Abstract
Cumulative estrogen concentration is an important determinant of the risk of developing breast cancer. Estrogen carcinogenesis is attributed to the combination of receptor-driven mitogenesis and DNA damage induced by quinonoid metabolites of estrogen. The present study was focused on developing an improved breast cancer prediction model using estrogen quinone-protein adduct concentrations. Blood samples from 152 breast cancer patients and 71 healthy women were collected, and albumin (Alb) and hemoglobin (Hb) adducts of estrogen-3,4-quinone and estrogen-2,3-quinone were extracted and evaluated as potential biomarkers of breast cancer. A multilayer perceptron (MLP) was used as the predictor model and the resultant prediction of breast cancer was more accurate than other existing detection methods. A MLP using the logarithm of the concentrations of the estrogen quinone-derived adducts (four input nodes, 10 hidden nodes, and one output node) was used to predict breast cancer risk with accuracy close to 100% and area under curve (AUC) close to one. The AUC value of one showed that both data sets were separable. We conclude that Alb and Hb adducts of estrogen quinones are promising biomarkers for the early detection of breast cancer.
Introduction
For more than half a century, early detection of breast cancer has been an important issue in cancer research. Pioneered by Egan [1] and then advanced by Wolberg & Mangasarian [2, 3], mammography is now a de facto technique for diagnosing the development of breast cancer and identifying the locations where a biopsy should be conducted. However, this technique relies heavily on visual inspection or computer aided pattern recognition techniques in order to recognize cancer cells. If cancer cells have not yet developed, diagnosis relies on biomarkers such as single nucleotide polymorphisms (SNPs) [4, 5], gene expression profiles [6], estrogen/progesterone receptors [7], DNA adducts [8], serum proteins [9, 10], and albumin and hemoglobin adducts of estrogen quinone [11, 12].
Cumulative estrogen concentration is an important determinant of the risk of developing breast cancer [13, 14]. Estrogen carcinogenesis is attributed to a combination of receptor-driven mitogenesis [15] and DNA damage induced by quinonoid metabolites of estrogen [16–19]. Metabolic activation of estrogen forms quinonoid metabolites that lead to the generation of promutagenic DNA lesions and the subsequent formation of oncogenic mutations derived from depurinating DNA adducts [20–23]. Cytochrome P450 1A1 and cytochrome P450 1B1 catalyze the oxidation of 17-estradiol (E2) to estrogen catechols including 2-hydroxyestradiol (2-OH-E2) and 4-hydroxyestradiol (4-OH-E2) [24–26]. Further conversion of 2-OH-E2 and 4-OH-E2 generates the respective estrogen quinones including estrogen-2,3-quinone (E2-2,3-Q) and estrogen-3,4-quinone (E2-3,4-Q) [27, 28]. These estrogen quinones are believed to play critical roles in the initiation of estrogen carcinogenesis [14, 18, 29].
In Taiwan, the onset of breast cancer tends to occur at a younger age than in western countries [30]. More than 50% of women diagnosed with breast cancers in Taiwan were found to be premenopausal [31] in contrast to approximately 25% of those in western populations. The incidence rate of breast cancer in Taiwanese women born after the 1960s is shifting toward that in Caucasian Americans. Environmental and dietary risk factors have been implicated in contributing to the increase in breast cancer in Taiwanese women. Development of biomarkers to identify individuals at high risk of developing breast cancer is a necessity. In our previous investigation [11, 12], we demonstrated that there are elevated levels of both the Alb and Hb adducts of E2-2,3-Q and E2-3,4-Q in breast cancer patients compared with those in healthy women. One of the unique features of this finding is that the ratio of Alb-E2-3,4-Q adducts to Alb-E2-2,3-Q adducts was 2:1 in breast cancer patients but 1:2 in healthy women, whereas the ratio of Hb-E2-3,4-Q adducts to Hb-E2-2,3-Q adducts in both breast cancer patients and healthy women was 2:1.
These recent findings suggest that Alb and Hb adducts of estrogen quinone could be used for early detection of breast cancer. A decision model could be developed based upon the concentration of the estrogen-quinone adducts. Visual inspection of scatter plots of the log-concentration values of E2-3,4-Q adducts versus E2-2,3-Q adducts of both Alb and Hb showed that the decision boundary of the model is unlikely to be a hyperplane. This suggested that a linear decision model such as logistic regression is not suitable for the decision model; instead, a non-linear decision model such as a multilayer perceptron would be more appropriate. In this report, we describe how a multilayer perceptron (MLP) with four input nodes, one hidden layer, and one output node could be applied to develop a decision model for breast cancer detection.
