Abstract
Ascaris lumbricoides and Necator americanus are soil-transmitted parasites with global geographic distribution, and they represent some of the most common and neglected infections in the world. Periodic treatment with mass drug administration (MDA) in endemic areas is the recommended action put forth by the World Health Organization. However, MDA can cause the selection of subpopulations that possess the genetic ability to overcome the mechanism of drug action. In fact, beta-tubulin gene mutations (codons 167, 198 and 200) are correlated with benzimidazole resistance in nematodes of veterinary importance. It is possible that these SNPs also have strong correlation with treatment resistance in the human geohelminths A. lumbricoides, Trichuris trichiura and hookworms. Here, we aimed to investigate the presence of some of these canonical molecular markers associated with parasite resistance to benzimidazole in N. americanus and A. lumbricoides collected from six Brazilian states. Nested-PCR and PCR-RFLP were used to detect mutations at codons 167 and 198 in 601 individual eggs of A. lumbricoides collected from 62 human stool samples; however, no mutations were found. Codons 198 and 200 were tested in 552 N. americanus eggs collected from 48 patients using the same methodology, which presented a relative frequency of 1.4% and 1.1%, respectively. The presence of these SNPs in N. americanus eggs is an important finding, indicating that with high benzimidazole drug pressure there is potential for benzimidazole resistance to be selected in this hookworm. However, at these low frequencies it does not indicate that there is at present any benzimidazole resistance problem. This is the first systematic study performed in South America, and the study yielded a landscape of the genetic variants in the beta-tubulin gene and anthelmintic resistance to soil-transmitted parasites detected by a simple, rapid and affordable genotyping assay of individual eggs.
Author summary
The soil-transmitted helminths Ascaris lumbricoides and the hookworms (Necator americanus and Ancylostoma duodenale) are the most prevalent intestinal helminths of humans. The standard approach for geohelminth control is large-scale preventive chemotherapy predominantly using benzimidazoles through mass drug administration. Nevertheless, this inexpensive and highly effective strategy can potentially select subpopulations of parasites that become resistant to treatment. In veterinary parasites, intensive reliance on the same anthelmintics has led to the emergence of benzimidazole resistance, but this is not yet proven to helminth parasites of humans. Here we describe a PCR-RFLP approach for single nucleotide polymorphisms (SNPs) detection at codons 167 and 198 in the beta-tubulin gene of A. lumbricoides, and SNPs at codons 198 and 200 in the beta-tubulin gene of N. americanus of Brazilian populations. A. lumbricoides samples showed no mutations in any codons, and mutation at codons 198 and 200 were observed at low frequencies in N. americanus. The observed data posit important public health issues: the mutations are a substrate for selective pressure and a long-term mass drug administration could allow to the selection of parasites resistant to benzimidazole in Brazil.
Introduction
Soil-transmitted helminth infections represent important, neglected, tropical diseases that affect approximately one-fourth of the global population. Notably, 820 million people are infected with Ascaris lumbricoides, and 439 million people are infected with hookworms (Necator americanus or Ancylostoma duodenale) [1, 2]. These parasites can cause abdominal pain, colic, diarrhea, nausea, vomiting and, in more severe cases, death of the host. Hookworm infection is the most common cause of iron deficiency anemia and protein-energy malnutrition in children living in developing countries [3]. The standard approach for geohelminth control, including hookworms and A. lumbricoides, is large-scale preventive chemotherapy predominantly using benzimidazoles through mass drug administration (MDA), based on the drug’s performance in overall reductions in prevalence and reductions in the extent and severity of infection [4]. Nevertheless, this inexpensive and highly effective strategy can potentially select subpopulations of parasites that become resistant to treatment.
Single nucleotide polymorphisms (SNPs) in the beta-tubulin gene at codons 167 (TTC, TTT/Phenylalanine → TAC, TAT/Tyrosine), 198 (GAG, GAA/Glutamic acid → GCG, GCA/Alanine) and 200 (TTC/ Phenylalanine → TAC/Tyrosine) have been linked to benzimidazole resistance in several helminths [5–7]. SNPs at codons 198 [8] and 200 [9, 10] have been reported in hookworms. These common resistance-associated polymorphisms are not frequently found in those populations that did not respond to treatment [9, 11]. Conversely, a mutation at codon 167 was detected at high frequencies in A. lumbricoides samples [9], while mutations at codon 198 and 200 have never been described for this species.
Brazil is a tropical developing country with high incidence of geohelminths infection. It is estimated that 10,448,507 pre-school-age and school-age children require preventive chemotherapy [12]. MDA has been performed by the government, according to the free attendance health policy guidelines [13]. Nonetheless, there is a lack of studies that have investigated the presence of molecular markers related to parasite resistance to the available drugs. Here, we evaluate the frequency of some of the canonical codons in N. americanus and A. lumbricoides that are involved in the process of benzimidazole resistance in specimens from six Brazilian states. To our knowledge, this is the first systematic study performed in South America that examines SNPs present in soil-transmitted helminth human infections associated with benzimidazoles resistance.
Methods
Ethical considerations
This work is approved by the Comitê de Ética em Pesquisa—COEP (CAAE 61047216.7.0000.5149) from Universidade Federal de Minas Gerais (UFMG). As we had used human feces obtained from commercial laboratories performing pathology analysis, an informed consent document was not required. We did not obtain any subject identification and the data were analyzed anonymously.
