Abstract
Alopecia with mental retardation syndrome is a rare autosomal recessive disorder characterized clinically by total or partial alopecia and mental retardation. In an effort to understand the molecular bases of this form of alopecia syndrome, large Pakistani consanguineous kindred with multiple affected individuals has been ascertained from a remote region in Pakistan. Genome wide scan mapped the disease locus on chromosome 3q26.33–q27.3. A maximum two-point LOD score of 3.05 (h=0.0) was obtained at marker D3S3583. Maximum multipoint LOD score exceeding 5.0, obtained with several markers, supported the linkage. Recombination events observed in affected individuals localized the disease locus between markers D3S1232 and D3S2436, spanning 11.49-cM region on chromosome 3q26.33–q27.3. Sequence analysis of a candidate gene ETS variant gene 5 from DNA samples of two affected individuals of the family revealed no mutation.
Introduction
Genetic conditions involving total or partial absence of hair may occur alone or in association with other anomalies as a part of syndrome. Isolated forms of alopecia include congenital atrichia (MIM 203655), Marie Unna hereditary hypotrichosis (MIM 146550), hypotrichosis simplex (MIM 605389), localized hereditary hypotrichosis (LAH, MIM 607903) and autosomal recessive hereditary hypotrichosis (AH, MIM 609167). All of these conditions have been described at molecular level and two causative genes including hairless (HR, MIM 602302) for congenital atrichia (Ahmad et al. 1998) and desmoglein 4 (MIM 607892) for LAH (Kljuic et al. 2003; Rafique et al. 2003) have been cloned.
Among the syndromic forms of alopecia, major defects associated with hair loss include total or partial anodontia, hyperkeratosis, impaired sweating, dwarfism, mental retardation, epilepsy, nail dystrophy, retinas pigmentation, etc. Alopecia with mental retardation syndrome (APMR, MIM 203650) is a rare autosomal recessive disorder characterized by complete hair loss and severe mental retardation.
In the present study we have studied a consanguineous family with the autosomal recessive form of APMR from a remote area of Pakistan. Genome wide scan led to the identification of a novel locus for this form of syndrome on chromosome 3q26.33–q27.3 flanked by markers D3S1232 and D3S2436.
Materials and methods
Family history and clinical findings
A large seven-generation Pakistani family (Fig. 1), segregating an autosomal recessive form of APMR syndrome, was ascertained in which four males and three females were affected. In individuals affected with APMR syndrome, hairs were completely absent by birth from all areas of normal hair growth including scalp, eyebrows, eyelashes, axillary and pubic hair. Affected individuals showed normal hearing, teeth and nails and no abnormalities in sweating were observed. In addition, all the affected individuals were similarly severely mentally retarded and were described as difficult to handle. Neurological examination of two of the affected individuals showed no abnormality; in particular, gross motor development was normal and they never received physiotherapeutic treatment. No other clinical signs including seizures were detected.
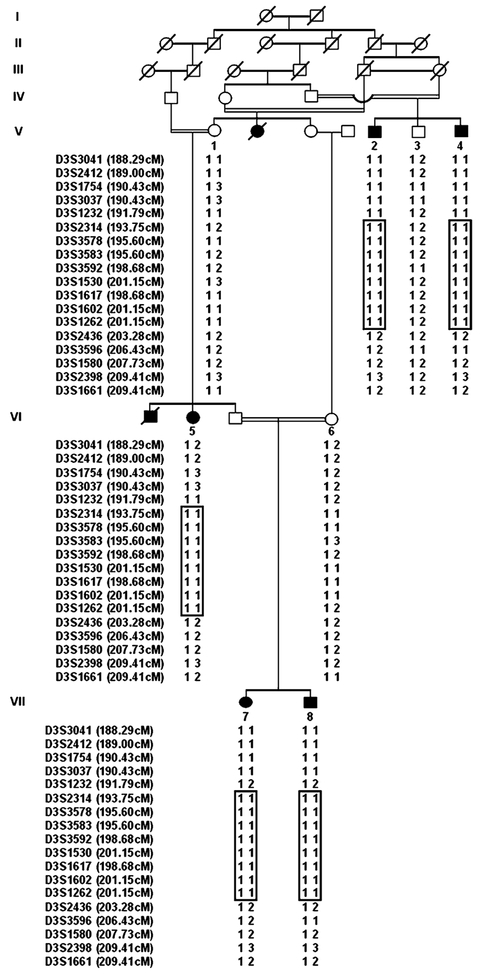
Fig. 1.
