Abstract
Purpose
The systemic treatment of cancer is primarily through the administration of complex chemotherapy protocols. To date, this knowledge has not been systematized, because of the lack of a consistent nomenclature and the variation in which regimens are documented. For example, recording of treatment events in electronic health record notes is often through shorthand and acronyms, limiting secondary use. A standardized hierarchic ontology of cancer treatments, mapped to standard nomenclatures, would be valuable to a variety of end users.
Methods
We leveraged the knowledge contained in a large wiki of hematology/oncology drugs and treatment regimens, HemOnc.org. Through algorithmic parsing, we created a hierarchic ontology of treatment concepts in the World Wide Web Consortium Web Ontology Language. We also mapped drug names to RxNorm codes and created optional filters to restrict the ontology by disease and/or drug class.
Results
As of December 2017, the main ontology includes 30,526 axioms (eg, doxorubicin is an anthracycline), 1,196 classes (eg, regimens used in the neoadjuvant treatment of human epidermal growth factor receptor 2–positive breast cancer, nitrogen mustards), and 1,728 individual entities. More than 13,000 of the axioms are annotations including RxNorm codes, drug synonyms, literature references, and direct links to published articles.
Conclusion
This approach represents, to our knowledge, the largest effort to date to systematically categorize and relate hematology/oncology drugs and regimens. The ontology can be used to reason individual components from regimens mentioned in electronic health records (eg, R-CHOP maps to rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) and also to probabilistically reconstruct regimens from individual drug components. These capabilities may be particularly valuable in the implementation of rapid-learning health systems on the basis of real-world evidence. The derived Web Ontology Language ontology is freely available for noncommercial use through the Creative Commons 4.0 Attribution-NonCommercial-ShareAlike license.
INTRODUCTION
The field of hematology/oncology is generally acknowledged for its complexity and encompasses many disparate diseases. Over the past 70 years, a large body of knowledge has evolved around single-drug and multidrug treatment regimens. Because of the complexity of many of these regimens, clinicians rarely document specific drugs and instead use commonly understood acronyms or shorthand. Unfortunately, this hampers the secondary use of electronic health records (EHRs), such as retrospective studies on the cause of specific adverse effects of specific drugs within regimens. Although several general purpose medical extraction natural language processing algorithms currently exist (eg, cTAKES1 and MedEx,2 both introduced in 2010; MedXN,3 introduced in 2014; and CLAMP,4 introduced in 2017), they typically rely on the presence of drug names, routes, and doses in the narrative. Recently, the complexity of the field has been increasing exponentially,5 aided by exploding scientific knowledge and a concomitant rapid escalation of drug approvals. For example, a recent estimate found more than 2,000 cancer immunotherapy drugs in the development pipeline.
The National Cancer Institute (NCI) has worked for many years to compile variables to ease both clinical application and research analysis in hematology/oncology. The main product of this work, the NCI Thesaurus6 (NCIT), does have formal representations of some chemotherapy regimens (eg, the R-CHOP regimen [rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone] is represented by NCIT code C9760). The NCIT also includes terms and properties, synonym details, relationships, and mappings. However, the relationships in the NCIT are limited to the component drug names and a small number of partially contextual assertions (eg, R-CHOP is used in the treatment of diffuse large B-cell lymphoma [DLBCL], whereas a fully contextual assertion would state that R-CHOP is used with curative intent in the treatment of previously untreated DLBCL). Additional details such as similarity to other regimens, use of alternate medications (eg, prednisolone substituted for prednisone in R-CHOP7), supportive medications (eg, filgrastim in R-CHOP8), regimen relevance (eg, m-BACOD [methotrexate, bleomycin, doxorubicin, cyclophosphamide, vincristine, dexamethasone] in the treatment of untreated DLBCL9 is obsolete), and literature references are not provided. We sought to improve the status quo by creating a formal domain ontology, as defined in the field of information science,10 based primarily on HemOnc.org content. Specifically, we sought to create a self-contained information model establishing the relationships between antineoplastic drugs and regimens and the contexts in which they are used.