Methods
Data
The study population was recruited in a suburban medical center in central Taiwan. Women with breast cancer and healthy female subjects were recruited between May 2009 and May 2012. All the subjects provided sufficient venous blood for protein adduct analyses and completed questionnaires regarding age, body mass index, occupation, disease history, cigarette smoking, alcohol consumption, and dietary habits. Of those recruited, 152 breast cancer patients (BCP) and 71 healthy controls (HC) without any history of cancer were enrolled in the study. None of the enrolled individuals had a history of cancer, alcohol use, smoking, or chemotherapy. The age range of the BCP group was from 16 to 79 and the age range of the HC group was from 23 to 69. Mean age was 39.3 for HC and 50.8 for BCP. Of those recruited, 84 BCP and 58 HC were premenopausal. The Institutional Review Board of the Changhua Christian Hospital reviewed and approved this study (CCH IRB No. 081219). Each subject provided her written informed consent before participating in the study.
For each subject, the Alb adducts of E2-2,3-Q and E2-3,4-Q were analyzed from the serum following the procedure outlined in [11] and the Hb adducts of E2-2,3-Q and E2-3,4-Q were extracted from the red blood cells following the procedure outlined in [12]. All cysteinyl adducts arising from estrogen quinones were assayed using the procedure described previously [11]. Briefly, after bringing protein samples to complete dryness, estrogen quinone-derived adducts were cleaved after reaction with trifluoroacetic acid and methane sulfonic acid and analyzed via gas chromatography and mass spectrometry (S1 Data).
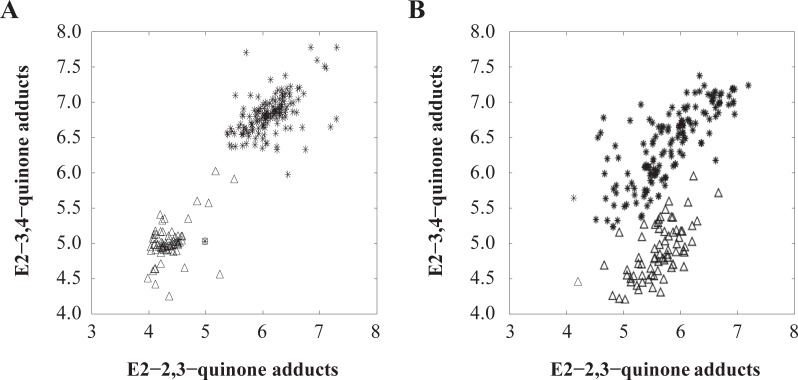
Fig 1 shows the scatter plots of the logarithm values of the E2-3,4-Q adduct concentrations plotted against those of the E2-2,3-Q adduct concentrations. Two observations are notable. First, models using Alb adducts or Hb adducts alone were unable to achieve 100% accuracy because both groups of data overlapped. Second, the concentration of Alb adducts of E2-3,4-Q was higher than that of E2-2,3-Q adducts in cancer patients with a ratio of approximately 2:1, whereas a ratio of 0.5 was observed in healthy controls, consistent with the finding in [31]. On the other hand, the levels of Hb adducts of E2-3,4-Q were higher than those of E2-2,3-Q adducts in both cancer patients and controls, with a ratio of approximately 2:1.
Fig 1. Scatter plots of the natural logarithm values of E2-3,4-Q and E2-2,3-Q adduct concentrations.
(A) Hemoglobin adducts. (B) Albumin adducts. The asterisks (*) represent the cancer patients while the triangles (Δ) represent the healthy controls.
Model
To construct the MLP model, we assumed that the training set was the set of blood samples obtained from the 223 subjects. Here, was the kth sample input and was the diagnostic result. If the kth subject had cancer, . Else, . To model the risk function, we applied a MLP with four input nodes, 10 hidden nodes, and one output node. The inputs to the MLP were the logarithm values of the following adduct concentrations.
Let be the parametric vector, where d ∈ R10 and c0 ∈ R are the weight vector and bias associated with the output node, respectively, and aj ∈ R4 and cj are the weight vector and bias associated with the jth hidden node, respectively. The output of the MLP, f (xk,w), with input xk is thus given by
| (1) |
where for j = 1,…, m.