Parasite material and DNA extraction
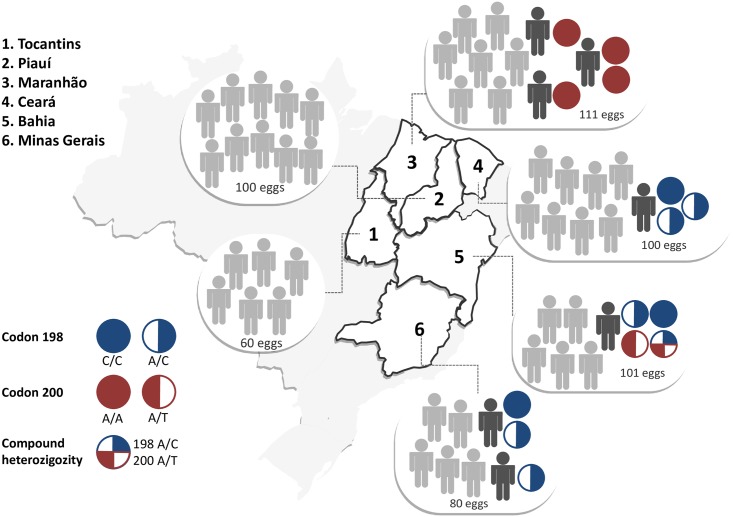
Human coproparasitological collection and screening analysis was performed in six Brazilian states. Positive samples for A. lumbricoides and N. americanus were stored in 10% formaldehyde for later molecular analysis. The initial isolation of eggs was performed according to Ritchie (1948) with modifications [14]. In summary, 2 ml of stool suspension was homogenized, filtered through gauze and transferred to a 15ml tube. Five ml of sulfuric ether was added and to the suspension and then stirred vigorously, followed by 1 minute centrifugation at 14,000 x g. The supernatant was discarded. Eggs were washed with in a new step by adding 500 μl of 1.0% and 5.0% of hypochlorite for 10 minutes to the N. americanus and A. lumbricoides samples, respectively. The material was centrifuged at 14,000 x g, and the supernatant was discarded. The eggs were washed again using 500 μl of ultrapure water, followed by centrifugation at 14,000 x g. The supernatant was then discarded. The pellet was resuspended in 100 μl of ultrapure water for N. americanus. A. lumbricoides eggs processing included the additional steps of a) incubation at 30 °C in 500 μl of 0.2 N sulphuric acid for 30 days (for larvae development), b) centrifugation at 14,000 x g and discard of the supernatant, c) washing (resuspension in 500 μl of ultrapure water, centrifugation at 14,000 x g and discard of the supernatant), d) incubation with 500 μl of 1.0% hypochlorite up to the point at which the outerlimiting membrane dissolved using a microscope for visual confirmation, e) repetition of steps a-c followed by the addition of 100 μl of ultrapure water. For DNA extraction, the eggs from both N. americanus and A. lumbricoides were observed under an optical microscope, individually pipetted into a volume of 1 μl and transferred to a 500 μl microcentrifuge tube containing 10 μl of buffer, as described by Lake and colleagues [15] and modified by Diawara and colleagues [9]. In total, 864 A. lumbricoides eggs from 64 patients and 552 N. americanus eggs from 48 patients, collected in six Brazilian states, were analyzed. None of these samples came from polyparasited patients. Table 1 shows the collection sites, as well as the number of patients and eggs of A. lumbricoides and N. americanus collected from each state.
Table 1. Collection sites, number of patients and eggs of A. lumbricoides and N. americanus used for the SNP genotyping associated with drug resistance.
| Ascaris lumbricoides | Necator americanus | |||||
|---|---|---|---|---|---|---|
| Patients | Total eggs | Eggs per patient (Minimum and maximum) | Patients | Total eggs | Eggs per patient (Minimum and maximum) | |
| Bahia | 7 | 70 | 12–16 | 6 | 101 | 11–21 |
| Ceará | 16 | 160 | 9–15 | 9 | 100 | 7–14 |
| Maranhão | 13 | 130 | 11–16 | 10 | 111 | 9–16 |
| Minas Gerais | 4 | 21 | 9–15 | 7 | 80 | 7–18 |
| Piauí | 10 | 100 | 8–15 | 10 | 100 | 5–12 |
| Tocantins | 12 | 120 | 10–14 | 6 | 60 | 7–12 |
| Total | 62 | 601 | 48 | 552 | ||
Primer design
All primers were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) based on beta-tubulin nucleotide sequences for N. americanus (EF392851.1) and A. lumbricoides (EU814697.1) from GenBank and WormBase ParaSite (N. americanus: PRJNA72135, Assembly GCA_000507365.1; A. lumbricoides: PRJEB4950, Assembly GCA_000951055.1) (Table 2).
Table 2. Primers used for analysis of mutations in the beta-tubulin gene of A. lumbricoides and N. americanus and their respective nucleotide changes (when applicable).
Positions where changes have been made are underlined.
| Species | Primer (5’- 3’) | Change | |
|---|---|---|---|
| N. americanus | Fa198/200Na | TTTCCGACACTGTGGTTGAG | |
| Fb198/200Na | **AATGCTACACTCTCTGTTCACCAGTT | ||
| Fm198Na | ACAGATGCGACCTTCTGTATT | A→C | |
| Fm200Na | ACAGATGAGACCTACTGTATT | T→A | |
| Rab198/200Na | GGGAATGGAACCATGTTGAC | ||
| A. lumbricoides | AltubF | ATGTGAGAAAATGCGGTCAT | |
| AltubR | GGTTGAGGTCTCCGTATGTG | ||
| Al167F | *GCGGTCATAGTTTTCAGGGTTT | ||
| Al198R | **CTCCGTATGTGGGATTTGTAAGC | ||
| AlMega167R | AACAACTGAGTACGAGCTCA | T→A | |
| AlMega198F | ACCGATGCAACCTTCTGCAT | A→C | |
Primers with M13 sequence forward* (CAGGAAACAGCTATGAC) and reverse** (GTAAAACGACGGCCAG) used for wider range in sequencing.
