Abstract
Hepatic steatosis is a multifactorial condition often observed in obese patients and a prelude to non-alcoholic fatty liver disease. Here we combine shotgun sequencing of faecal metagenomes with molecular phenomics (hepatic transcriptome, plasma and urine metabolomes) in two well-characterized cohorts of morbidly obese women recruited to the FLORINASH study. We reveal molecular networks linking gut microbiome and host phenome to hepatic steatosis. Patients with steatosis have low microbial gene richness and increased genetic potential for processing of dietary lipids and endotoxin biosynthesis (notably from Proteobacteria), hepatic inflammation and dysregulation of aromatic and branched-chain amino acid (AAA and BCAA) metabolism. We demonstrated that faecal microbiota transplants and chronic treatment with phenylacetic acid (PAA), a microbial product of AAA metabolism, successfully trigger steatosis and BCAA metabolism. Molecular phenomic signatures were predictive (AUC = 87%) and consistent with the gut microbiome making an impact on the steatosis phenome (>75% shared variation) and, therefore, actionable via microbiome-based therapies.
Introduction
Hepatic steatosis is a multi-factorial phenotype common to several chronic conditions such as insulin resistance, atherosclerosis and fatty liver disease, with increasing worldwide prevalence related to the obesity epidemic1–5. The gut microbiota recently emerged as a pivotal transducer of environmental influences (i.e., dietary components, drug treatments) to exert protective or detrimental effects on several host tissues and systems, including regulation of intermediary metabolism, liver function and cardiovascular disorders, either directly via translocation or indirectly through microbial metabolism or function in metabolic disorders6–8,9–11. Rodent studies demonstrated the role of the gut microbiome in liver disease and to the stratification of Type 2 diabetes (T2D) and cardiovascular disorders (CVD). Microbiome-associated factors involve, for instance, bacterial lipopolysaccharides (LPS) or methylamines such as trimethylamine (TMA) and trimethylamine N-oxide (TMAO)12,13 playing a role in the development of insulin resistance and atherosclerosis6,14,15. Hepatic steatosis is a shared mechanism for the development of T2D and CVD in humans in both non-alcoholic and virus-associated fatty liver disease16 but the physiological mechanisms behind this interplay remain poorly understood17,18.
Here, we take advantage of the advances in high-throughput sequencing and phenotyping technologies to characterize in humans physiological mechanisms responsible for the integrated interactions between signals from the gut metagenome and the host molecular phenome (a comprehensive set of molecular phenotypes useful to identify subgroups of patients17) of hepatic steatosis. We introduce a unique integrative multi-omics and precision medicine approach combining shotgun metagenomics, liver transcriptomics, metabolomics in plasma and urine and clinical phenotyping to reveal the molecular mechanisms and multi-scalar interactions involved in the physiology of steatosis in a new cohort of non-diabetic obese women we recruited as part of the FLORINASH consortium.
In-depth analyses of faecal metagenomics and phenomics reveal a robust signature highlighting a tight crosstalk between the microbiome, host gene expression and metabolism in hepatic steatosis involving low microbial gene richness19 (MGR) and imbalances in aromatic amino acid (AAA) and branched-chain amino acid (BCAA) metabolism20,21. Based on the results obtained in our clinical study, we then demonstrated a causal role of the microbiota-produced metabolite most strongly associated with steatosis, namely phenylacetic acid (PAA), in the triggering of the hepatic steatosis phenome by faecal microbiota transplants (FMT) and by testing PAA on primary cultures of human hepatocytes and in mice.
Results
Cohort design and identification of clinical confounders
To characterize the hepatic steatosis phenome, we established two unique and independent cohorts of women – negative for viral hepatitis – from Italy and Spain who elected for bariatric surgery. We focused on morbid obesity in non-diabetic women to examine liver steatosis variability. In particular, we excluded patients with known T2D to avoid the confounding influence of long-term hyperglycemia22 or medications such as metformin23,24 on the microbiome (see Methods for full inclusion and exclusion criteria). The degree of hepatic fat was defined according to the joint guidelines from the European Associations for the study of Liver, Obesity and Diabetes (EASL, EASO, EASD, see Methods)25,26,27. Given the impact of the microbiome on insulin resistance6,28, we also performed oral glucose tolerance tests (OGTT) and euglycemic hyperinsulinemic clamps (EHC) (Supplementary Table 1). Clinical phenotypes were complemented by faecal metagenomics and molecular phenomics (plasma and urine metabolomes and liver transcriptomes) for association studies29,30. We then devised a data-driven hypothesis generation and validation strategy (Fig. 1). We first identified age, cohort and BMI as confounders, while all other clinical variables were mediators or had no effect on generalized linear models (Supplementary Fig. 1, Supplementary Table 2). These three confounders were taken into account in all subsequent partial Spearman’s rank-based correlation (pSRC) patterns across clinical variables.
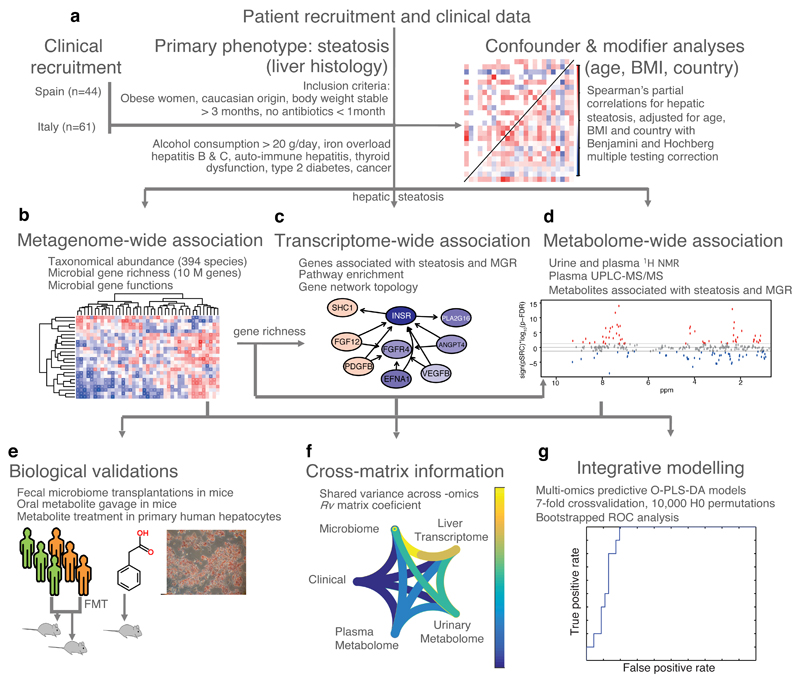
Figure 1. Flowchart showing approach used for the integration of clinical, molecular phenomics and metagenomic information and biological validations.