METHODS
Data Source
Our source, HemOnc.org, has been previously described.11 Briefly, HemOnc.org was created in 2011 and is now the largest freely available wiki Web site of drugs and regimens relevant to hematology/oncology. HemOnc.org has become an increasingly used resource, with more than 170,000 visitors from 179 countries within the past year. The site includes a large number of single- and multidrug regimens, including details concerning antineoplastic drug administration instructions, regimen variants, and associated supportive medications. All included regimens are referenced, with direct links to the original manuscript, PubMed or conference abstract, and PubMed Central version (when available). The content of HemOnc.org is the property of HemOnc.org LLC, which was cofounded by two of the authors (P.C.Y. and J.L.W.) in 2017. Although we considered using other data sources of chemotherapy regimens (eg, National Comprehensive Cancer Network or CancerTherapyAdvisor.com), these sources either are not machine readable (eg, National Comprehensive Cancer Network guidelines are PDF based) and/or are proprietary.
HemOnc.org Web Crawler for Existing Metadata
HemOnc.org is structured in MediaWiki with the semantic mediawiki extension.12 This format allows for structured categorization of pages and sustained connections via hyperlinks. This creates an implicit ontology that the project extracted and enhanced. A Web crawler was created that started at the top-level page index and traversed every page to gather data and analyze connections. The first step of this process was filtering redirect pages, which allow people to use multiple search terms to find the same page (eg, "AML" redirects to "Acute Myeloid Leukemia"). This involved tracing the page request and verifying that it was a direct connection with no redirects or diversions. The remaining pages were then run through a recursive function that traverses hyperlinks until they lead to a dead end or to a familiar page. Sequential storage of all new connections (ie, memoization13) was further used to increase efficiency and reduce repeats. The Web crawling function consisted of three sections: identification of all possible connections, categorization of those connections, and calling a function on all new connections. After identifying the categories that each concept falls under, those categories are rerun through a function to identify their lineage. The result was a general structure where the base classes and axioms (relationships) made up the ontology.
Parsing Treatment Regimens
Once the page-level structure had been identified, recursion was used again to identify pages including cancer regimens. The structure of the Web site was used to guide the program through the extraction of the following eight items: regimen names; regimen links, if part of a multipart regimen; regimen context (eg, neoadjuvant, adjuvant, salvage, first-line metastatic); regimen type (eg, chemotherapy, immunotherapy, chemoradiotherapy); drugs contained within the regimens; reference shorthand (eg, Smith et al 2010); study names if available (eg, CALGB [Cancer and Leukemia Group B] 9732,14 DSHNHL [German High-Grade Non-Hodgkin Lymphoma Study Group], RICOVER-6015); and URLs for the research article(s). Drugs were additionally assigned one of four properties (ie, antineoplastic, CNS directed, immunosuppressive, or supportive component), depending on the context in which they were used. The parsing process was simplified through the creation of a function that once given the page and expected structure around the keyword, identified the keyword by doing multiple searches that gradually identified the result. Regimens that span multiple cancer conditions and/or contexts were archived separately to gather all variants and later combined into a single concept with multiple parents.
Drug Indexing
To increase the standardization of the ontology and to allow integration with other data sources, we mapped HemOnc.org medication names to RxNorm16 codes, which are widely used by the international standards community and are required for EHR certification under the meaningful use regulations.17 RxNorm codes were added to all drugs within the ontology, when they were available. This was done through the use of the RxNorm application programming interface, which the program sent requests to for every identified drug. The program searched for the commercial name, the technical name, and the compound name if applicable; drugs that returned no match were manually reviewed.
Ontology Filtering
After the creation of the overarching ontology, it became apparent that the ontology must be filtered into manageable facets for specific research purposes, such as the incorporation of rules engines. To start the filtration process, the goal needs to be defined and vital data need to be specified. For the goals of this project, the team determined that vital information for a cancer type includes all conditions, regimens, and accompanying drugs that are contained in any of the regimens. The class structure for drugs will remain only for those drugs that are used in any regimen that is used for the specific cancer type. As a demonstration of this capability, we filtered the ontology by two restriction dimensions: restriction by drug class and restriction by disease subtype. We illustrate these filters by restricting to the class of anti-CD20 antibody-containing regimens and the disease DLBCL (described in Results).