Wild-type and mutated plasmid constructs
Controls were constructed for the absence (wild-type) and presence of the mutation for each codon for each species. To construct a wild-type control allele for codons 198 and 200 (N198/200Na), PCR amplification was performed with the primers Fa198/200Na + Rab198/200Na (325 bp) and genomic DNA from N. americanus. The primers AltubF + AltubR (596 base pairs, bp) and DNA collected and pooled from A. lumbricoides eggs were used to construct the wild-type control allele for both codons 167 (N167Al) and 198 (N198Al). PCR amplifications for the wild-type controls were performed using GoTaq Green Master Mix (Promega, USA), with a final concentration of 0.2 μM for each primer, according to the following program: 95°C for 5 min, 30 cycles at 95°C for 30 s, 60°C for 45 s, 72°C for 60 s and a final step of 72°C for 8 min.
To construct the mutated controls for codons 167 (M167Al for A. lumbricoides), 198 (M198Na for N. americanus; M198Al for A. lumbricoides) and 200 (M200Na for N. americanus), site-directed mutagenesis was performed using the Megaprimer-PCR technique. First, for N. americanus the wild-type N198/200Na was used as a template for codons 198 and 200 with the primer combinations Fm198Na + Rab198/200Na (263 bp) and Fm200Na + Rab198/200Na (263 bp), respectively. For A. lumbricoides, the wild-type N167Al DNA template was used to perform the megaprimer-PCR for mutated codon 167 with the primers AlMega167R + AltubF (146 bp), while AlMega198F + AltubR was used to perform megaprimer for mutated control 198 (94 bp). The Fm198Na, Fm200Na, AlMega167R and AlMega198F primers were designed to introduce a mismatch that mimics the mutated sequence corresponding to each codon (Table 2). The site-directed mutagenesis was performed using Kapa HiFi polymerase (Kapa Biosystems, USA) following the manufacturer instructions, and a final concentration of 0.2 μM for each primer, with cycling conditions of 95°C for 3 min, 30 cycles at 98°C for 20 s, 50°C for 45 s, 72°C for 45 s and a final step of 72°C for 8 min. The reaction products were subjected to electrophoresis on 1.0% agarose gels (w/v) (Midsci, USA) stained with GelRed (Biotium, USA). The fragments were excised from the gel, purified (Illustra GFX PCR DNA and Gel Band Purification Kit, GE Healthcare, UK), and measured for concentration. Approximately 20 ng of the first reaction product was used as a megaprimer in combination with 1 μM final concentration of the primers Fa198/200Na for mutated M198Na and M200Na (respectively mutated codon 198 and 200, 325 bp each) for N. americanus, 1 μM of primer AltubR and 1 μM of AltubF for M167Al and M198Al, respectively (596 pb each), using Kapa HiFi polymerase (Kapa Biosystems, USA) according to the manufacturer instructions with cycling conditions of 95°C for 3 min, 30 cycles at 98°C for 20 s, 60°C for 45 s, 72°C for 45 s and a final step of 72°C for 8 min. The fragments were subsequently cloned using the pGEM-T Easy Vector System (Promega, USA), transformed into XL1-blue cells (Phoneutria, Brazil) and recovered via miniprep (Wizard Plus Miniprep DNA Purification System, Promega, USA). The clones were sequenced, and the absence/presence of the mutations was successfully confirmed.
Genotyping of individual samples
In silico analysis of A. lumbricoides and N. americanus beta-tubulin nucleotide sequences retrieved from GenBank and WormBase ParaSite databases were performed using the NEBcutter V2.0 tool (http://www.labtools.us/nebcutter-v2-0/) in order to search for restriction sites that could be used to distinguish between the mutated and the wild type alleles (of 198 and 200 for N. americanus; and 167 and 198 for A. lumbricoides) so that a PCR-RFLP approach could be employed. The enzymes RsaI (Promega, USA) and BmsI (Thermo Fisher Scientific, USA) were chosen to differentiate between mutated and unmutated codons 167 and 198 of A. lumbricoides. Sites for enzymes Alw26I and HpyAV (Thermo Fisher Scientific, USA) were present in the N. americanus sequence and therefore used to distinguish between mutated and unmutated alleles for the codons 198 and 200 of N. americanus, respectively. A suitable enzyme that could differentiate between mutated and unmutated codon 200 of A. lumbricoides was not found. This same situation was observed for codon 167 of N. americanus.
For N. americanus, an initial PCR was performed using one primer pair to amplify the codons 198 and 200 (Fa198/200Na + Rab198/200Na; 325 bp) using GoTaq Green Master Mix (Promega, USA), with 0.2 μM of each primer and 4.2 μl of the buffer containing the DNA of a single egg. The cycling conditions followed the same parameters as described for the wild-type control synthesis. A semi-nested PCR was performed using 1 μl from the first PCR reaction employing primers Fb198/200Na + Rab198/200Na (315 pb), using the same PCR conditions as described above. One μl of the products from the second reaction was digested at 37°C for 1 hour with 1 unit of enzyme (Alw26I and HpyAV for the codons 198 and 200) in a total volume of 15 μl. The products were subjected to electrophoresis in a 6.0% polyacrylamide gel (w/v) stained with GelRed (Biotium, USA).