a, Confounder and modifier analysis performed using linear models on the FLORINASH clinical markers identified three confounders: age, BMI and country (n = 105). Subsequent analyses were performed using partial Spearman’s rank-based correlation (pSRC) coefficients adjusted for age, BMI and country and corrected for multiple testing using the Benjamini and Hochberg criterion (p-FDR). b, Metagenome-wide and phenome-wide association of taxonomic abundance data with clinical markers (n = 56 patients, pSRC, p-FDR < 0.05). c, Network analysis of hepatic transcriptome (n = 56 patients, pSRC, p-FDR < 0.05). d, Metabolome-Wide Association Study based on plasma (n = 56) and urine (n = 102, pSRC, p-FDR < 0.05) 1H-NMR spectra. e, In vitro and in vivo pre-clinical validation protocols. f, Integrative comparison analysis using Rv coefficients (n = 56). g, Predictive performance of an O-PLS-DA model integrating all metagenomic and phenomic modalities for prediction of non-alcoholic fatty liver (no hepatic steatosis, score = 0, n = 10 vs. steatosis, score > 0, n = 46) in ROC curves. All tests are two-sided.
Metagenomic signatures of hepatic steatosis
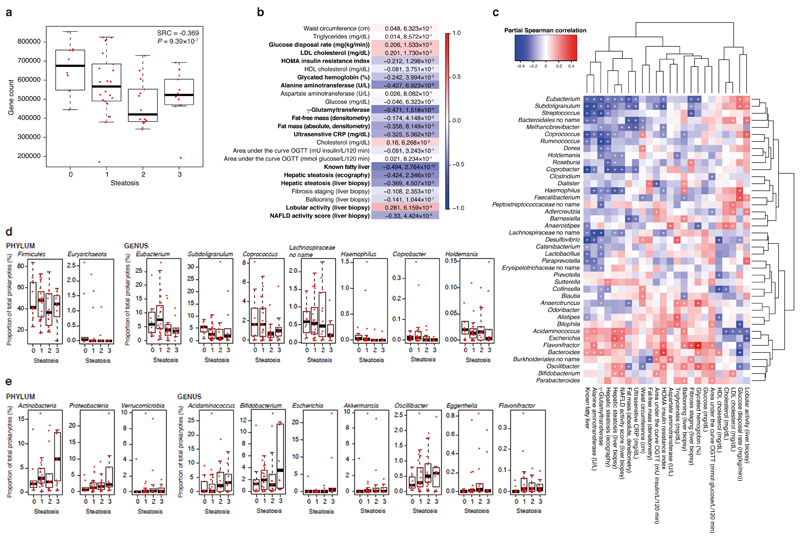
To obtain detailed taxonomic and functional information in hepatic steatosis for the faecal microbiome, we sequenced the patients’ faecal metagenome and data were processed using our in-house pipeline, performing QC checks, filtering, and binning of reads into taxonomic kingdoms (Supplementary Table 3, Supplementary Fig. 2); metagenome assembly31, gene prediction and clustering32,33, functional annotation of gene clusters and comparison with the HMP Integrated Gene Catalog (IGC)34 were performed. A total of 19,140,155 predicted genes were identified, which formed 3,902,787 gene clusters. Of these genes, 2,320,286 mapped to the IGC, while 1,582,501 were novel (90 % query length, 95 % identity; Supplementary Table 4). We derived gene counts, i.e., a measure of MGR, based on average values obtained from 30 samplings of 7 million randomly sampled reads that mapped to genes8, resulting in a mean of 558,246 ± 154,249 genes across the samples (Supplementary Table 5), which is in the same order of magnitude as previous reports19. Remarkably MGR was significantly anti-correlated with hepatic steatosis (Fig. 2a; liver steatosis 0 665,063 ± 126,062 vs liver steatosis 3 517,989 ± 126,062 genes, n = 10 patients both groups, P = 0.03 Wilcoxon rank sum test) and with a number of markers of liver function, including γ-glutamyltransferase, alanine aminotransferase and inflammation (C-reactive protein) as well as with echography-assessed liver steatosis (Fig. 2b). Our data demonstrate for the first time the association of MGR with liver steatosis in a BMI-adjusted context, reinforcing previous observations for body weight and liver cirrhosis19.
Figure 2. Association between liver steatosis, microbial gene richness (MGR) and metagenomic data in obese women.
a, Boxplots showing that MGR was significantly anti-correlated with liver steatosis using Spearman’s rank-based correlation, adjusted for age, BMI and sex (pSRC, two-sided, n = 56). b, Heatmap showing the significant correlation of MGR with clinical data (n = 56, pSRC, two-sided, p-FDR < 0.05 values shown). c, Heatmap showing the association of genus-level abundance data with clinical data. (n = 56, pSRC, two-sided, + p-FDR < 0.05). (d-e) Boxplots showing prokaryotic taxa significantly associated with hepatic steatosis. d, Prokaryotic taxa significantly anti-correlated with liver steatosis at the phylum and genus levels (n = 56, pSRC, two-sided, p-FDR < 0.05, see Supplementary Table S6 for exact and BH-adjusted p-values). e, Prokaryotic taxa significantly correlated with liver steatosis at the phylum and genus levels (n = 56, pSRC, two-sided, p-FDR < 0.05 see Supplementary Table S6 for exact and BH-adjusted p-values). For all panels, n = 56, groups as no liver steatosis = 10; liver steatosis 1 = 22; liver steatosis 2 = 14; liver steatosis 3 = 10. All boxplots are median + interquartile range, error bars are 1.5 times interquartile range.
To determine whether specific microbes were responsible for this correlation, we assessed the abundance of prokaryotes within the metagenomes. Several taxa were significantly associated with liver steatosis and other related clinical parameters (Fig. 2c–e, Supplementary Fig. 3a, Supplementary Fig. 4, Supplementary Table 6): at the phylum level Proteobacteria, Actinobacteria and Verrucomicrobia were significantly correlated with liver steatosis, while Firmicutes and Euryarchaeota were significantly anti-correlated, whereas species diversity (calculated using the Chao1 estimator) was not correlated with liver steatosis (Supplementary Fig. 3b).
We next investigated associations between microbial function, by mapping our microbial gene catalog onto KEGG modules, and clinical phenotypes, thus revealing positive associations of hepatic steatosis with microbial carbohydrate, lipid and amino acid metabolism (Supplementary Fig. 5). These data suggest a change in microbial metabolism may contribute to liver health in morbidly obese women. Of particular relevance, LPS and peptidoglycan biosynthesis was significantly correlated with liver steatosis (Supplementary Fig. 5); this increase in LPS biosynthetic potential being consistent with an increased representation of Gram-negative Proteobacteria in steatosis, as observed in rodents35. These pathway-level analyses also highlight an increase in bacterial biosynthetic potential for fatty acids and sugars and various amino acids including BCAAs (Val, Leu and Ile) and AAAs (Trp, Tyr and Phe) associated with steatosis and insulin resistance; this increase in BCAA biosynthesis further confirming previous reports in obesity and insulin resistance contexts28,36.