Output and Validation
The output of the parser was stored in the World Wide Web Consortium Web Ontology Language (OWL) format.18 OWL was selected because of its widespread use in the ontology community as well as its compatibility with the Protégé ontology browser.19 Our team subsequently used Protégé (version 5.2.0) to validate and interact with the results. Axiomatic relationships between ontology concepts were visualized using the OntoGraf Protégé plugin.20
Regarding general availability, the entire OWL ontology and/or subontologies filtered by the parameters we have described are freely available to noncommercial users through the Creative Commons 4.0 Attribution-NonCommercial-ShareAlike license21; commercial uses will be considered on a case-by-case basis.
RESULTS
A total of 613 (79%) of 753 Web pages, representing 284,000 lines (27.1 million characters) of content, were parsed by the algorithm. Excluded Web pages did not have regimen or drug content (eg, general reference pages about the site). As of December 5, 2017, the main HemOnc.org ontology includes 30,526 axioms, 1,196 classes (categories of regimens, drugs, devices, and diseases), and 1,728 entities (individual regimens/drugs). It furthermore contains 4,439 drug-to-regimen relationships, of which 3,334 are antineoplastic components, 34 are immunosuppressive components, 88 are CNS therapy components, and 983 are supportive medication components. There are 402 links tying multipart regimens together (eg, cisplatin plus radiotherapy has seven possible preceding treatments, depending on the context). The ontology contains 3,790 literature references, all but nine of which have an accompanying reference URL, for a total of 7,571 literature references and URLs for regimens. Finally, it includes 338 RxNorm codes for drugs; the remainder that were not successfully mapped were manually reviewed and were either at the investigational stage or newly approved and not yet accepted by the RxNorm ontology. There are four parent classes in the HemOnc.org ontology: drug index, device index, disease index, and regimen index. As an example of the complex polyhierarchy of the ontology, categorizations of drug index and examples are listed in Table 1. Numbers of regimens categorized by the major axes regimens by class and regimens by context are listed in Tables 2 and 3.
Table 1.
Subclasses of the Drug Index Class With Representative Examples
Table 2.
Distinct Treatment Regimens by Class

Table 3.
Distinct Chemotherapy Regimens by Therapeutic Context

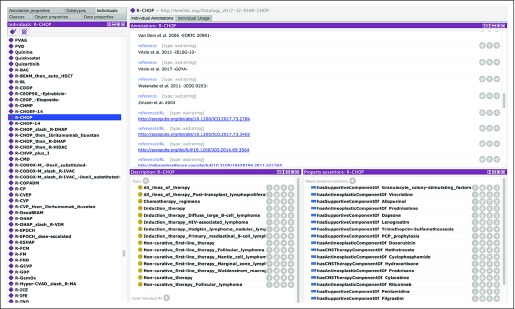
Application of the filters we have described resulted in smaller but substantial subontologies. For example, restriction to anti-CD20 antibody-containing regimens began with the eight drugs shown in Figure 1. The ontology remains substantial, because any regimen that has one or more of these eight drugs will be included, along with its context(s). For example, there are 152 regimens within the ontology that contain the drug rituximab; one of these, the R-CHOP regimen, is shown in Figure 2. A tabular representation of the drug components of the R-CHOP regimen is shown in Table 4. This regimen is found in 10 different contexts: as induction therapy for DLBCL, HIV-associated lymphoma, nodular lymphocyte-predominant Hodgkin lymphoma, and primary mediastinal B-cell lymphoma; as noncurative first-line therapy for follicular lymphoma, mantle cell lymphoma, marginal zone lymphoma, and Waldenström macroglobulinemia; as noncurative therapy (ie, administered other than in first line) for follicular lymphoma; and in all lines of therapy for post-transplantation lymphoproliferative disorder. More information on the ontology is available in the Data Supplement.
Fig 1.
Screenshot of Protégé with the class “anti-CD20 antibodies” loaded. The left panel shows the other antibody medication classes as well as several other subclasses of drug index. The bottom right panel shows the eight instances of anti-CD20 antibodies included on HemOnc.org (seven approved and one investigational).
Fig 2.