For A. lumbricoides, an initial PCR was performed using one primer pair to amplify the codons 167 and 198 (AltubF + AltuR; 596 bp). One μl of the PCR product was used as a template for a nested PCR with Al167F + Al98R (608 pb). The second reaction showed a larger amplicon because the primers had M13 adapters (Table 2). The PCRs and digestions performed for A. lumbricoides were performed at the same volumes and conditions as used for N. americanus, except for the enzymes used. The enzymes RsaI and BmsI were used for digestion of codons 167 and 198, respectively. The products were subjected to electrophoresis in a 1.5% agarose gel (w/v) stained with GelRed (Biotium, USA). The expected fragment sizes after the digestion of each genotype of each codon are listed in Table 3. Samples with positive PCR-RFLP mutations were sequenced for confirmation.
Table 3. Expected sizes of DNA fragments for the three possible genotypes of each codon of the beta-tubulin gene of A. lumbricoides and N. americanus after digestion with the appropriate restriction enzymes.
| Codon | Genotype | Fragments (bp) | |
|---|---|---|---|
| A. lumbricoides | 167 | Mutated homozygous (TAC/TAC) | 404, 139, 65 |
| Unmutated homozygous (TTC/TTC) | 543, 65 | ||
| Heterozygous (TTC/TAC) | 543, 404, 139, 65 | ||
| 198 | Mutated homozygous (GCA/GCA) | 500, 108 | |
| Unmutated homozygous (GAA/GAA) | 608 | ||
| Heterozygous (GAA/GCA) | 608, 500, 108 | ||
| N. americanus | 198 | Mutated homozygous (GCG/GCG) | 315 |
| Unmutated homozygous (GAG/GAG) | 262, 53 | ||
| Heterozygous (GAG/GCG) | 315, 262, 53 | ||
| 200 | Mutated homozygous (TAC/TAC) | 315 | |
| Unmutated homozygous (TTC/TTC) | 242, 73 | ||
| Heterozygous (TTC/TAC) | 315, 242, 73 |
A negative control sample was included in all amplification runs. Extreme care was taken to avoid contamination with another sample and/or with the control plasmids. Tubes containing the control plasmids were handled in a different room where the DNA samples were manipulated, and filter tips were used in all procedures.
Fig 1 shows the methodological scheme adopted for the analysis of all the SNPs in this work.
Fig 1. Schematic representation of the PCR-RFLP methodology for single cell screening of beta-tubulin polymorphisms of A) codons 167 and 198 (A. lumbricoides) and B) codons 198 and 200 (N. americanus).
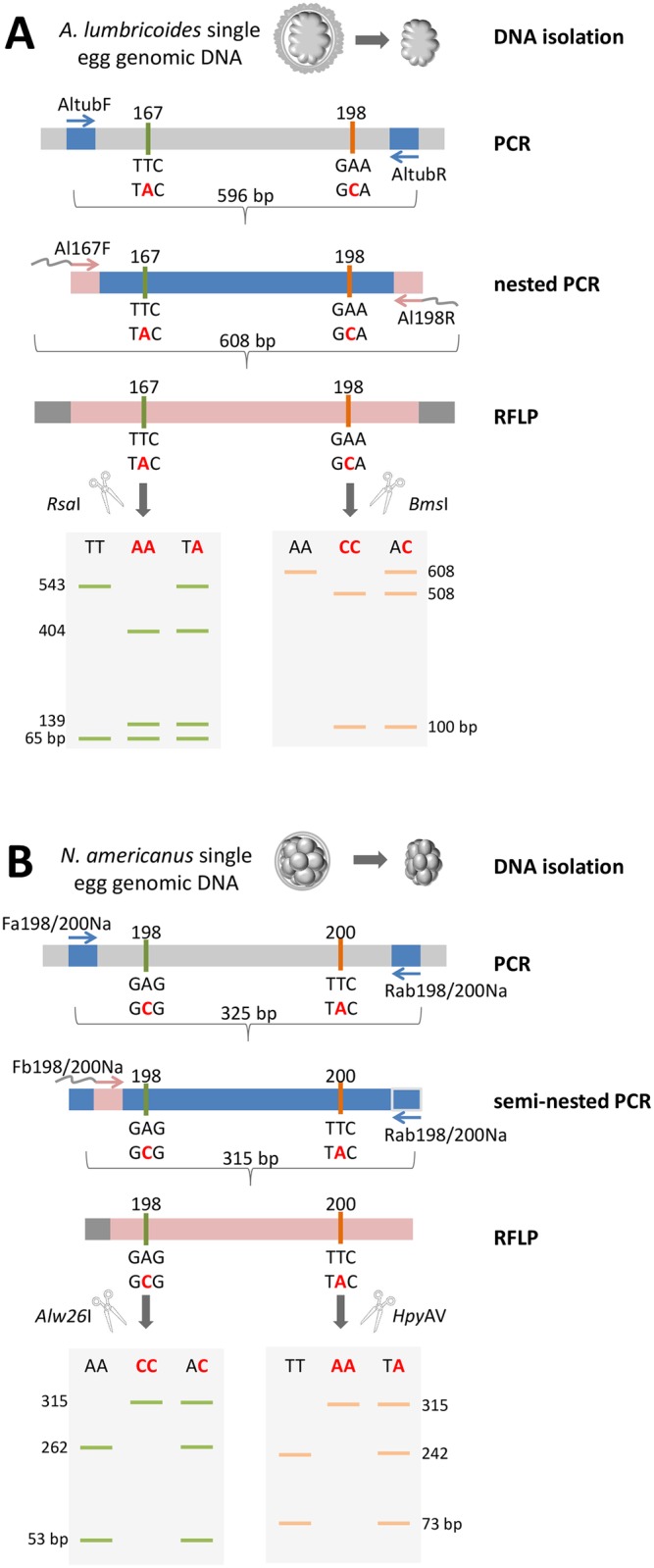
PCRs were performed using the DNA isolated from a single egg as a template, followed by a nested (for A. lumbricoides) or semi-nested PCR (N. americanus). PCR-RFLP was carried out using the selected restrictions enzymes to differ between the mutated and wild-type alleles, as observed by the different sizes of fragments detected in the gel electrophoresis. The gray tails of primers Al167F, Al198R and Fb198/200Na represents the M13 universal primers added to the sequences. The numbers in the gels represent the size of the fragments in base pairs observed after the PCR-RFLP digestion.