Impact of the microbiome on the hepatic steatosis phenome
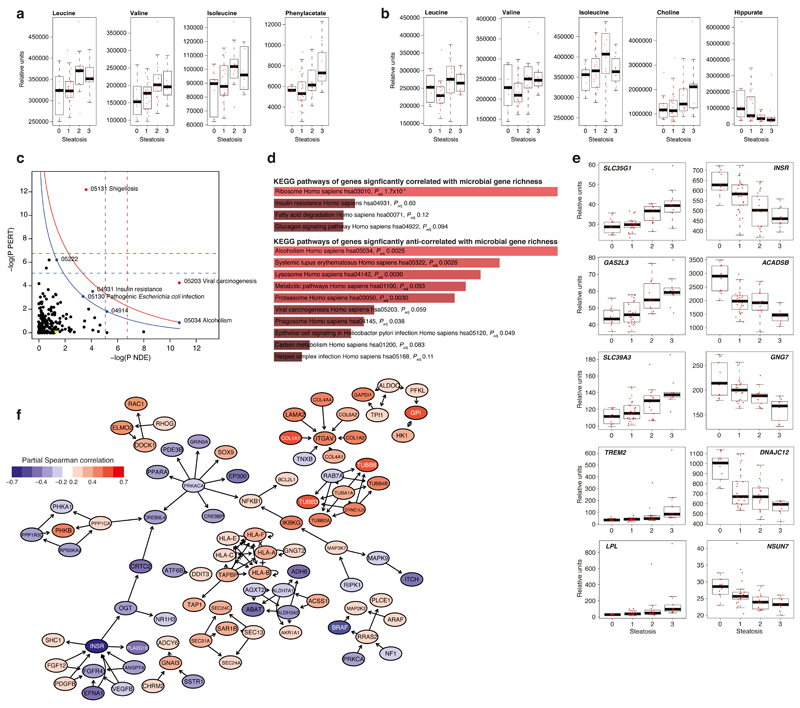
To reveal metabolic phenotypes possibly involved in steatosis related to the gut microbiome and liver steatosis, we performed metabolic profiling of urine and plasma by 1H-NMR spectroscopy. A metabolome-wide association study (MWAS)30 resulted in 124 metabolite signals in urine and 80 in plasma correlated with hepatic steatosis and associated clinical traits (Supplementary Fig. 6, 7). Strikingly, the majority of liver steatosis-associated metabolites in plasma and urine were also associated with low MGR (Fig. 3a, b, Supplementary Fig. 8, Supplementary Table 7). Among the top liver steatosis metabolites (also associated with low MGR), we observed a significant correlation with BCAAs in plasma (leucine p-FDR = 4.69×10-5; valine p-FDR = 1.72×10-4; isoleucine p-FDR = 9.72×10-5, Fig. 3a) and a significant increase in urine (leucine p-FDR = 6.1×10-4; valine p-FDR = 1.73×10-3; isoleucine p-FDR = 0.024, Fig. 3b) consistent with reports in obese patients20,28. Plasma choline and phosphocholine were not anti-correlated with liver steatosis for the 56 patients (but were anti-correlated in the larger cohort, n=102, Supplementary Fig. 9), whereas increased choline excretion was observed in liver steatosis (Fig. 3b), which is consistent with previous reports regarding choline bioavailability6,37. Remarkably, urinary hippurate was associated with MGR, echoing similar associations recently observed with Shannon diversity index obtained from 16S rRNA gene sequence profiling38. Among the microbial–mammalian co-metabolites significantly associated with steatosis and low MGR, plasma PAA (p-FDR = 4.69×10-5) showed the strongest positive association (Fig. 3a). High MGR observed in non-steatotic patients was significantly correlated with a number of gut-derived microbial metabolites, such as urinary phenylacetylglutamine (p-FDR = 3.10×10-9), plasma acetate (p-FDR = 0.009) and TMAO (p-FDR = 0.006) (Supplementary Table 7), a microbial-host co-metabolite playing a role in insulin resistance and atherosclerosis6,14,15. We further confirmed that TMAO, but not TMA, was marginally anti-correlated with steatosis by UPLC-MS/MS using isotopically-labelled standards39,40, which is consistent with recent reports on the role of TMAO in metabolic homeostasis15,41 (Supplementary Table 8). Altogether, these results suggest for the first time the existence of a metabolic phenotype associated with hepatic steatosis and low MGR, pinpointing elevated BCAAs, AAAs and microbial metabolites coupled to a potential imbalance in hepatic oxidation and conjugation of those microbial substrates.
Figure 3. Association of metabolomic and transcriptomic data with liver steatosis and microbial gene richness (MGR).
(a-b) Metabolome-wide association with steatosis (n = 56, pSRC, two-sided, p-FDR < 0.05, See Supplementary Table 7 for exact and BH-adjusted p-values). Boxplots showing plasma (a) and urinary (b) metabolites most significantly partially correlated with liver steatosis. (c-f) Transcriptome-wide association with steatosis and MGR (n=56, pSRC, two-sided, p-FDR < 0.05, see Supplementary Table 9-10 for exact and BH-adjusted p-values) c, SPIA evidence plot for the 2,277 genes significantly associated with liver steatosis and MGR. The pathways above the oblique lines are significant (< 0.2) after Bonferroni correction of the global P values (pG, obtained by combining the pPERT and pNDE using the normal inversion method, in red) and after a FDR correction of the global P values, pG (in blue). The yellow node represents the KEGG pathway ‘Non-alcoholic fatty liver disease (NAFLD) – Homo sapiens (human)’; 05222, Small cell lung cancer; 4914, Progesterone-mediated oocyte maturation. d, Enrichr (KEGG pathway) analysis of the hepatic genes significantly correlated and anti-correlated with MGR. e, Boxplots showing the ten hepatic genes most significantly correlated and anti-correlated with liver steatosis. f, Topological analysis of the KEGG networks resulting from hepatic steatosis – MGR intersecting genes showing the genes with the highest betweenness centrality43,44 (blue, anti-correlated with steatosis, red correlated with steasosis). For all panels, groups are: no liver steatosis = 10; liver steatosis 1 = 22; liver steatosis 2 = 14; liver steatosis 3 = 10 for all panels. Boxplots are median, with interquartile range and 1.5 times interquartile range.