To obtain the parametric vector w, we applied the gradient descent, minimizing the objective function given by
| (2) |
where the summation term is the mean squared error and the last term is the weight decay. The weight vector w is thus updated recursively by the following equation.
| (3) |
where μ is the step size and ∇wV (w) is the gradient vector. In our study, α = 0.0004, μ = 0.02, and the total number of updates in (3) was 50000.
After the training was completed, the MLP was used to classify the samples. Let be the class label of the kth sample.
| (4) |
where t ∈ [0, 1] is called the decision threshold. The kth subject was classified as a cancer patient if = 1. Otherwise, the kth subject was classified as healthy. The training error rate was defined as the total misclassification over the size of training samples. Clearly, with different values of t, the error rate will be different. Usually, researchers arbitrarily set this value to 0.5.
In our analysis, we set the value of t in a different way. To determine the value of t, we attempted all the possible values of t from 0, 0.01, and so on, to 1. For each value of t, the training error rate was recorded. The value of t was then set to the one with the minimum training error rate and denoted as topt. As shown in the analysis, there could have been a range of values that gave the same minimum training error rate if the MLP gave the same minimum training error rate, topt = (tmin + tmax) /2 for all t ∈ [tmin, tmax].
Results
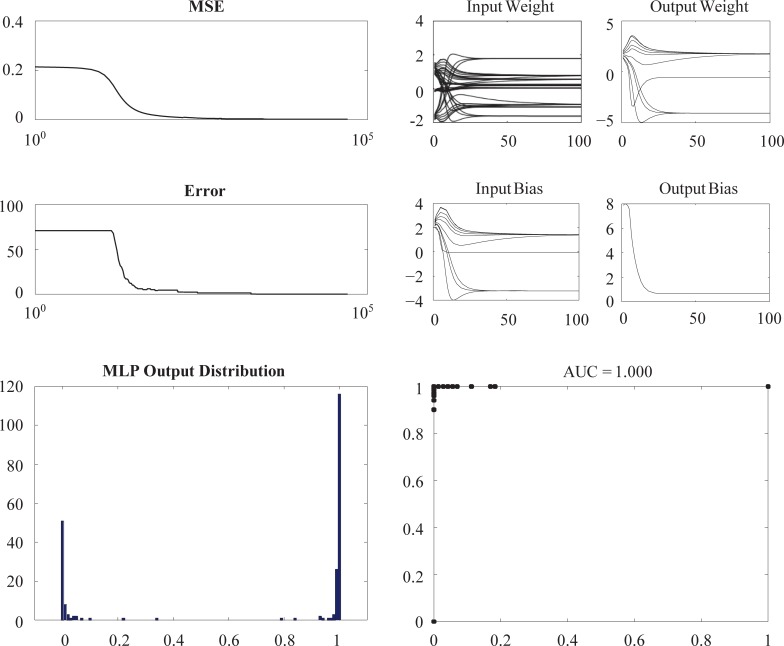
Fig 2 shows the results of a typical training that used all four adducts of estrogen quinone. The results shown in the top panels indicate that the mean square error (MSE), the error rate (i.e. misclassification rate), and the parameters converged. Both the MSE and the error rate converged to zero. The right bottom panel shows that changing the threshold value t from 0 to 1 resulted in an area under curve (AUC) value of one. This indicated that the two sets of data were indeed separable.
Fig 2. Typical training results.
Another four MLPs, each consisting of different combinations of three adducts, were generated based on the same procedure as for the MLP that used four adducts. The AUC analytical results are depicted in Table 1. For reference, the result of using all four adducts is included as Case 1. The results of Case 1, Case 2, and Case 5 revealed that HB adducts of E2-2,3-Q and Alb adducts of E2-3,4-Q are potential biomarkers for breast cancer detection. We also generated scatter plots of the logarithm values of the adduct concentrations; these demonstrated that while the two sets of data were separable, their set boundaries were very close to each other. Thus, MLPs generated using these two adducts may be sensitive to data error.
Table 1. AUC analysis for five combinations of adducts.