Results
We analyzed a total of 1153 geohelminths individual DNA samples from eggs belonging to 110 independent human feces samples collected from six Brazilian states. No mutations were detected in all 601 A. lumbricoides eggs analyzed for codons 167 and 198 of beta-tubulin gene. Mutation at codon 198 of N. americanus was observed in three localities: 1) Ceará, with 3.0% positivity (3/100): one homozygous and two heterozygous eggs from the same patient; 2) Minas Gerais, with 3.75% positivity (3/80): one patient had one heterozygous and one homozygous egg (nine eggs evaluated), and another patient had one heterozygous egg (18 eggs analyzed); and 3) Bahia, with 2.0% positivity (2/101): one patient with one homozygous and one heterozygous egg (20 eggs analyzed). Considering all the regions analyzed, 1.4% (8/552) of the eggs examined were found to have the mutation at codon 198.
The tests performed examining codon 200 of N. americanus showed the presence of the mutation in two states. In Maranhão there was 3.6% (4/111) positivity: one homozygous egg from one patient who had 12 eggs evaluated; another homozygous egg from one patient who had 11 eggs evaluated, and two homozygous eggs from one patient with 10 genotyped eggs. In Bahia there was 2% (2/101) positivity: two heterozygous eggs, one of which was also heterozygous at codon 198, from a patient with 20 eggs evaluated. Therefore, the percentage of examined eggs positive for the mutation at codon 200 was 1.1% (6/552). It is noteworthy that beta-tubulin gene from an egg (from a Bahia patient) was heterozygous for codon 198 and codon 200.
S1 Fig shows a representative image of polyacrylamide gel of the PCR-RFLP for N. americanus. S2 Fig shows a representative image of agarose gel of the PCR-RFLP for A. lumbricoides. Fig 2 illustrates the genotypes found for codons 198 and 200 in 552 N. americanus egg samples collected from six Brazilian states.
Fig 2. Spectrum of mutations of codons 198 and 200 in 552 N. americanus samples from six Brazilian regions.
Three localities had SNPs at codon 198, representing a 1.4% (8/552) positivity: 1) Ceará with one homozygous and two heterozygous eggs from a same patient (3.0% positivity); 2) Minas Gerais with two heterozygous and one homozygous egg from two patients (3.75% positivity); and 3) Bahia with one homozygous and one heterozygous egg from one patient (2.0% positivity). The overall percentage of egg positivity for the mutation at codon 198 was 1.4% (8/552). For codon 200, the total percentage of egg positivity was 1.1% (6/552), detected in Maranhão (four homozygous eggs from three patients, positivity of 3.6%) and Bahia (two heterozygous eggs from one patient 2.0% of positivity).
Discussion
Treatment with benzimidazoles is the foremost approach for soil-transmitted helminth control; however, the indiscriminate use of these drugs in a target population selects naturally resistant parasites capable of surviving exposure to the drug and can produce resistant offspring [4, 16, 17]. This selective pressure has been associated with the occurrence of SNPs in codons 167, 198 and 200 of the beta-tubulin gene of several helminths [5, 18].
We evaluated the frequency of some of these canonical SNPs in A. lumbricoides and N. americanus collected from six Brazilian states by PCR- RFLP technique. Mutations related to resistance to benzimidazoles have been detected by diverse techniques for helminths [10, 19] and fungi [20]. In the present work, a PCR- RFLP molecular screening test was performed to detect mutations in the beta-tubulin gene of A. lumbricoides and N. americanus, and controls were synthesized for absence and presence of each SNP analyzed in both species. The PCR- RFLP technique requires the creation or disruption of a restriction endonuclease’s cleavage site by the mutation. In some cases, however, commercially endonucleases are not capable to detect the nucleotide change, as observed for the codons 167 of N. americanus and 200 of A. lumbricoides. A wide range of molecular techniques may be employed for the analysis of these SNPs. Methodologies based on qPCR and sequencing have been described for analysis of SNPs in the beta-tubulin gene [19, 21]. Rashwan and colleagues [8] developed a genotyping assay of SNPs on the N. americanus beta-tubulin gene using the SmartAmp2 method. We therefore aim to overcome this limitation by analyzing these SNPs using ARMS-PCR or tetraprimer ARMS-PCR, as performed by our group for A. caninum [10, 22, 23].