To identify hepatic molecular mechanisms associated with the gut microbiome, we complemented our phenome coverage by profiling hepatic (liver biopsy) transcriptomes from the same set of patients. We identified 3,386 and 3,201 genes significantly positively and negatively correlated, respectively, with liver steatosis (pSRC p-FDR < 0.05) (Supplementary Table 9). Furthermore, 3,581 human genes significantly correlated (p-FDR < 0.05) with MGR: the pathways associated with the 1,776 genes significantly positively correlated with MGR largely matched those significantly anti-correlated with steatosis (Supplementary Fig. 10). However, the 1,805 anti-correlated with MGR matched those positively associated with hepatic steatosis (Supplementary Fig. 10), consistent with an anti-correlation between liver steatosis and MGR (Supplementary Table 9, Supplementary Table 10).
To generate molecular hypotheses that could be useful for microbiota-related next-generation therapeutic strategies we performed a hepatic signalling pathway impact analysis (SPIA42, see Methods) including the 2,277 genes intersecting the liver steatosis and MGR-associated genes. In particular hepatic genes associate with non-specific pathways involved in the core immune response to clearance of viral and bacterial (Proteobacteria, Gram-negative) infections (i.e., viral carcinogenesis; pathogenic Escherichia coli infection, shigellosis), alcoholism and insulin resistance (Fig. 3c). Enrichment analyses (see Methods) of the hepatic genes significantly associated with MGR further highlighted a significant (p-FDR < 0.2) over-representation of KEGG pathways associated with the proteasome, phagosome, insulin resistance, glucagon signalling and non-specific responses to microbial (Gram-negative, viral) infections (Fig. 3d). Among the overlapping genes co-associated with hepatic steatosis and low MGR, LPL (lipoprotein lipase) was among the most correlated with hepatic steatosis, while ACADSB (short/branched chain acyl-CoA dehydrogenase) and INSR (insulin receptor) were the most anti-correlated (Fig. 3e), suggesting a molecular basis for the observation that individuals with low MGR have a reduced capacity to respond to insulin exemplified by decreased glucose disposal rate (during the EHC) and increased HOMA-IR (as shown in Fig. 2b and previously reported in ref 28).
We further complemented our analyses of the hepatic transcriptome by assessing the topology of a directional network made of 2,277 genes significantly associated with liver steatosis and low MGR mapped onto KEGG pathways involved in liver disease, by aggregating all the KEGG networks with at latest one gene in common with the genes included in the NAFLD pathway. To analyse the topology of this resulting network, we computed shortest paths between the significant genes and derived the betweenness centrality metric43,44, i.e., the number of shortest paths passing through a particular gene product, to evaluate how central these genes are in the network. Ranking of betweenness centrality from highest to lowest showed CREB3L4, PRKACA, CRTC2, OGT, INSR, NFKB1, PPP1CA, IKBKG, MAP3K7, MAPK9, ITGAV, RRAS2, RPS6KA2, PHKA1, PHKB, BRAF, ALDOC, PFKL, EFNA1, FGF12, ANGPT4, PDGFB, VEGFB, FGFR4, MAP2K2, TAPBP, ALDH3A2, ALDH7A1, GPI and GNAI3 to be the 30 genes having most control over the network. In particular, betweenness centrality further highlights clusters of central genes channelling a high proportion of the shortest paths involving cAMP-related genes (CREB3L4, PRKACA, CRTC2), innate immunity (Nuclear Factor Kappa B subunit 1, NFKB1) and INSR amongst others (Fig. 3f). Overall, hepatic gene expression is concordant with the metabolic signature obtained in plasma and urine showing elevated BCAAs concomitantly associated with low MGR, liver steatosis and insulin resistance, highlighting the interconnection among these three parameters. Genetic manipulation of INSR in the hepatocyte displayed a NAFLD phenotype45,46 and the gut microbiome has recently been shown in rodents to interfere with INSR activation in the liver47. These results provide in humans a validation of numerous rodent-based hypotheses.
Steatosis-associated microbiota and microbial metabolites modulate the steatosis phenome
Our results document a strong contribution of the gut microbiome to the hepatic steatosis phenome. The increased microbial capacity for metabolism of BCAAs and metabolism of AAAs such as phenylalanine, tyrosine and tryptophan in liver steatosis (Supplementary Fig. 5) – phenylalanine metabolism resulting in PAA production – is supported by circulating metabolic markers (Fig. 3a-b), suggesting potentially causal mechanisms involving the microbiome in the steatosis phenome. In particular, our results strengthen the contribution of the gut microbiome to increased levels of circulating BCAAs in the host28,36 – a metabolic phenotype gaining a central role in metabolic disorders20. This disruption of the gut–liver axis is further exemplified by the increase in inflammatory response, ER stress and phagosome pathways associated with a decrease in insulin signalling and small-molecule catabolic processes, conceivably altogether contributing to impaired BCAA and AAA metabolism as well as detoxification of liver steatosis-associated microbial compounds.
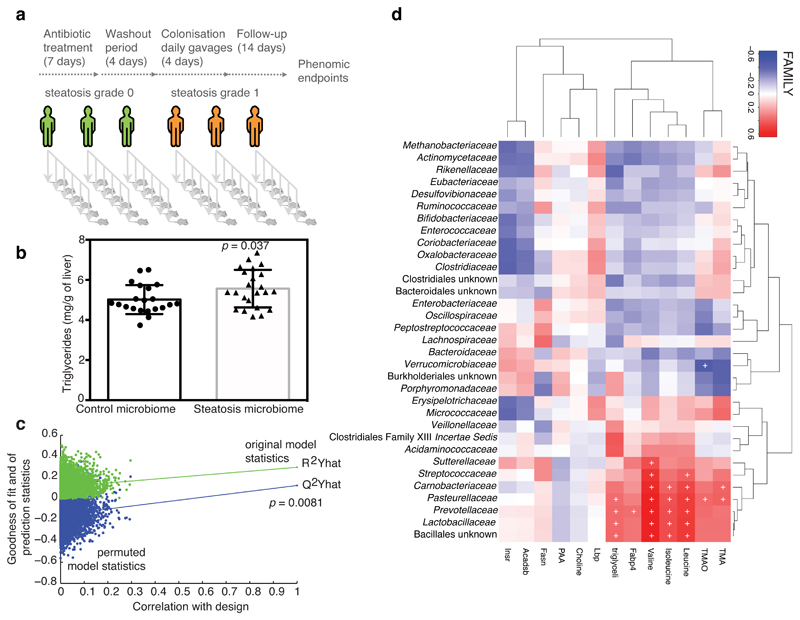
We then tested whether faecal microbial communities from donors with hepatic steatosis (steatosis grade 3) could trigger steatosis molecular mechanisms to recipient mice when compared with samples from donors with no hepatic steatosis (grade 0) (Fig. 4a). Donors with hepatic steatosis (n=3, steatosis grade 3) were randomly selected. Among subjects without hepatic steatosis (n=3, grade 0), we chose those that were similar in age, BMI, and fasting glucose to those with steatosis. For instance, fasting glucose was 87.3 ± 16.7 mg/dL in subjects without steatosis and 97.3 ± 6.4 mg/dL in the steatosis group (P = 0.39). After a short antibiotic treatment and wash-out period and four consecutive daily faecal microbiota transplantations (FMTs as in 48,49), the recipient mice were fed a chow diet for 2 weeks. In the former group, this procedure resulted in a moderate but rapid accumulation of hepatic triglycerides (Fig. 4b). We also observed an increased Fabp4 expression and plasma valine concentration compared with mice that received samples from patients without liver steatosis (Supplementary Fig. 11a,b), showing the general impact of the steatosis-associated microbiota from human donors on mouse liver lipid accumulation. By permutation testing seven-fold cross-validated O-PLS models using the donor human microbiome composition, we could successfully predict recipient mouse phenome responses, especially for steatosis, hepatic triglyceride content, Fabp4 and plasma valine levels (Fig. 4c and Supplementary Fig. 11c-e, 1,000 random permutations, see Methods), highlighting the statistical robustness of the prediction between human donor microbiomes and recipient mouse phenome. We then derived significant associations between the donor microbiota composition and the mouse phenome, showing that the steatosis-associated microbiota influences multiple patterns of association with hepatic triglycerides, circulating BCAAs and TMAO (Fig. 4d). Similar, yet weaker associations were also observed between the mouse phenome and recipient mouse microbiota evaluated by faecal 16S rRNA gene amplicon analysis (Supplementary Fig. 12). The rapid hepatic lipid accumulation suggested a causal role of the human faecal microbiota in the triggering of hepatic steatosis which over a long-term period could lead to a highly significant liver lipid depot further contributing to hepatic insulin resistance.
