatively correlated with reproductive efficiency. Future work will be needed to verify the absence or presence of this haplotype in the cycle 1 population of Brahman- and Nellore-influenced, crossbred cows.
The critical region from 43 to 50 Mb on BTA 5 has been associated with many traits in B. indicus–B. taurus crossbred cattle, including age at puberty (Hawken et al., 2012), reproductive efficiency (McDaneld et al., 2014), udder characteristics (Tolleson et al., 2017), and growth traits such as live weight and hump score (Bolormaa et al., 2013) and percent intramuscular fat, back fat, and mature hip height (Bolormaa et al., 2014). This gene-rich region contains several candidate genes previously implicated in physiological processes associated with reproduction (Fortes et al., 2011; Beltman et al., 2013). There are genes involved in immune response (IFNG, IL22, LYZ2), apoptotic processes (TMBIM4), protein dephosphorylation (PPM1H, PTPRR), signal transduction (SRGAP1, RAB3IP, KCNMB4), DNA replication and RNA processing (HELB, XPOT), and processes potentially directly influencing reproduction such as cellular response to hormone stimulus (GRIP1, MDM2), and regulation of intracellular estrogen receptor signaling pathways (CNOT2).
Although there are no previously reported SNP directly associated with beef cow stayability in B. indicus–influenced cattle, Saatchi and Garrick (2016) recently identified two QTL on BTA 6 associated with stayability in Simmental cattle but did not observe QTL on BTA 5 or 9 as in the current analyses. These QTL were located on BTA 6 at 40 and 71 Mb and did not concur with SNP associations observed in this study. Hamidi Hay and Roberts (2017) investigated longevity as a continuous trait, measuring the number of months from first calving until disposal. Cows were culled if they failed to become pregnant or failed to wean a calf (Roberts et al., 2016). It should be noted that these B. taurus composite cows were part of a long-term study of supplemental feeding during postweaning development and winter grazing. There was a trend (P < 0.07) for the interaction of dam treatment and heifer treatment to affect pregnancy rate and the proportion of cows retained in the herd at 2.2 and 5.2 yr of age. After correcting the longevity trait for contemporary group and the fixed effects of the two treatments, Hamidi Hay and Roberts (2017) reported five SNPs associated with cow survivability in B. taurus composite cows on BTA 1, 3, 9, 19, and 25. Although the average age for cow disposal in their study was less than 4 yr, compared with 4.3, 3.2, 3.2, 3.7, and 3.5 yr for criteria 1 to 5, respectively, in the current study, there appears to be limited correspondence in the GWAS results. There were no similarities in the location of significant SNP. It is possible that this is because their population is strictly of B. taurus origin or it may be that the environmental effects of the two supplementation treatments masked expression of the natural genetic variation in the phenotype.
Lack of significant associations was likely due to the combination of small sample size and high degree of environmental influence on the phenotypes. Heritability for stayability to 6 yr of age has been estimated to be low-to-moderate in both B. taurus and B. indicus cattle. Heritability was estimated to be 0.18, 0.18, and 0.15 by Snelling et al. (1995), Brigham et al. (2007), and Jamrozik et al. (2013), respectively, in B. taurus cattle using threshold models. Success rates for cows in these studies ranged from 38% to 62% dependent on breed. Heritability for stayability in B. indicus cattle was estimated at 0.12, 0.22, 0.19, 0.10, and 0.19 using threshold models (Silva et al., 2003; Van Melis et al., 2007; Eler et al., 2014; Cavani et al., 2015; Guarini et al., 2015). Success rates for cows in these studies ranged from 29% to 31%. In the current study, success rates for criteria 2 and 3 were comparable with those found in the B. taurus studies, whereas success rates for criteria 1, 4, and 5 were much higher (Table 2) and probably too high to detect genetic variation for the traits in such a small population.
Stayability traits are expected to be largely influenced by environmental factors, so low-to-moderate heritability estimates are expected (Jamrozik et al., 2013). Martinez et al. (2005) found that the heritability for stayability for calving or weaning (h2 = 0.35 and 0.21, respectively) was greater than for stayability to a defined age of 6 yr (h2 = 0.17), indicating that selection for these definitions of stayability would be more effective. Even with low-to-moderate heritability estimates, selection for stayability is possible and warranted (Hudson and Van Vleck, 1981; Martinez et al., 2005; Van Melis et al., 2007; Jamrozik et al., 2013; Rizzo et al., 2015).