None of the individual DNA samples of A. lumbricoides presented mutation at codon 167. This mutation was initially described in Haemonchus contortus and later reported in Teladorsagia circumcincta, cyathostomes, Haemonchus placei and Trichuris trichiuria [24]. Notably, a mutation at codon 167 with a distinct elevated frequency was detected in A. lumbricoides collected from different endemic areas [9]. This finding was not replicated for codon 167 [25] or for any of the three canonical codons in an expanded drug-resistant sampling in Africa [26]. Schwenkenbecher and colleagues [19] detected a very-low-frequency mutation at codon 167 and 200 in hookworms collected from children who received treatment periodically, suggesting that these data may be associated with possible qPCR experimental error. Ishii and colleagues [27] detected a high frequency (61.2%) of this mutation in cyathostomines from Paraná, Brazil. Contrary to the results for most species, polymorphisms at codon 167 were more frequent than mutations at codon 200 in cyathostomes [28].
Analysis of 110 feces samples from patients from Sri Lanka infected with N. americanus also showed positive results for the mutation at codon 198, but the frequency per helminth was not detected due to the approach of pooling larvae samples instead of searching for SNPs individually [8]. A. caninum adults and eggs collected from dogs in the United States showed no mutation at codon 198 [29]; similarly, no mutation was observed in a large number of A. caninum (327 adult worms) obtained from two different states in Brazil [10]. A lack of investigation at codon 198 of the beta-tubulin gene may lead to an underestimation of the reported frequencies. Therefore, it is likely that this mutation has not been extensively described in human hookworms because of the scarcity of available data, rather than being absent in these worms.
The mutation at codon 200 in N. americanus presented a relative frequency of 1.1% in the Brazilian samples. A frequency of 2.3% of individual eggs analyzed in this species was detected in patients from Kenya; but not detected in samples from Haiti, Panama [9] and Sri Lanka [8]. The highest frequency of this SNP was reported in 36% N. americanus samples from Haiti where patients had been submitted to MDA [30].
N. americanus Brazilian population showed homozygous and heterozygous alleles for codons 198 and 200. Curiously, one egg showed mutation in both codons demonstrating compound heterozygosity. None of the samples evaluated in this study exhibited concurrently homozygous mutations at both codons, as observed for other helminths. [31, 32].
Eighty nine eggs of N. americanus and 163 eggs of A. lumbricoides did not result in PCR amplifications (results not counted in the sampling number). Three hypotheses might may have accounted for the amplification failure: 1) another mutation (not related to resistance) at the primer’s sequence preventing its annealing and further amplification; 2) sample loss during egg transfer into the lysis buffer; and 3) damage of samples conserved with formaldehyde. The third hypothesis appears to be more likely, since the degradation of DNA in the presence of this preservative has been reported [33, 34].
The medical records of the patients used in the present study were not available, which was a limitation in our analysis. Information regarding the presence of anthelmintic treatment and the drug periodicity would potentially clarify the correlation between SNPs in beta-tubulin gene and benzimidazole resistance in those humans. The absence and low frequency of the selected mutations herein raises the hypothesis that the studied populations did not undergo a mass treatment, or if they did, the periodicity of the chemoprophylaxis was not enough to confer high levels of mutated alleles.
The PCR-RFLP approach, largely used in multi-organism DNA genotyping for decades, has proved to be an affordable, easy and high-throughput screening assay to detect mutations in a substantial number of individual helminths eggs. A. lumbricoides individual samples showed no mutations at the tested beta-tubulin codons 167 and 198. Low frequencies of mutations at codons 198 and 200 of N. americanus individual eggs were observed. In a scenario of highly indiscriminate use of albendazoles, the establishment of drug resistance in N. americanus populations might likely arise. We suggest the inclusion of a comprehensive number of samples encompassing additional geographical regions, to investigate the genetic landscape of those mutations in populations subjected to MDA. The understanding of the genetic components in the dynamics of drug resistance is essential: early monitoring increases the likelihood of delaying the establishment of resistant parasites in the populations.
Supporting information
Lanes 1 to 3 contain undigested PCR products using the plasmid controls (1: unmutated plasmid, 2: mutated plasmid, and 3: mutated and unmutated plasmid mix). Lanes 4 to 6 contain PCR digestion products using the control plasmids (4: unmutated plasmid, 5: mutated plasmid, and 6: mutated and unmutated plasmid mix). Lanes 7 to 18 contain PCR products using DNA from N. americanus. Each image is a polyacrylamide gel (6%) that was stained with GelRed (Biotium, USA). MW: 50 bp molecular weight ladder. Expected fragments sizes: codon 198—unmutated: 262 + 53 bp, mutated: 315 bp; and codon 200—unmutated: 242 + 73, mutated: 315 bp.
(TIFF)
Lanes 1 to 3 contain PCR digestion products using the control plasmids (1: mutated plasmid, 2: mutated and unmutated plasmid mix, 3: unmutated plasmid). Lanes 4 to 7 contain PCR products using DNA from A. lumbricoides. Each image is an agarose gel (1,5%) that was stained with GelRed (Biotium, USA). MW: 100 bp molecular weight ladder. Expected fragments sizes: codon 167—unmutated: 543 + 65 bp, mutated: 404 + 139 + 65 bp; codon 198—unmutated: 608 bp, mutated: 500 + 108 bp.