Figure 4. Transfer of steatotic and metabolic phenotypes to mice through FMT of material from patients with liver steatosis grade 3.
a, FMT protocol. b, Hepatic triglycerides in recipient mice (control microbiome, n = 21 and steatosis microbiome, n = 23, t test, 2-sided). c, Permutation tests for goodness of fit (R2) and prediction (Q2) parameters obtained from a seven-fold cross-validated O-PLS regression model quantitatively predicting recipient mouse hepatic lipid accumulation from human donor microbiome composition (n = 44). d, Heatmap showing the association between human donor microbiota and recipient mouse phenome (n = 44). Predictivity of O-PLS models is validated through 10,000 random permutations of the class membership variable and assessing the significance of the goodness-of-fit (R2, explained variance) and goodness-of-prediction (Q2, predicted variance) parameters. The horizontal axis corresponds to the correlation between the original class membership (on the right) and the permuted class membership (10,000 permutations on the left of the plot). The vertical axis corresponds to the R2 (green dots) and Q2 (blue dots) coefficients. Data obtained from FMT protocols performed independently with faecal material from three patients with liver steatosis (grade 3, >66% steatosis) and three control patients (grade 0, <5% steatosis), Data are mean ± s.e.m., * p < 0.05.
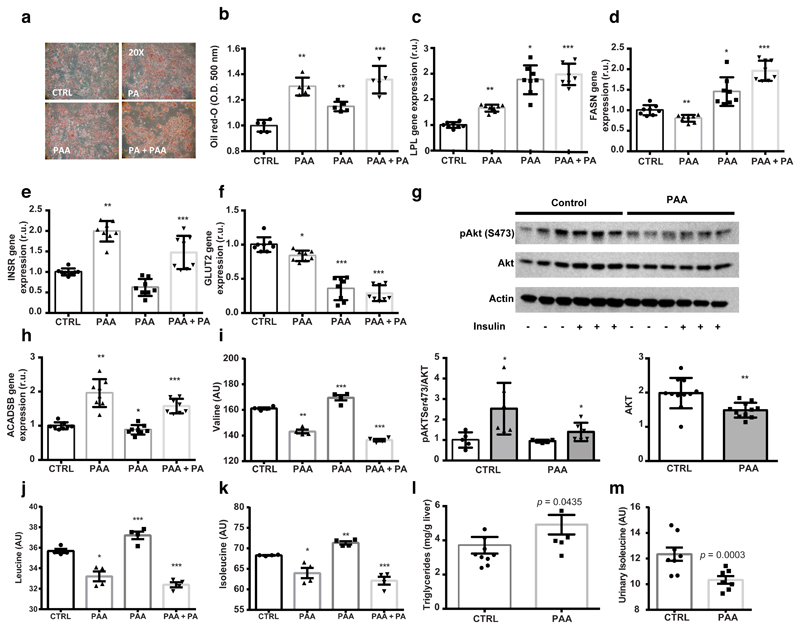
To highlight the potential of novel microbial compounds to directly affect the hepatic steatosis phenome, we selected PAA due to the convergence of metagenomic and metabolomic observations: i) there is increased abundance of microbial gene pathways associated with its production in metagenomic sequences (Supplementary Fig. 5), and ii) it is the strongest microbial metabolite associated with steatosis in our MWAS models (Fig. 3a). We compared its effects with the effects of palmitic acid, a free fatty acid known to trigger hepatic steatosis in human primary hepatocytes50, using a full factorial design. We assessed lipid accumulation, expression of genes involved in steatosis as well as BCAA metabolism and consumption. PAA initiates molecular mechanisms leading to triglyceride accumulation in human primary hepatocytes in synergy with palmitic acid (Fig. 5a-b) and induces expression of lipid metabolism genes (LPL and FASN, Fig. 5c-d). PAA induced INSR expression contrary to palmitic acid and participated in the reduction of GLUT2 expression (Fig. 5e-f). We next investigated AKT phosphorylation, which was significantly lowered by PAA, suggesting PAA reduces the response to insulin (Fig. 5g). PAA increased ACADSB expression (Fig. 5h) and resulted in an increased utilization of BCAA from the cell medium (Fig. 5i-k). We then treated mice with PAA for 2 weeks and confirmed the increase in hepatic triglycerides and decrease in excreted isoleucine (Fig. 5l-m). These results suggest that PAA, as one of our top hepatic steatosis-associated microbial metabolites, significantly increases hepatic BCAA utilization and hepatic lipid accumulation.
Figure 5. Microbial PAA induces liver steatosis and BCAA use in primary human hepatocytes and mice.
a, Micrographs of primary human hepatocytes stained with oil Red-O (representative images from n = 5 independent batches). b, Quantification of lipid accumulation (n = 5 independent batches, two-sided t test). c, LPL expression in hepatocytes. d, FASN expression in hepatocytes. e, INSR expression in hepatocytes. f, GLUT2 expression in hepatocytes. g, AKT phosphorylation in hepatocytes. h, ACADSB expression in hepatocytes. i, Valine in hepatocyte cell medium. j, Leucine in hepatocyte cell medium. k, Isoleucine in hepatocyte cell medium (c-k) n = 8 independent batches, two-sided t test. l, Hepatic triglycerides in PAA-treated mice (control, n = 10, PAA, n = 7, Mann-Whitney test, one-sided). m, Isoleucine in urine from PAA-treated mice (control, n = 10, PAA, n = 7, Mann-Whitney test, one-sided). Data are mean ± s.e.m., * p < 0.05, ** p < 0.01, *** p < 0.001. Abbreviations: CTRL, control group; PAA, phenylacetic acid treatment group; PA, palmitic acid treatment group; PA+PAA, palmitic acid and phenylacetic acid treatment group.