Screenshot of Protégé with the regimen concept “R-CHOP” (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) loaded. The left panel shows some other entities, in alphabetic order. The top right panel shows some of the 36 references and 36 reference URL annotations for R-CHOP. The bottom middle panel shows the 15 types to which R-CHOP belongs, which are primarily contextual. The bottom right panel shows all drugs that are found in one or more of the R-CHOP regimens, including supportive medications and intrathecal prophylaxis (eg, methotrexate).
Table 4.
Expansion of the Regimen Concept R-CHOP Into Its Component Medications
DISCUSSION
Using HemOnc.org as a fairly comprehensive source, this project works to create chemotherapy ontologies, with one broad ontology and a filtering mechanism that can focus on specific key drugs, drug classes, regimens, and regimen classes and their relation to the rest of the ontology. Therefore, the ontologies allow for the mapping of the interrelation between regimens, drugs, and general categories to contextualize hematology/oncology regimens. The information made available through the ontology may be useful to a variety of users, including practicing oncologists, trainees, creators of consensus guidelines, insurance providers, and terminology experts. In particular, the resulting ontologies could support the use of retrospective studies and data mining of real-world evidence (RWE) to identify links between drugs, drug categories, conditions, and regimens. As an illustration, the ontology can be used in two important use cases: the decomposition of regimens referred to only by acronym or reference in narrative EHR text into their component drugs and the composition of regimens from component drug references in narrative or structured EHR data. As an example of the former, a reasoning system built on the HemOnc.org ontology could take the following phrase, “The patient received R-CHOP per the RICOVER-60 protocol,” and determine that the patient received dose-dense R-CHOP with granulocyte colony-stimulating factor support (R-CHOP-14), as described by Pfreundschuh et al.15 As an example of the latter, a system could take mentions of the individual drugs gemcitabine and cisplatin from the EHR of a patient with pancreatic cancer and infer that the patient received the regimen cisplatin plus gemcitabine as first-line treatment for advanced pancreatic cancer per a small number of possible protocols.22-24
A system that can recognize drug names and regimens to this level of specificity could help make use of the abundant RWE available within EHR systems. For example, 3,952 patients at Vanderbilt University Medical Center have the terms R-CHOP, RCHOP, CHOP-R, or CHOPR present in their EHR, whereas only 1,560 (39%) of these have rituximab or the brand name Rituxan (Genentech, South San Francisco, CA) in their EHR. Although some of these may represent false positives (eg, “R-CHOP was considered but not given due to frailty”), it is highly likely that many of them are true positives; natural language processing negation algorithms such as NegEx25 could be used in conjunction with our ontology to reduce the risk of false positives. Precisely detecting regimens and their component drugs within EHRs could help physicians and researchers detect patients’ therapeutic histories. Extraction of these rich data could help with discovering potential new indications for drugs that have been used in an off-label fashion26,27 and/or have had off-target effects,28 help in the detection of drug resistance patterns across populations of patients, and associate regimens with adverse effect profiles—all focuses of US Food and Drug Administration efforts to take advantage of RWE.29 Toward this end, we are working to integrate parts of the ontology into DeepPhe, which was recently described.30 The recognition of drug regimen names could also help current patients. Clinical trials have stringent inclusion and exclusion criteria, which can include a history of exposure to one or more particular chemotherapy regimens. By better recognizing the therapies a patient has received, a system built on our ontology could help with identifying appropriate clinical trials for hematology/oncology patients at scale and could complement clinical trial matching efforts such as MatchMiner.31
An inherent challenge in creating this ontology is our ever-changing understanding of cancer and hematologic diseases. Therefore, it is important for such an ontology to be updated frequently and remain dynamic rather than static. Our ontology is highly dynamic as it draws on data from HemOnc.org, which is updated by direct contribution or indirect suggestions to contributors on a daily or weekly basis. A limitation of this approach is that HemOnc.org may have biases in content coverage and/or completeness; the user is referred to the Web site tutorial32 for a discussion of the approach to curation. Despite this limitation, it has been shown that crowd-sourced knowledge bases increase in their comprehensiveness over time, including in the medical domain.33,34 In this light, our ontology is primed to remain up to date as new relationships and concepts are introduced into the field of oncology. New concepts, relations, instances, and axioms can be integrated within the ontology as information is added to the wiki.