SNP included in the genomic relationship matrix were not removed when evaluated for associations because SNP on the Illumina BovineSNP50v1 chip are relatively sparse and are common variants, so few of the SNP are likely to be causative (Wiggans et al., 2016). However, due to proximal contamination, the GWAS may be underpowered (Listgarten et al., 2012). Furthermore, Boyle et al. (2017) have recently proposed an “omnigenic” model for complex traits in which all genes expressed in relevant cells have very small effects on phenotypic variation because gene regulatory networks are interconnected. Thus, regardless of population size, SNP with nonzero effects on stayability may never reach genome-wide significance.
A tendency for B. indicus–influenced cattle to experience greater rates of reproductive failure early in life vs. straight B. taurus cattle (Chenoweth, 1994) is reflected in the definitions of stayability applied in studies using B. indicus–influenced cattle. In multiple studies of reproductive performance of Brazilian Nellore cattle, stayability has been defined as a cow’s ability to produce three calves by 76 mo (Guarini et al., 2015; Rizzo et al., 2015). This was referenced as being the earliest age at which a cow was expected to produce three calves, given that most cows first calve at 32 mo, and assuming that three calves were the break-even point for Brazilian producers (Rizzo et al., 2015). Similar definitions have been used in the analysis of Brahman cattle, where stayability represented a cow’s ability to produce three calves by 6 yr (Cavani et al., 2015). Other Brazilian studies with Nellore cows used a system of defining stayability where a cow qualifies as meeting the stayability threshold only if she successfully and successively produces a calf every year until a given age, generally 6 yr (Silva et al., 2003; Van Melis et al., 2007; Santana et al., 2013; Eler et al., 2014). This final definition closely follows the definition most commonly used in the United States and more accurately reflects the goals of a typical American producer. Although not the purpose of the current study, future economic analyses are warranted to determine the optimal ages for stayability benchmarks in B. indicus–influenced cattle.
As the average age of a herd increases, herd productivity is expected to peak as well. Older, productive cows demonstrate an increase in percentage calf crop born and weaned and in total kg of calf weaned (Cundiff et al., 1992). Maintaining productive cows for longer increases economic returns and increases cow value (Garcia et al., 2014). Bos indicus–B. taurus crossbred cows have been reported to have increased reproductive longevity (Riley et al., 2001; Thrift and Thrift, 2003). Bos indicus–B. taurus cows are more likely to survive culling due to decreased rates of dystocia and decreased tooth loss at advanced ages (Riley et al., 2001; Thrift and Thrift, 2003). Furthermore, Nellore-sired cows have been shown to maintain udder integrity and to have increased survivability and overall lifetime productivity than other B. indicus– or B. taurus–sired females (Riley et al., 2001). In a Brazilian study estimating the influence of popular Nellore founders on the current top 1% of Nellore sires for stayability EPD, the bull Karvadi was the most influential bull and contributed an estimated 8.2% of the genetics in the population subset, mainly through his son Chummak (Marcondes et al., 2007). Karvadi is the great great grand-sire of two of the F1 donor cows through his son Chummak, and an F1 bull that contributed to four ET families and an NS family through his son Chakkar. However, there was no evidence that cows from those families performed differently for stayability (P > 0.05).
Beef cow stayability is an important yet complicated measure of cow reproduction and productivity. The large number of sources of variation associated with the trait makes it difficult for producers to select for and for geneticists to understand. It has been shown herein that there is potential to identify genomic regions associated with a complex trait such as stayability in B. indicus–B. taurus crossbred cows. It has been the long-term goal that the McGregor Genomics herd be used to understand genetic factors influencing cow lifetime productivity traits and to identify important variants that may be applied in the development of genomic selection tools for tropically adapted beef breeds. As the median age of the cycle 1 herd increases, future analyses will focus on reproductive performance to later ages and lifetime productivity.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Animal Science online.
Footnotes
1This project was supported in part by National Research Initiative Competitive Grant no. 2008-35205-18767 from the USDA National Institute of Food and Agriculture. BNE is a USDA National Needs Fellow supported by 2014-38420-21835. Authors acknowledge the efforts of B. D. Johnson, M. D. Freedman, and all personnel at the Texas A&M AgriLife Research Center at McGregor.