Integrative data crosstalk and steatosis signatures
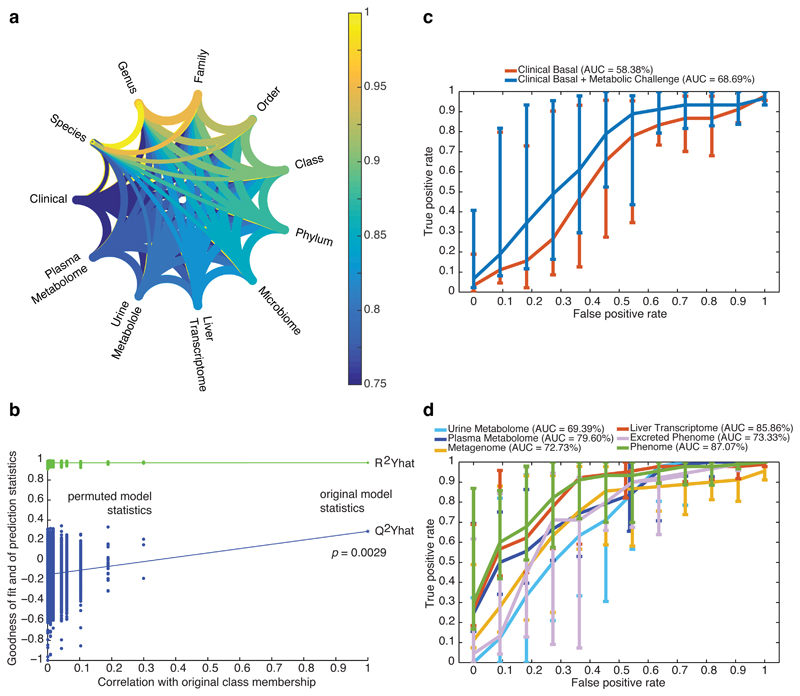
We finally quantified the crosstalk among gut microbiome, clinical phenotypes, liver transcriptome, urine and plasma metabolomes by estimating the proportion of shared variation amongst the different tables through Rv coefficients (Fig. 6, see Methods). A high proportion of information (79–97%) was shared between matching datasets (Fig. 6a, Supplementary Table 11), suggesting a strong similarity between metagenomic and phenomic data; the weakest (79.44%) being between urinary metabolome and clinical parameters. The metagenomic data shared 92–93% similarity with clinical parameters, liver transcriptome and plasma metabolome, while they only shared 74.68% with the urinary metabolome. This statistical crosstalk analysis suggests that, although metagenomic and phenomic data have strong similarity, there is still information attached to each original dataset which, if pooled together, could result in a robust signature.
Figure 6. Phenome-wide crosstalk and predictive modelling.
a, Metagenome–phenome matrix correlation network computed for the patients with matching metagenomic and phenomic profiles (n = 56) using the modified Rv correlation matrix coefficient. Each phenomic table corresponds to a node and edges represent the relationships between tables, i.e., the per cent of shared information, derived from the Rv2 matrix correlation coefficient corresponding to the proportion of variance shared by the two tables – which like a squared Pearson’s correlation coefficient (r2) – corresponds to the proportion of explained variance between two variables. b, Discriminative power of a supervised multivariate model (OPLS-DA) fitted with patients with matching metagenomic and phenomic profiles (n = 56) to predict new samples, using random permutation testing (10,000 iterations). (c-d), Performance of classification of liver steatosis status (n = 10, vs. others, n = 46) based on clinical data (c) or matching molecular phenomic and gut metagenomic profiles (d). ROC curves (mean + 95% confidence interval) were obtained for the cross-validated model predictions derived from the O-PLS-DA model, reaching an AUC of 87.07%, corresponding to the successful prediction rate. Groups for all panels are: no steatosis (grade 0), n = 10; steatosis (grades 1-3), n = 46. Data are mean ± pointwise confidence bounds.
We then built a multivariate model integrating metagenomic, transcriptomic and metabolomic information by fitting an orthogonal partial least squares discriminant analysis (O-PLS-DA) and tested its ability to correctly predict new samples during a seven-fold cross-validation through random permutation testing (Fig. 6b, 10,000 random permutations, P = 0.0029). We derived a bootstrapped Receiver-Operator Characteristic (ROC) curve for the cross-validated models illustrating the ability of the model to correctly predict new samples (AUC=87%, Fig. 6c, Supplementary Table 12) of the binary prediction of steatosis (i.e., steatosis vs. no steatosis) using cross-validated scores derived from seven-fold cross-validation of the O-PLS-DA model (see Methods), thereby confirming the joint predictive power of molecular phenomics and metagenomics. The predictive power of the phenome model is driven by the hepatic transcriptome (AUC 85%) that directly relates to the affected organ, but the excreted phenome and plasma metabolome both reach 73% and 79%, respectively. This AUC is particularly relevant as the non-invasive basal clinical data yielded 58%, which only increases through addition of more invasive metabolic challenges (OGTT and EHC, AUC 69%). Altogether, these predictive models based on molecular phenomics and metagenomics further support the idea that these molecular signatures used to generate hypotheses are robust and ultimately suggesting that the link tethering the microbiome to hepatic steatosis is robust too.
Discussion
In this study, we performed an in-depth clinical characterization of well-phenotyped non-diabetic obese women from Spain and Italy. We then reveal molecular networks between the gut microbiome and the hepatic steatosis phenome in this population of morbidly obese women, through computational integration of individual metagenomes, metabolomes and hepatic transcriptomes with histological steatosis scores. The robustness of our phenome signatures and the experimental follow-ups show that hepatic steatosis is negatively associated with MGR and the microbiome contributes to the steatosis phenome. The striking association between low MGR and hepatic steatosis is consistent with clinical and preclinical results confirming the role of the microbiome in rodent models7 of non-alcoholic fatty liver disease and the role of MGR in metabolic disease19,51.