Being able to keep up with the rapidly growing and evolving field of hematology/oncology is quite challenging for clinicians.5 Our ontology will be able to adapt and grow as changes are made to HemOnc.org; however, keeping track of the growing amount of evidence and literature upon which the ontology is based is nearly impossible for an individual clinician. Critically, our ontology captures the evidence and support for the regimen and drug instances found within. The practice and growth of hematology/oncology relies on comparing evidence of new therapeutics with former strategies, whether directly through randomized controlled trials or through other indirect means. By tracking the references associated with particular instances, our ontology preserves the provenance of knowledge.
Our work has several limitations. In particular, we do not yet account for variants of chemotherapy regimens on the basis of dosages or substitution of similar drugs. Even a seemingly uniform concept such as R-CHOP can in fact be quite complex. As summarized in Table 4, only some of the component medications have an unambiguous route and dose. Vincristine and prednisone dosing in particular is variable across published regimens and in common practice. Nevertheless, a formal ontologic representation of this concept can enable rational decomposition from acronym representation as well as synthesis from component medications. We elected to map to RxNorm codes so as to increase the standardization of the content; however, a sizeable minority of medications did not have an assigned RxNorm code. It is possible that some of these are available in other structured databases (eg, the National Drug Code Directory35 or the National Drug File Reference Terminology)36; future work will seek to search these databases for drugs lacking RxNorm codes.
We do not yet account for most nomenclature variation. For example, the terms R-CHOP, RCHOP, CHOP-R, and CHOPR are potentially interchangeable but not formally accounted for as aliases in the present ontology. Conversely, the regimens R-CHOP and R-CHOP-14 (as described earlier in Discussion) are distinct in the ontology, as are CHOEP (etoposide plus CHOP) and EPOCH (infusional etoposide, prednisone, vincristine, cyclophosphamide, and doxorubicin), which contain the same antineoplastic drugs but have different dosing and infusion parameters. In parallel with this work, we are formulating a standardized chemotherapy regimen nomenclature, including synonyms, which can then be introduced into the ontology. We also do not account for the linkage of multipart protocols. For example, the RICOVER-60 protocol previously mentioned has a prephase of vincristine and prednisone administered for 1 week before the main R-CHOP portion; other protocols such as GIMEMA AIDA-2000 (Gruppo Italiano per le Malattie Ematologiche dell’Adulto All-Trans-Retinoic Acid and Idarubicin) for acute promyelocytic leukemia are much more complex.37 In its current state, the HemOnc.org ontology could be used to link linear protocols such as RICOVER-60 but not branching protocols such as AIDA-2000, which uses risk-adapted consolidation therapy. Future work will include formalization of the links between multipart protocols. The fact that the ontology is not yet finished, in a sense, describes its emergent (ie, bottom-up) and conceptual properties, akin to efforts such as the Systematized Nomenclature of Medicine–Clinical Terms.38,39 Whether evolution toward a realist ontology, such as the Gene Ontology,40 occurs will depend on our clinical and research use cases and those of our users.
In conclusion, we have presented an initial attempt to formalize the relationships between medications, regimens, and treatment contexts in hematology/oncology. The resultant large ontology can be used for a number of applications and will be iteratively improved over time. The product is freely available for noncommercial use and represents a substantial step forward in the formalization of hematology/oncology treatments.
ACKNOWLEDGMENT
We thank Richard Moldwin and Peter Gabriel for valuable input and critical appraisal. We also thank the DeepPhe team members, in particular Sean Finan and Guergana Savova, for their input. Finally, we thank the current licensees of the HemOnc.org ontology.
Footnotes
Supported in part by National Institutes of Health (NIH) Grants No. L30 CA171123, U24 CA184407, and U24 CA194215. This work was conducted using the Protégé resource, which is supported by Grant No. GM10331601 from the NIH National Institute of General Medical Sciences.