LITERATURE CITED
- Beltman M. E., Forde N., Lonergan P., and Crowe M. A.. 2013. Altered endometrial immune gene expression in beef heifers with retarded embryos. Reprod. Fertil. Dev. 25:966–970. doi:10.1071/RD12232 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300. [Google Scholar]
- BIF 2016. Guidelines for uniform beef improvement programs. 9th rev. ed Prairie (MS): Beef Improvement Federation; – [accessed August 28, 2017]. https://beefimprovement.org/library-2/bif-guidelines. [Google Scholar]
- Bolormaa S., Pryce J. E., Kemper K., Savin K., Hayes B. J., Barendse W., Zhang Y., Reich C. M., Mason B. A., Bunch R. J.,. et al. 2013. Accuracy of prediction of genomic breeding values for residual feed intake and carcass and meat quality traits in Bos taurus, Bos indicus, and composite beef cattle. J. Anim. Sci. 91:3088–3104. doi:10.2527/jas.2012-5827 [DOI] [PubMed] [Google Scholar]
- Bolormaa S., Pryce J. E., Reverter A., Zhang Y., Barendse W., Kemper K., Tier B., Savin K., Hayes B. J., and Goddard M. E.. 2014. A multi-trait, meta-analysis for detecting pleiotropic polymorphisms for stature, fatness and reproduction in beef cattle. PLoS Genet. 10:e1004198. doi:10.1371/journal.pgen.1004198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E. A., Li Y. I., and Pritchard J. K.. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169:1177–1186. doi:10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigham B. W., Speidel S. E., Enns R. M., and Garrick D. J.. 2007. Stayability to alternate ages. Proc. West. Sec. Amer. Soc. Anim. Sci. 58:27–30. [Google Scholar]
- Cavani L., Garcia D. A., Carreño L. O., Ono R. K., Pires M. P., Farah M. M., Ventura H. T., Millen D. D., and Fonseca R.. 2015. Estimates of genetic parameters for reproductive traits in Brahman cattle breed. J. Anim. Sci. 93:3287–3291. doi:10.2527/jas.2015-8970 [DOI] [PubMed] [Google Scholar]
- Chenoweth P. J. 1994. Aspects of reproduction in female Bos indicus cattle: a review. Aust. Vet. J. 71:422–426. doi:10.1111/j.1751-0813.1994.tb00961.x [DOI] [PubMed] [Google Scholar]
- Cundiff L. V., Núñez-Dominguez R., Dickerson G. E., Gregory K. E., and Koch R. M.. 1992. Heterosis for lifetime production in Hereford, Angus, Shorthorn, and crossbred cows. J. Anim. Sci. 70:2397–2410. doi:10.2527/1992.7082397x [DOI] [PubMed] [Google Scholar]
- Eler J. P., Bignardi A. B., Ferraz J. B., and Santana M. L. Jr. 2014. Genetic relationships among traits related to reproduction and growth of Nellore females. Theriogenology 82:708–714. doi:10.1016/j.theriogenology.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Fortes M. R., Reverter A., Nagaraj S. H., Zhang Y., Jonsson N. N., Barris W., Lehnert S., Boe-Hansen G. B., and Hawken R. J.. 2011. A single nucleotide polymorphism-derived regulatory gene network underlying puberty in 2 tropical breeds of beef cattle. J. Anim. Sci. 89:1669–1683. doi:10.2527/jas.2010-3681 [DOI] [PubMed] [Google Scholar]
- Garcia J., Herring A. D., Riley D. G., Sanders J. O., and Anderson D. P.. 2014. Economic analysis of cow longevity. Proc. West. Sec. Amer. Soc. Anim. Sci. 65:83–86. [Google Scholar]
- Gregory K. E., Laster D. B., Cundiff L. V., Smith G. M., and Koch R. M.. 1979. Characterization of biological types of cattle—cycle III: II. Growth rate and puberty in females. J. Anim. Sci. 49:461–471. doi:10.2527/jas1979.492461x [Google Scholar]
- Guarini A. R., Neves H. H., Schenkel F. S., Carvalheiro R., Oliveira J. A., and Queiroz S. A.. 2015. Genetic relationship among reproductive traits in Nellore cattle. Animal 9:760–765. doi:10.1017/S1751731114003103 [DOI] [PubMed] [Google Scholar]