We then functionally characterized an increased gut microbial amino-acid metabolism in steatotic subjects that has a profound impact on their liver transcriptome, biofluid metabolomes and liver fat accumulation, leading eventually to fatty liver. We found an anti-correlation pattern between steatosis and MGR was valid for the most significant steatosis-associated genes and metabolites, thereby suggesting that the reduction in MGR is a key factor that imbalances microbiome metabolic pathways leading to a steatosis-associated phenome, as observed for obesity19,51. From this tight crosstalk, we further depict a coordinated disruption of the gut–liver axis in hepatic steatosis that manifests itself across the faecal metagenome, hepatic transcriptome and biofluid metabolome. For instance, the increased Proteobacteria frequency in hepatic steatosis is mirrored by an increase in microbial-associated functional pathways related to endotoxin production and immune response in steatotic patients – both at the hepatic and circulating levels. Our study further confirms the impact of LPS and putatively other microbial products on liver lipid accumulation in humans52, as previously proposed in rodent models35.
By integrating numerous biological measurements, our data analysis strategy implemented a detailed functional analysis of the patient faecal metagenomes and molecular phenomes, offering novel insights for the integrative physiology of hepatic steatosis. For instance, the increased microbial potential for BCAA production, a phenomenon already reported for insulin resistance and obesity28,36, is mirrored by an increase in the BCAA pool in biofluids. Also, our bioinformatic analysis of metagenomic sequences combined with metabolomic data suggested a direct role for microbial degradation of AAAs into PAA in patients with steatosis. Our preclinical studies in rodents and primary culture of human hepatocytes corroborated the role of this metabolite, amongst others, as an example of a microbially-related metabolite involved in hepatic steatosis. By subsequently focussing on a unique microbiome-associated feature such as PAA, which was selected through converging patterns observed in microbial gene functions and biofluid metabolomes, we identified a novel mechanism by which the microbiome facilitates steatosis, via increased BCAA utilisation and AAA metabolism. Whilst acknowledging the complexity of the microbiome–host interplay, it should be noted that although PAA is an exemplar metabolite highlighted in our human dataset, its effects are here limited to triggering steatosis-associated molecular mechanisms and it is unlikely to be the sole player in steatosis. The PAA effects are most likely part of a much broader, multifactorial process orchestrated by the microbiome and involving many factors that warrant further studies.
The demonstration that the faecal microbiota obtained from patients with steatosis (grade 3, >66%) initiated hepatic lipid accumulation and affected the phenome of recipient mice through FMTs reinforces the causal role of the microbiota in steatosis. Not only did the human donor microbiota from patients with steatosis trigger hepatic triglyceride accumulation in recipient mice, but it also affected their circulating metabolome and hepatic transcriptome, through an increase in circulating valine levels and an increased expression of genes involved in lipid metabolism. Moreover, the characteristics of the donor microbiota predicts the extent of the phenomic response in the donor mice, which echoes recent reports on the prodromal role of the microbiota for metabolic response to diet in animal models15,53 and humans54. The successful replication of the steatotic phenotype using human donor material for FMT in mice represents a key translational link between metagenomic studies in patients with NAFLD, hepatic fibrosis or cirrhosis8,55,56 and previous FMT studies that had only been established for NAFLD with mouse donors7.
Altogether, we propose a model in which the microbiome orchestrates three possibly complementary contributions to hepatic steatosis in obesity: i) reduced MGR – indicative of deleterious changes in microbiome functions – can trigger steatosis and increase the BCAA pool; ii) microbially-produced PAA and possibly other related metabolites facilitate hepatic lipid accumulation via a synergetic increase in BCAA utilization in the TCA cycle; and iii) microbially-associated factors such as LPS induce inflammation in hepatocytes.
Similar to Qin et al. (2014) who studied the faecal metagenome of liver cirrhosis patients8, our data indicate a slight shift of the faecal microbiome in patients with steatosis to one more similar to that found in the human small intestine and oral cavity. For example, patients with steatosis had fewer Lachnospiraceae and Ruminococcaceae responsible for butyrate production and were enriched in Acidaminococcus and Escherichia spp. Bacteroides spp. were associated with insulin resistance, concordant with observations from Pedersen et al.28, who showed Bacteroides vulgatus was one of the main species contributing to insulin resistance, and circulating levels of BCAAs in humans28.
In conclusion, this work offers a unique clinical resource and integrated analysis of metagenomics with molecular phenomics of hepatic steatosis in non-diabetic obese women coupled with experimental validations in cellular and animal models. Not only does our work further validate previous studies in humans55, but it also confirms hypotheses formulated in rodent models, such as the role of LPS, in which the gut microbiome was shown to influence gene pathways involved in the immune system and metabolic disorders (i.e., inflammation impacting host metabolism7,35,57). Ultimately, this integrated database and modelling approach also suggests new potentially causal mechanisms in hepatic steatosis involving BCAA- and AAA-derived metabolites. Our investigations further support the view that the molecular crosstalk between the microbiome and its human host is of utmost importance for patient health and highlights the need for integrative analyses of metagenomes and broad-sense phenomes58,59. Our study establishes a comprehensive understanding of the microbial factors affecting human metabolic disease states for precision medicine, thereby laying the groundwork for targeted FMT therapies and pharmacotherapies to promote hepatic metabolic homeostasis.
Online Content
Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Material
Acknowledgements
We thank Professor Nigel J. Gooderham for critical reading of the manuscript. This work was supported by EU-FP7 FLORINASH (Health-F2-2009-241913) to RB, MF, JMFR, FF, CP, EH and JKN. This work used the computing resources of the UK MEDical BIOinformatics partnership – aggregation, integration, visualisation and analysis of large, complex data (UK MED-BIO), which is supported by the Medical Research Council (grant number MR/L01632X/1). LH is in receipt of an MRC Intermediate Research Fellowship in Data Science (MR/L01632X/1, UK MED-BIO). This work was also supported by funding to RB (Région Midi-Pyrénées 2009-2014 RPV09003BBA), Agence Nationale de la Recherche ANR 09-GEN TRANSFLORAP & GAD 08-2_378258), to MF (Ministry of University (MIUR) Progetti di Ricerca di Interesse Nazionale (PRIN) protocol number 2015MPESJS_004, Ministry of Health Ricerca Finalizzata RF-2011-02349921, Fondazione Roma call for Non-Communicable Diseases NCD 2014), to JMFR (Ministry of health FIS project 15/01934, CIBERobn Pathophysiology of Obesity and Nutrition and FEDER funds) and to MED (EU METACARDIS under agreement HEALTH-F4-2012-305312, Neuron II under agreement 291840 and the MRC MR/M501797/1).
Footnotes
Competing financial interests. L.H., J.-M.F.-R., M.F., R.H.B., J.L.L., E.H., J.K.N., C.P., R.B. and M.-E.D. are named as co-inventors on pending patents held by INSERM Transfert, INSERM, University of Rome Tor Vergata, University of Girona and Imperial College on non-alcoholic fatty liver disease diagnostics and have the right to receive royalty payments for inventions or discoveries related to non-alcoholic fatty liver disease diagnostics.