AUTHOR CONTRIBUTIONS
Conception and design: Andrew M. Malty, Peter C. Yang, Jeremy L. Warner
Financial support: Jeremy L. Warner
Provision of study material or patients: Jeremy L. Warner
Collection and assembly of data: Andrew M. Malty, Sandeep K. Jain, Peter C. Yang, Jeremy L. Warner
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Andrew M. Malty
No relationship to disclose
Sandeep K. Jain
No relationship to disclose
Peter C. Yang
Stock and Other Ownership Interests: Merck, Pfizer, Cyclacel
Other Relationship: HemOnc.org
Krysten Harvey
No relationship to disclose
Jeremy L. Warner
Stock and Other Ownership Interests: HemOnc.org
REFERENCES
- 1.Savova GK, Masanz JJ, Ogren PV, et al. : Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J Am Med Inform Assoc 17:507-513, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H, Stenner SP, Doan S, et al. : MedEx: A medication information extraction system for clinical narratives. J Am Med Inform Assoc 17:19-24, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohn S, Clark C, Halgrim SR, et al. : MedXN: An open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc 21:858-865, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soysal E, Wang J, Jiang M, et al. : CLAMP: A toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc [epub ahead of print on November 24, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rioth MJ, Osterman TJ, Warner JL: Advances in website information resources to aid in clinical practice. Am Soc Clin Oncol Educ Book 35:e608-e615, 2015 [DOI] [PubMed] [Google Scholar]
- 6.Sioutos N, de Coronado S, Haber MW, et al. : NCI Thesaurus: A semantic model integrating cancer-related clinical and molecular information. J Biomed Inform 40:30-43, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Fridrik MA, Jaeger U, Petzer A, et al. : Cardiotoxicity with rituximab, cyclophosphamide, non-pegylated liposomal doxorubicin, vincristine and prednisolone compared to rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisolone in frontline treatment of patients with diffuse large B-cell lymphoma: A randomised phase-III study from the Austrian Cancer Drug Therapy Working Group [Arbeitsgemeinschaft Medikamentöse Tumortherapie AGMT](NHL-14). Eur J Cancer 58:112-121, 2016 [DOI] [PubMed] [Google Scholar]
- 8.Balducci L, Al-Halawani H, Charu V, et al. : Elderly cancer patients receiving chemotherapy benefit from first-cycle pegfilgrastim. Oncologist 12:1416-1424, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Skarin AT, Canellos GP, Rosenthal DS, et al. : Improved prognosis of diffuse histiocytic and undifferentiated lymphoma by use of high dose methotrexate alternating with standard agents (M-BACOD). J Clin Oncol 1:91-98, 1983 [DOI] [PubMed] [Google Scholar]
- 10.Gruber TR: A translation approach to portable ontology specifications. Knowl Acquis 5:199-220, 1993 [Google Scholar]
- 11.Warner JL, Cowan AJ, Hall AC, et al. : HemOnc.org: A collaborative online knowledge platform for oncology professionals. J Oncol Pract 11:e336-e350, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz I, Decker S, Allemang D, et al. (eds): The Semantic Web: ISWC 2006. http://link.springer.com/10.1007/11926078
- 13.Norvig P: Techniques for automatic memoization with applications to context-free parsing. Comput Linguist 17:91-98, 1991 [Google Scholar]
- 14.Niell HB, Herndon JE, II, Miller AA, et al. : Randomized phase III intergroup trial of etoposide and cisplatin with or without paclitaxel and granulocyte colony-stimulating factor in patients with extensive-stage small-cell lung cancer: Cancer and Leukemia Group B Trial 9732. J Clin Oncol 23:3752-3759, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Pfreundschuh M, Schubert J, Ziepert M, et al. : Six versus eight cycles of bi-weekly CHOP-14 with or without rituximab in elderly patients with aggressive CD20+ B-cell lymphomas: A randomised controlled trial (RICOVER-60). Lancet Oncol 9:105-116, 2008 [DOI] [PubMed] [Google Scholar]
- 16.Nelson SJ, Zeng K, Kilbourne J, et al. : Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 18:441-448, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Office of the National Coordinator of Health Information Technology : 2015 Edition Health IT Certification Criteria final rule. https://www.healthit.gov/policy-researchers-implementers/2015-edition-final-rule