- Hamidi Hay E., and Roberts A.. 2017. Genomic prediction and genome-wide association analysis of female longevity in a composite beef cattle breed. J. Anim. Sci. 95:1467–1471. doi:10.2527/jas.2016.1355 [DOI] [PubMed] [Google Scholar]
- Hawken R. J., Zhang Y. D., Fortes M. R., Collis E., Barris W. C., Corbet N. J., Williams P. J., Fordyce G., Holroyd R. G., Walkley J. R.,. et al. 2012. Genome-wide association studies of female reproduction in tropically adapted beef cattle. J. Anim. Sci. 90:1398–1410. doi:10.2527/jas.2011-4410 [DOI] [PubMed] [Google Scholar]
- Hearnshaw H., Arthur P., Barlow R., Kohun P., and Darnell R.. 1994. Evaluation of Bos indicus and Bos taurus straightbreds and crosses. II. Post-weaning growth, puberty, and pelvic size of heifers. Aust. J. Agr. Res. 45:795–805. doi:10.1071/AR9940795 [Google Scholar]
- Hudson G. F. S. and Van Vleck L. D.. 1981. Relationship between production and stayability in Holstein cattle. J. Dairy Sci. 64:2246–2250. doi:10.3168/jds.S0022-0302(81)82836-6. [Google Scholar]
- Hulsman Hanna L. L., Garrick D. J., Gill C. A., Herring A. D., Riggs P. K., Miller R. K., Sanders J. O., and Riley D. G.. 2014. Genome-wide association study of temperament and tenderness using different Bayesian approaches in a Nellore–Angus crossbred population. Livest. Sci. 161:17–27. doi:10.1016/j.livsci.2013.12.012 [Google Scholar]
- Jamrozik J., McGrath S., Kemp R. A., and Miller S. P.. 2013. Estimates of genetic parameters for stayability to consecutive calvings of Canadian Simmentals by random regression models. J. Anim. Sci. 91:3634–3643. doi:10.2527/jas.2012-6126 [DOI] [PubMed] [Google Scholar]
- Listgarten J., Lippert C., Kadie C. M., Davidson R. I., Eskin E., and Heckerman D.. 2012. Improved linear mixed models for genome-wide association studies. Nat. Methods 9:525–526. doi:10.1038/nmeth.2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., Hunter D. J., McCarthy M. I., Ramos E. M., Cardon L. R., Chakravarti A.,. et al. 2009. Finding the missing heritability of complex diseases. Nature 461:747–753. doi:10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcondes C. R., Vozzi P. A., Araújo R. O., Glória W. P., and Lôbo R. B.. 2007. Contribuição dos efeitos de genearcas e de famílias sobre a probabilidade de permanência em rebanhos da raça Nellore. Arq. Bras. Med. Vet. Zootec. 59:977–982. [Google Scholar]
- Martinez G. E., Koch R. M., Cundiff L. V., Gregory K. E., Kachman S. D., and Van Vleck L. D.. 2005. Genetic parameters for stayability, stayability at calving, and stayability at weaning to specified ages for Hereford cows. J. Anim. Sci. 83:2033–2042. doi:10.2527/2005.8392033x [DOI] [PubMed] [Google Scholar]
- McDaneld T. G., Kuehn L. A., Thomas M. G., Snelling W. M., Smith T. P., Pollak E. J., Cole J. B., and Keele J. W.. 2014. Genomewide association study of reproductive efficiency in female cattle. J. Anim. Sci. 92:1945–1957. doi:10.2527/jas.2012-6807 [DOI] [PubMed] [Google Scholar]
- Plasse D., Warnick A. C., and Koger M.. 1968. Reproductive behavior of Bos indicus females in a subtropical environment. I. Puberty and ovulation frequency in Brahman and Brahman x British heifers. J. Anim. Sci. 27:94–100. [DOI] [PubMed] [Google Scholar]
- Psaros K. M., McDaneld T. G., Kuehn L. A., Snelling W. M., and Keele J. W.. 2015. Evaluation of single nucleotide polymorphisms in chromosomal regions impacting pregnancy status in cattle. J. Anim. Sci. 93:978–987. doi:10.2527/jas.2014-8509 [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J.,. et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi:10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley D. G., Chase C. C. Jr, Coleman S. W., Olson T. A., and Randel R. D.. 2010. Evaluation of tropically adapted straightbred and crossbred beef cattle: heifer age and size at first conception and characteristics of their first calves. J. Anim. Sci. 88:3173–3182. doi:10.2527/jas.2009-2573 [DOI] [PubMed] [Google Scholar]