(TIFF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work received financial support from Pró-Reitoria de Pesquisa da Universidade Federal de Minas Gerais (PRPq; https://www.ufmg.br/prpq/; received by EMLR), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq Grant number 470968/2014-1; http://cnpq.br/; received by EMLR) and Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG; Grant number APQ-02417-16; http://www.fapemig.br/; received by EMLR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Adu-Gyasi D, Asante KP, Frempong MT, Gyasi DK, Iddrisu LF, Ankrah L, et al. Epidemiology of soil transmitted Helminth infections in the middle-belt of Ghana, Africa. Parasite Epidemiol Control. 2018;3(3): e00071 10.1016/j.parepi.2018.e00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pullan RL, Smith JL, Jasrasaria R, Brooker SJ. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit Vectors. 2014;7: 37 10.1186/1756-3305-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humphries D, Mosites E, Otchere J, Twum WA, Woo L, Jones-Sanpei H, et al. Epidemiology of hookworm infection in Kintampo North Municipality, Ghana: patterns of malaria coinfection, anemia, and albendazole treatment failure. Am J Trop Med Hyg. 2011;84(5): 792–800. 10.4269/ajtmh.2011.11-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albonico M, Savioli L. Hookworm: a neglected resurgent infection. Bmj. 2017;359: j4813 10.1136/bmj.j4813 [DOI] [PubMed] [Google Scholar]
- 5.Kwa MS, Veenstra JG, Roos MH. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol Biochem Parasitol. 1994;63(2): 299–303. [DOI] [PubMed] [Google Scholar]
- 6.Prichard R. Genetic variability following selection of Haemonchus contortus with anthelmintics. Trends Parasitol. 2001;17(9): 445–453. [DOI] [PubMed] [Google Scholar]
- 7.Ghisi M, Kaminsky R, Maser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet Parasitol. 2007;144(3–4): 313–320. 10.1016/j.vetpar.2006.10.003 [DOI] [PubMed] [Google Scholar]
- 8.Rashwan N, Bourguinat C, Keller K, Gunawardena NK, de Silva N, Prichard R. Isothermal Diagnostic Assays for Monitoring Single Nucleotide Polymorphisms in Necator americanus Associated with Benzimidazole Drug Resistance. PLoS Negl Trop Dis. 2016;10(12): e0005113 10.1371/journal.pntd.0005113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diawara A, Halpenny CM, Churcher TS, Mwandawiro C, Kihara J, Kaplan RM, et al. Association between response to albendazole treatment and beta-tubulin genotype frequencies in soil-transmitted helminths. PLoS Negl Trop Dis. 2013;7(5): e2247 10.1371/journal.pntd.0002247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furtado LF, Bello AC, dos Santos HA, Carvalho MR, Rabelo EM. First identification of the F200Y SNP in the beta-tubulin gene linked to benzimidazole resistance in Ancylostoma caninum. Vet Parasitol. 2014;206(3–4): 313–316. 10.1016/j.vetpar.2014.10.021 [DOI] [PubMed] [Google Scholar]
- 11.Albonico M, Wright V, Bickle Q. Molecular analysis of the beta-tubulin gene of human hookworms as a basis for possible benzimidazole resistance on Pemba Island. Mol Biochem Parasitol. 2004;134(2): 281–284. 10.1016/j.molbiopara.2003.12.008 [DOI] [PubMed] [Google Scholar]
- 12.WHO. Schistosomiasis and soil-transmitted helminthiases: number of people treated in 2016. Weekly epidemiological record. Department of control of neglected tropical diseases, 2017 8 December 2017. Report No. 49.
- 13.Trevisol DJ, Traebert J, Schuelter-Trevisol F. Brazil’s Family Health Strategy. N Engl J Med. 2015;373(13):1277. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie LS. An ether sedimentation technique for routine stool examinations. Bull U S Army Med Dep. 1948;8(4): 326 [PubMed] [Google Scholar]
- 15.Lake SL, Matthews JB, Kaplan RM, Hodgkinson JE. Determination of genomic DNA sequences for beta-tubulin isotype 1 from multiple species of cyathostomin and detection of resistance alleles in third-stage larvae from horses with naturally acquired infections. Parasit Vectors. 2009;2 Suppl 2: S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elard L, Sauve C, Humbert JF. Fitness of benzimidazole-resistant and -susceptible worms of Teladorsagia circumcincta, a nematode parasite of small ruminants. Parasitology. 1998;117 (Pt 6): 571–578. [DOI] [PubMed] [Google Scholar]
- 17.Prichard RK, Basanez MG, Boatin BA, McCarthy JS, Garcia HH, Yang GJ, et al. A research agenda for helminth diseases of humans: intervention for control and elimination. PLoS Negl Trop Dis. 2012;6(4): e1549 10.1371/journal.pntd.0001549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prichard RK. Markers for benzimidazole resistance in human parasitic nematodes? Parasitology. 2007;134(Pt 8): 1087–1092. 10.1017/S003118200700008X [DOI] [PubMed] [Google Scholar]
- 19.Schwenkenbecher JM, Albonico M, Bickle Q, Kaplan RM. Characterization of beta-tubulin genes in hookworms and investigation of resistance-associated mutations using real-time PCR. Mol Biochem Parasitol. 2007;156(2): 167–174. 10.1016/j.molbiopara.2007.07.019 [DOI] [PubMed] [Google Scholar]