- 18.W3C : OWL 2 Web Ontology Language document overview (ed 2). https://www.w3.org/TR/owl2-overview/
- 19.Musen MA; Protégé Team : The Protégé project: A look back and a look forward. AI Matters 1:4-12, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falconer S: OntoGraf: Protege wiki. https://protegewiki.stanford.edu/wiki/OntoGraf
- 21.Creative Commons : Attribution-NonCommercial-ShareAlike 4.0 International: CC BY-NC-SA 4.0. https://creativecommons.org/licenses/by-nc-sa/4.0/legalcode
- 22.Heinemann V, Quietzsch D, Gieseler F, et al. : Randomized phase III trial of gemcitabine plus cisplatin compared with gemcitabine alone in advanced pancreatic cancer. J Clin Oncol 24:3946-3952, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Cascinu S, Berardi R, Labianca R, et al. : Cetuximab plus gemcitabine and cisplatin compared with gemcitabine and cisplatin alone in patients with advanced pancreatic cancer: A randomised, multicentre, phase II trial. Lancet Oncol 9:39-44, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Colucci G, Labianca R, Di Costanzo F, et al. : Randomized phase III trial of gemcitabine plus cisplatin compared with single-agent gemcitabine as first-line treatment of patients with advanced pancreatic cancer: The GIP-1 study. J Clin Oncol 28:1645-1651, 2010 [DOI] [PubMed] [Google Scholar]
- 25.Chapman WW, Bridewell W, Hanbury P, et al. : A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 34:301-310, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Levêque D: Off-label use of anticancer drugs. Lancet Oncol 9:1102-1107, 2008 [DOI] [PubMed] [Google Scholar]
- 27.Van Allen EM, Miyake T, Gunn N, et al. : Off-label use of rituximab in a multipayer insurance system. J Oncol Pract 7:76-79, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H, Aldrich MC, Chen Q, et al. : Validating drug repurposing signals using electronic health records: A case study of metformin associated with reduced cancer mortality. J Am Med Inform Assoc 22:179-191, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman RE, Anderson SA, Dal Pan GJ, et al. : Real-world evidence: What is it and what can it tell us? N Engl J Med 375:2293-2297, 2016 [DOI] [PubMed] [Google Scholar]
- 30.Savova GK, Tseytlin E, Finan S, et al. : DeepPhe: A natural language processing system for extracting cancer phenotypes from clinical records. Cancer Res 77:e115-e118, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsay J, Del Vecchio Fitz C, Zwiesler Z, et al. : MatchMiner: An open source computational platform for real-time matching of cancer patients to precision medicine clinical trials using genomic and clinical criteria. https://www.biorxiv.org/content/early/2017/10/23/199489
- 32.HemOnc.org : Tutorial: A hematology oncology wiki. https://hemonc.org/wiki/Tutorial
- 33.Clauson KA, Polen HH, Boulos MN, et al. : Scope, completeness, and accuracy of drug information in Wikipedia. Ann Pharmacother 42:1814-1821, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Kräenbring J, Monzon Penza T, Gutmann J, et al. : Accuracy and completeness of drug information in Wikipedia: a comparison with standard textbooks of pharmacology. PLoS One 9:e106930, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.US Food and Drug Administration : National Drug Code Directory. https://www.accessdata.fda.gov/scripts/cder/ndc/default.cfm
- 36.US National Library of Medicine : National Drug File: Reference terminology source information. https://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/NDFRT/
- 37.Lo-Coco F, Avvisati G, Vignetti M, et al. : Front-line treatment of acute promyelocytic leukemia with AIDA induction followed by risk-adapted consolidation for adults younger than 61 years: Results of the AIDA-2000 trial of the GIMEMA Group. Blood 116:3171-3179, 2010 [DOI] [PubMed] [Google Scholar]
- 38.Stearns MQ, Price C, Spackman KA, et al. : SNOMED clinical terms: overview of the development process and project status. Proc AMIA Symp 662-666, 2001 [PMC free article] [PubMed] [Google Scholar]
- 39.Schulz S, Suntisrivaraporn B, Baader F, et al. : SNOMED reaching its adolescence: Ontologists’ and logicians’ health check. Int J Med Inform 78:S86-S94, 2009 (suppl 1) [DOI] [PubMed] [Google Scholar]
- 40.Ashburner M, Ball CA, Blake JA, et al. : Gene ontology: Tool for the unification of biology. Nat Genet 25:25-29, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]