- Riley D. G., Sanders J. O., Knutson R. E., and Lunt D. K.. 2001. Comparison of F1 Bos indicus x Hereford cows in central Texas: II. Udder, mouth, longevity, and lifetime productivity. J. Anim. Sci. 79:1439–1449. doi:10.2527/2001.7961439x [DOI] [PubMed] [Google Scholar]
- Rizzo E. C., Neto F. R., Diaz I. D., Dias M. M., Costa R. B., Ventura H. T., Oliveira H. N., and Falcão A. J.. 2015. Genetic association of productive and reproductive traits with stayability in Nellore cattle: analysis using Bayesian models. Genet. Mol. Res. 14:14956–14966. doi:10.4238/2015.November.24.3 [DOI] [PubMed] [Google Scholar]
- Roberts A. J., Funston R. N., Grings E. E., and Petersen M. K.. 2016. Triennial reproduction symposium: beef heifer development and lifetime productivity in rangeland-based production systems. J. Anim. Sci. 94:2705–2715. doi:10.2527/jas.2016-0435 [DOI] [PubMed] [Google Scholar]
- Rogers L. F. 1972. Economics of replacement rates in commercial beef herds. J. Anim. Sci. 34:921–925. doi:10.2527/jas1972.346921x [Google Scholar]
- Saatchi M., and Garrick D. J.. 2016. 032 Quantitative trait loci and candidate genes associated with heifer pregnancy rate and stayability in beef cattle. J. Anim. Sci. 94:15–15. doi:10.2527/msasas2016-032 [Google Scholar]
- Santana M. L. Jr, Eler J. P., Bignardi A. B., and Ferraz J. B.. 2013. Genetic associations among average annual productivity, growth traits, and stayability: a parallel between Nellore and composite beef cattle. J. Anim. Sci. 91:2566–2574. doi:10.2527/jas.2012-5856 [DOI] [PubMed] [Google Scholar]
- Silva J. A. V., Eler J. P., Ferraz J. B. S., Golden B. L., and Oliveira H. N.. 2003. Heritability estimate for stayability in Nellore cows. Livest. Prod. Sci. 79:97–101. doi:10.1016/S0301-6226(02)00149-5 [Google Scholar]
- Snelling W. M., Golden B. L., and Bourdon R. M.. 1995. Within-herd genetic analyses of stayability of beef females. J. Anim. Sci. 73:993–1001. doi:10.2527/1995.734993x [DOI] [PubMed] [Google Scholar]
- Thallman R. M., Cundiff L. V., Gregory K. E., and Koch R. M.. 1999. Germplasm evaluation in beef cattle–cycle IV: postweaning growth and puberty of heifers. J. Anim. Sci. 77:2651–2659. [DOI] [PubMed] [Google Scholar]
- Thrift F. A., and Thrift T. A.. 2003. Review: longevity attributes of Bos indicus x Bos taurus crossbred cows. Prof. Anim. Sci. 19:329–341. doi:10.15232/S1080-7446(15)31438-8 [Google Scholar]
- Tolleson M. W., Gill C. A., Herring A. D., Riggs P. K., Sawyer J. E., Sanders J. O., and Riley D. G.. 2017. Association of udder traits with single nucleotide polymorphisms in crossbred Bos indicus–Bos taurus cows. J. Anim. Sci. 95:2399–2407. doi:10.2527/jas.2017.1475 [DOI] [PubMed] [Google Scholar]
- Van Melis M. H., Eler J. P., Oliveira H. N., Rosa G. J., Silva J. A. II, Ferraz J. B., and Pereira E.. 2007. Study of stayability in Nellore cows using a threshold model. J. Anim. Sci. 85:1780–1786. doi:10.2527/jas.2005-608 [DOI] [PubMed] [Google Scholar]
- Wiggans G. R., Cooper T. A., VanRaden P. M., Van Tassell C. P., Bickhart D. M., and Sonstegard T. S.. 2016. Increasing the number of single nucleotide polymorphisms used in genomic evaluation of dairy cattle. J. Dairy Sci. 99:4504–4511. doi:10.3168/jds.2015-10456 [DOI] [PubMed] [Google Scholar]
- Zhou X., and Stephens M.. 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44:821–824. doi:10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.