- 20.Fan J, Luo Y, Michailides TJ, Guo L. Simultaneous quantification of alleles E198A and H6Y in the beta-tubulin gene conferring benzimidazole resistance in Monilinia fructicola using a duplex real-time (TaqMan) PCR. Pest Manag Sci. 2014;70(2): 245–251. 10.1002/ps.3549 [DOI] [PubMed] [Google Scholar]
- 21.Milhes M, Guillerm M, Robin M, Eichstadt M, Roy C, Grisez C, et al. A real-time PCR approach to identify anthelmintic-resistant nematodes in sheep farms. Parasitol Res. 2017;116(3): 909–920. 10.1007/s00436-016-5364-z [DOI] [PubMed] [Google Scholar]
- 22.Furtado LF, Rabelo EM. Molecular analysis of the F167Y SNP in the beta-tubulin gene by screening genotypes of two Ancylostoma caninum populations. Vet Parasitol. 2015;210(1–2): 114–117. 10.1016/j.vetpar.2015.03.018 [DOI] [PubMed] [Google Scholar]
- 23.Furtado LFV, Alves WP, Moreira TB, Costa LM Junior, Miranda RRC, Rabelo EML. Standardization and application of the tetraprimer ARMS-PCR technique for screening of the E198A SNP in the beta-tubulin gene of hookworm populations in Brazil. Vet Parasitol. 2016;224: 65–67. 10.1016/j.vetpar.2016.05.013 [DOI] [PubMed] [Google Scholar]
- 24.Furtado LFV, de Paiva Bello ACP, Rabelo EML. Benzimidazole resistance in helminths: From problem to diagnosis. Acta Trop. 2016;162: 95–102. 10.1016/j.actatropica.2016.06.021 [DOI] [PubMed] [Google Scholar]
- 25.Diawara A, Drake LJ, Suswillo RR, Kihara J, Bundy DA, Scott ME, et al. Assays to detect beta-tubulin codon 200 polymorphism in Trichuris trichiura and Ascaris lumbricoides. PLoS Negl Trop Dis. 2009;3(3): e397 10.1371/journal.pntd.0000397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krucken J, Fraundorfer K, Mugisha JC, Ramunke S, Sifft KC, Geus D, et al. Reduced efficacy of albendazole against Ascaris lumbricoides in Rwandan schoolchildren. Int J Parasitol Drugs Drug Resist. 2017;7(3): 262–271. 10.1016/j.ijpddr.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii JB, Arenal A, Felix A, Yoshitani U, Beech R, Molento MB. Diagnosis of resistance alleles in codon 167 of the beta-tubulin (Cya-tbb-1) gene from third-stage larvae of horse cyathostomins. Res Vet Sci. 2017;115: 92–95. 10.1016/j.rvsc.2017.01.019 [DOI] [PubMed] [Google Scholar]
- 28.Blackhall WJ, Kuzmina T, von Samson-Himmelstjerna G. Beta-Tubulin genotypes in six species of cyathostomins from anthelmintic-naive Przewalski and benzimidazole-resistant brood horses in Ukraine. Parasitol Res. 2011;109(4): 1199–1203. 10.1007/s00436-011-2426-0 [DOI] [PubMed] [Google Scholar]
- 29.Schwenkenbecher JM, Kaplan RM. Real-time PCR assays for monitoring benzimidazole resistance-associated mutations in Ancylostoma caninum. Exp Parasitol. 2009;122(1): 6–10. 10.1016/j.exppara.2009.01.006 [DOI] [PubMed] [Google Scholar]
- 30.Diawara A, Schwenkenbecher JM, Kaplan RM, Prichard RK. Molecular and biological diagnostic tests for monitoring benzimidazole resistance in human soil-transmitted helminths. Am J Trop Med Hyg. 2013;88(6): 1052–1061. 10.4269/ajtmh.12-0484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Lourdes Mottier M, Prichard RK. Genetic analysis of a relationship between macrocyclic lactone and benzimidazole anthelmintic selection on Haemonchus contortus. Pharmacogenet Genomics. 2008;18(2): 129–140. 10.1097/FPC.0b013e3282f4711d [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Gasser RB, Yang X, Yin F, Zhao G, Bao M, et al. Two benzimidazole resistance-associated SNPs in the isotype-1 beta-tubulin gene predominate in Haemonchus contortus populations from eight regions in China. Int J Parasitol Drugs Drug Resist. 2016;6(3): 199–206. 10.1016/j.ijpddr.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campos PF, Gilbert TM. DNA extraction from formalin-fixed material. Methods Mol Biol. 2012;840: 81–85. 10.1007/978-1-61779-516-9_11 [DOI] [PubMed] [Google Scholar]
- 34.Guyard A, Boyez A, Pujals A, Robe C, Tran Van Nhieu J, Allory Y, et al. DNA degrades during storage in formalin-fixed and paraffin-embedded tissue blocks. Virchows Arch. 2017;471(4): 491–500. 10.1007/s00428-017-2213-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lanes 1 to 3 contain undigested PCR products using the plasmid controls (1: unmutated plasmid, 2: mutated plasmid, and 3: mutated and unmutated plasmid mix). Lanes 4 to 6 contain PCR digestion products using the control plasmids (4: unmutated plasmid, 5: mutated plasmid, and 6: mutated and unmutated plasmid mix). Lanes 7 to 18 contain PCR products using DNA from N. americanus. Each image is a polyacrylamide gel (6%) that was stained with GelRed (Biotium, USA). MW: 50 bp molecular weight ladder. Expected fragments sizes: codon 198—unmutated: 262 + 53 bp, mutated: 315 bp; and codon 200—unmutated: 242 + 73, mutated: 315 bp.
(TIFF)
Lanes 1 to 3 contain PCR digestion products using the control plasmids (1: mutated plasmid, 2: mutated and unmutated plasmid mix, 3: unmutated plasmid). Lanes 4 to 7 contain PCR products using DNA from A. lumbricoides. Each image is an agarose gel (1,5%) that was stained with GelRed (Biotium, USA). MW: 100 bp molecular weight ladder. Expected fragments sizes: codon 167—unmutated: 543 + 65 bp, mutated: 404 + 139 + 65 bp; codon 198—unmutated: 608 bp, mutated: 500 + 108 bp.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.