Pedigree of family with APMR syndrome over seven generations. Filled symbols represent affected subjects. Clear symbols represent unaffected individuals. The disease-associated haplotypes are shown beneath each symbol. Haplotypes, generated by SIMWALK2, are displayed in bars
Genotyping and linkage analysis
Genotyping was performed as described previously (Rafique et al. 2003). Two-point LOD score was calculated using MLINK of the FASTLINK computer package (Cottingham et al. 1993). Multipoint linkage analysis was performed using ALLEGRO (Gudbjartsson et al. 2002). For the analysis an autosomal recessive mode of inheritance with complete penetrance and a disease allele frequency of 0.001 were assumed. Equal allele frequencies were used for the markers in the analysis.
Results
After linkage to known genes involved in the related phenotype, were excluded, a genome scan was performed using the DNA samples of four affected individuals (V-5, V-7, VII-1, VII-2). In the course of screening 358 markers, spaced at approximately 10 cM intervals, from the Marshfield screening set (version 10.0), four markers (D3S1262, D4S2361, D10S564, D12S1301) were found to be homozygous in all four affected subjects and were tested further in rest of the family members. One of these markers, D3S1262, was found to be homozygous in all the five affected members of the family, indicating linkage of APMR syndrome to chromosome 3.
For fine mapping, 36 additional markers were selected from Marshfield genetic map (http://www.marshmed.org/genetics). After genotyping all the family members with these makers, the data were analyzed using two-point and multipoint linkage analysis. Table 1 summarizes the two-point LOD scores obtained for fine mapping markers. The two-point analysis generated a maximum LOD score of 3.05 (θ=0) for marker
Table 1.
Two-point logarithm of odds ratio score results between the APMR locus and chromosome 3 markers
| Marker |
LOD score at recombination fraction θ = | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 0.02 | 0.04 | 0.05 | 0.1 | 0.2 | 0.3 | |
| D3S3041 | −1.44 | 0.45 | 0.66 | 0.71 | 0.77 | 0.61 | 0.37 |
| D3S2412 | −1.44 | 0.455 | 0.66 | 0.71 | 0.77 | 0.61 | 0.37 |
| D3S1754 | −1.60 | 0.69 | 0.88 | 0.92 | 0.94 | 0.71 | 0.42 |
| D3S3037 | −1.60 | 0.69 | 0.88 | 0.92 | 0.94 | 0.71 | 0.42 |
| D3S1232 | −2.07 | 0.02 | 0.24 | 0.30 | 0.39 | 0.30 | 0.14 |
| D3S2314 | 2.01 | 1.87 | 1.75 | 1.68 | 1.37 | 0.80 | 0.37 |
| D3S3578 | 1.33 | 1.26 | 1.18 | 1.14 | 0.95 | 0.60 | 0.30 |
| D3S3583 | 3.05 | 2.87 | 2.70 | 2.62 | 2.20 | 1.41 | 0.74 |
| D3S3592 | 1.71 | 1.59 | 1.48 | 1.43 | 1.15 | 0.66 | 0.30 |
| D3S1530 | 2.50 | 2.34 | 2.18 | 2.11 | 1.73 | 1.03 | 0.48 |
| D3S1617 | 1.33 | 1.26 | 1.18 | 1.14 | 0.95 | 0.60 | 0.30 |
| D3S1602 | 1.33 | 1.26 | 1.18 | 1.14 | 0.95 | 0.60 | 0.30 |
| D3S1262 | 1.83 | 1.73 | 1.64 | 1.59 | 1.35 | 0.90 | 0.50 |
| D3S2436 | −2.48 | −0.81 | −0.53 | −0.44 | −0.20 | −0.04 | −0.01 |
| D3S3596 | −4.34 | −2.83 | −1.98 | −1.72 | −0.95 | −0.35 | −0.12 |
| D3S1580 | −2.48 | −0.81 | −0.53 | −0.44 | −0.20 | −0.04 | −0.01 |
| D3S2398 | −1.42 | 0.211 | 0.44 | 0.51 | 0.63 | 0.53 | 0.31 |
| D3S1661 | −2.56 | −0.86 | −0.57 | −0.47 | −0.20 | −0.02 | 0.00 |
D3S3583. Multipoint analysis supported linkage to this region, with maximum LOD score exceeding 5.0 at markers D3S3578, D3S3583, D3S3592, D3S1530, D3S1617 and D3S1602. The three-unit support for the APMR locus contained an 11.49-cM region which spanned from marker D3S1232 to D3S2436.