Author Contributions. R.B., J.-M.F.-R., M.F., F.L., F.F., C.P., E.H. and J.K.N. designed the study and supervised all parts of the project. R.B. is the project leader and chaired the consortium. M.-E.D. led data integration and elaborated the primary interpretation of analytical outcomes with L.H., in close collaboration with M.F., J.-M.F.-R. and R.B. L.H. implemented the microarray data analysis workflow, C.T. and M.W. developed the data repository. J.A. developed the metagenomic data analysis pipeline in collaboration with L.H., and S.A.B. supervised the development of the data repository and the pipeline. L.H., J.A., R.H.B., and M.-E.D. performed data analyses. J.-M.F.-R. and M.F. designed the clinical protocol and oversaw the clinical activities. M.C., F.D., I.C., O.P., P.G., J.P., G.X. and W.R. recruited and phenotyped patients, collected biological samples and physiological data. M.S., V.A. and V.B.-B. performed RNA and DNA extractions, R.B. and M.S. supervised DNA sequencing and gene profiling. J.L.L. and J.M.M.-N. performed cell culture experiments, F.B., E.A., J.Cha. and C.H. performed animal work. R.H.B., J.Chi. and L.M.-G. performed metabolic profiling of plasma and urine by 1H-NMR, E.H. and J.K.N. supervised metabolic profiling. A.M. performed methylamine quantifications. L.H. and M.-E.D. drafted the first versions of the paper with critical and substantial contributions from M.F., J.-M.F.-R. and R.B. All authors provided support and constructive criticism throughout the project and approved the final version.
References
- 1.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 2.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 3.Meex RCR, Watt MJ. Hepatokines: linking nonalcoholic fatty liver disease and insulin resistance. Nat Rev Endocrinol. 2017;13:509–520. doi: 10.1038/nrendo.2017.56. [DOI] [PubMed] [Google Scholar]
- 4.Petersen MC, Vatner DF, Shulman GI. Regulation of hepatic glucose metabolism in health and disease. Nat Rev Endocrinol. 2017;13:572–587. doi: 10.1038/nrendo.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams LA, Anstee QM, Tilg H, Targher G. Non-alcoholic fatty liver disease and its relationship with cardiovascular disease and other extrahepatic diseases. Gut. 2017;66:1138–1153. doi: 10.1136/gutjnl-2017-313884. [DOI] [PubMed] [Google Scholar]
- 6.Dumas M-E, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Roy T, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2012 doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 8.Qin N, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341 doi: 10.1126/science.1241214. 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA. 2012;109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoyles L, et al. Metabolic retroconversion of trimethylamine N-oxide and the gut microbiota. bioRxiv. 2017 doi: 10.1101/225581. 225581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumas M-E, et al. Microbial-Host Co-metabolites Are Prodromal Markers Predicting Phenotypic Heterogeneity in Behavior, Obesity, and Impaired Glucose Tolerance. Cell Rep. 2017;20:136–148. doi: 10.1016/j.celrep.2017.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonardo A, et al. Fatty liver is associated with an increased risk of diabetes and cardiovascular disease - Evidence from three different disease models: NAFLD, HCV and HIV. World J Gastroenterol. 2016;22:9674–9693. doi: 10.3748/wjg.v22.i44.9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 18.Dumas M-E, Kinross J, Nicholson JK. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and Fatty liver disease. Gastroenterology. 2014;146:46–62. doi: 10.1053/j.gastro.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Le Chatelier E, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 20.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang C, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med. 2016;22:421–426. doi: 10.1038/nm.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 23.Forslund K, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 25.European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD)European Association for the Study of Obesity (EASO) EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. Diabetologia. 2016;59:1121–1140. doi: 10.1007/s00125-016-3902-y. [DOI] [PubMed] [Google Scholar]
- 26.European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD)European Association for the Study of Obesity (EASO) EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J Hepatol. 2016;64:1388–1402. doi: 10.1016/j.jhep.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen HK, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 29.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 30.Holmes E, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 32.Besemer J. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132–e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 35.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 36.Shoaie S, et al. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab. 2015;22:320–331. doi: 10.1016/j.cmet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Spencer MD, et al. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pallister T, et al. Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-13722-4. 13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadar H, et al. A multiplexed targeted assay for high-throughput quantitative analysis of serum methylamines by ultra performance liquid chromatography coupled to high resolution mass spectrometry. Arch Biochem Biophys. 2016;597:12–20. doi: 10.1016/j.abb.2016.03.029. [DOI] [PubMed] [Google Scholar]
- 40.Plovier H, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 41.Schugar RC, et al. The TMAO-Producing Enzyme Flavin-Containing Monooxygenase 3 Regulates Obesity and the Beiging of White Adipose Tissue. Cell Rep. 2017;19:2451–2461. doi: 10.1016/j.celrep.2017.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarca AL, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidovic L, et al. A metabolomic and systems biology perspective on the brain of the fragile X syndrome mouse model. Genome Res. 2011;21:2190–2202. doi: 10.1101/gr.116764.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Martinez A, et al. MetaboSignal: a network-based approach for topological analysis of metabotype regulation via metabolic and signaling pathways. Bioinformatics. 2017;33:773–775. doi: 10.1093/bioinformatics/btw697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biddinger SB, et al. Hepatic insulin resistance directly promotes formation of cholesterol gallstones. Nat Med. 2008;14:778–782. doi: 10.1038/nm1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michael MD, et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- 47.Fujisaka S, et al. Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest. 2016;126 doi: 10.1172/JCI86674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicolas S, et al. Transfer of dysbiotic gut microbiota has beneficial effects on host liver metabolism. Mol Syst Biol. 2017;13:921. doi: 10.15252/msb.20167356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grasset E, et al. A Specific Gut Microbiota Dysbiosis of Type 2 Diabetic Mice Induces GLP-1 Resistance through an Enteric NO-Dependent and Gut-Brain Axis Mechanism. Cell Metab. 2017;25:1075–1090.e5. doi: 10.1016/j.cmet.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 50.Latorre J, et al. Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int J Obes (Lond) 2017;41:620–630. doi: 10.1038/ijo.2017.21. [DOI] [PubMed] [Google Scholar]
- 51.Cotillard A, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 52.Sharifnia T, et al. Hepatic TLR4 signaling in obese NAFLD. Am J Physiol Gastrointest Liver Physiol. 2015;309:G270–8. doi: 10.1152/ajpgi.00304.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thaiss CA, et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature. 2016;540:540–551. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 54.Zeevi D, et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 55.Boursier J, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loomba R, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017;25:1054–1062.e5. doi: 10.1016/j.cmet.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serino M, et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61:543–553. doi: 10.1136/gutjnl-2011-301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Claesson MJ, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 59.Falony G, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.