Haplotypes using SIMWALK2 were constructed to determine the critical recombination events. The disease haplotype (region of homozygosity) was the same region contained within the three-unit support interval. This region corresponds to a physical map distance of 5.4 Mb (International Human Genome Sequence Consortium 2001—http://genome.ucsc.edu/cgi-bin/hgGateway).
Discussion
In the present study we have assigned the gene for autosomal recessive alopecia and mental retardation syndrome (APMR) to chromosome 3q26.33–q27.3. Significant evidence of linkage to this chromosomal region was found with maximum two-point and multipoint LOD score exceeding 3.0 with several markers.
Previously, we have reported the identification of an AH (MIM 609167) locus on the same chromosomal region 3q26.33–q27.3 between markers D3S3730 (191.79 cM) and D3S1314 (212.61 cM) (Aslam et al. 2004). Patients in this family showed typical features of hereditary hypotrichosis with no other associated abnormality. The APMR locus reported in the present study share 11.49-cM region with AH locus. However, the clinical findings in the two families are different. Although the two genes in close proximity might cause APMR and AH, it is also possible that the two conditions are caused by different mutations in the same gene.
Through a database search we have identified a number of genes between markers D3S1232 and D3S2436, spanning 5.4-Mb region. Human ETS variant gene 5 (ETV5, MIM 601600), which is involved in the regulation of transcription, is located in this region. All 12 coding exons of ETV5 gene were sequenced in two affected individuals and an unaffected family member; however, no disease causing mutation was detected. The identification of APMR locus is the first step in identifying the underlying gene for alopecia and mental retardation.
Acknowledgments
We wish to thank the family members for their cooperation. The work presented was funded by Higher Education Commission (HEC), Islamabad, Pakistan. Peter John was supported by indigenous PhD fellowships from Higher Education Commission, Islamabad, Pakistan.
Contributor Information
Syed Muhammad S. Naqvi, Department of Biochemistry, University of Arid Agriculture, Rawalpindi, Pakistan
Suzanne M. Leal, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA
References
- Ahmad W, Faiyaz ul Haque M, Brancolini V, Tsou HC, ul Haque S, Lam H, Aita VM, Owen J, deBlaquiere M, Frank J, Cserhalmi-Friedman PB, Leask A, McGrath JA, Peacocke M, Ahmad M, Ott J, Christiano AM (1998) Alopecia universalis associated with a mutation in the human hairless gene. Science 279:720–724 [DOI] [PubMed] [Google Scholar]
- Aslam M, Chahrour MH, Razzaq A, Haque S, Yan K, Leal SM, Ahmad W (2004) A novel locus for autosomal recessive form of hypotrichosis maps to chromosome 3q26.33–q27.3. J Med Genet 41:849–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham RW Jr, Jonasson K, Frigge ML, Kong A (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2002) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Kljuic A, Bazzi H, Sundberg JP, Martinez-Mir A, O’Shaughnessy R, Mahoney MG, Levy M, Montagutelli X, Ahmad W, Aita VM, Gordon D, Uitto J, Whiting D, Ott J, Fischer S, Gilliam TC, Jahoda CA, Morris RJ, Panteleyev AA, Nguyen VT, Christiano AM (2003) Desmoglein 4 in hair follicle differentiation and epidermal adhesion: evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell 13:249–260 [DOI] [PubMed] [Google Scholar]
- Rafique MA, Ansar M, Jamal SM, Malik S, Sohail M, Faiyaz-Ul-Haque M, Haque S, Leal SM, Ahmad W (2003) A locus for hereditary hypotrichosis localized to human chromosome 18q21.1. Eur J Hum Genet 11:623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]