All 223 samples (152 BCP samples and 71 HC samples) were used to obtain the MLP for prediction.
| Case | Hemoglobin Adducts | Albumin Adducts | AUC | ||
|---|---|---|---|---|---|
| 1 | E2-3,4-Q | E2-2,3-Q | E2-3,4-Q | E2-2,3-Q | 1.000 |
| 2 | – | E2-2,3-Q | E2-3,4-Q | E2-2,3-Q | 1.000 |
| 3 | E2-3,4-Q | – | E2-3,4-Q | E2-2,3-Q | 0.999 |
| 4 | E2-3,4-Q | E2-2,3-Q | – | E2-2,3-Q | 0.999 |
| 5 | E2-3,4-Q | E2-2,3-Q | E2-3,4-Q | – | 1.000 |
To validate the models, cross-validation was applied. Samples of BCP and HC were both randomly partitioned into two sets. The training set consisted of 80 percent of the samples (121 BCP samples and 56 HC samples) and the testing set consisted of the remaining samples. The MLP was trained using only the training set.
The AUC was obtained by changing the value of t from 0 to 1. The error rate and AUC were analyzed for both the training and testing sets. The process was repeated 20 times in all five cases as depicted in Table 1. Each time, a new training set was randomly generated. Five MLPs were obtained for the corresponding cases. For each MLP, the threshold value topt was obtained via the method described earlier. The average MSE, the average AUC, and their standard deviation values (shown inside parentheses) are shown in Table 2. These results showed that using the four adducts as biomarkers yielded superior accuracy in breast cancer detection compared with the results obtained using other biomarkers (Table 3).
Table 2. Validation results.
| Case | Train Error (SD) | Test Error (SD) | Train AUC (SD) | Test AUC (SD) |
|---|---|---|---|---|
| 1 | 0 (0) | 0.0011 (0.0049) | 1 (0) | 1 (0) |
| 2 | 0 (0) | 0.0011 (0.0049) | 1 (0) | 1 (0) |
| 3 | 0 (0) | 0.0033 (0.0146) | 1 (0) | 0.9998 (0.0010) |
| 4 | 0.0059 (0.0130) | 0.0120 (0.0131) | 0.9992 (0.0010) | 0.9995 (0.0011) |
| 5 | 0.0028 (0.0045) | 0.0120 (0.0131) | 0.9999 (0.0001) | 0.9997 (0.0011) |
SD, Standard error.
Table 3. Comparisons with previously reported results.
| References | Biomarkers (No.) | Model | Test Error | AUC |
|---|---|---|---|---|
| This study | Adducts of estrogen quinone (4) | MLP | 0.0011 | 1 |
| [4] | SNP (1) | NB | 0.33 | – |
| [4] | SNP (2) | DT | 0.32 | – |
| [4] | SNP (3) | SVM | 0.31 | – |
| [9] | Serum proteins, Age, Race (100) | DF | – | 0.84 |
| [10] | Serum proteins (3) | BMA | 0.15 | 0.82 |
| [6] | Gene expression | PLSR | 0.205 | 0.88 |
| [32] | SNP from GWAS | KNN | 0.3975 | – |
| [33] | Gene expression (42) | SVM | – | 0.7879 |
| [5] | SNP (200) | SVM | 0.0395 | 0.94 |
| [34] | Mammogram | RF | 0.0838 | 0.938 |
| NC | 0.0859 | 0.962 | ||
| KNN | 0.0644 | 0.967 |
BMA, Bayesian modeling averaging; DF, Data fusion; DT, Decision tress
GWAS, Genome-wide association study; KNN, K-nearest neighbors
MLP, Multilayer perceptron; NB, Niäve Bayes; NC, Nearest centroid
PLSR, Partial least square regression; RF, Random forest; SNP, Single nucleotide polymorphism
SVM, Support vector machine.
Discussion
The high incidence rate of breast cancer in Taiwanese women emphasizes the need for better and more suitable screening and diagnostic technologies. In addition to the utility of mammography screening for early detection of breast cancer [1–3], recent studies have revealed the potential application of serum and plasma protein-based screening assays for diseases including prostate cancer, ovarian cancer, and breast cancer [10, 35, 36].
In this study, we aimed to develop a screening method with high sensitivity, specificity, and positive-predictive value for detecting breast cancer using blood protein adducts of estrogen quinones. Using MLPs, we were able to predict breast cancer risk based on the natural logarithm values of the estrogen quinone-protein adduct concentrations with an accuracy close to 100% and an AUC value close to one. The prediction results obtained using MLP with estrogen quinone-protein adducts were more accurate than those of other models [5, 6, 9, 10, 33]. In addition to the superior accuracy of our model compared with previously reported results of breast cancer prediction, the AUC value of one we obtained revealed that both data sets (cancer patients and healthy controls) were separable. Our findings strongly support the use of Alb and Hb adducts of estrogen quinone as biomarkers for early detection of breast cancer. These biomarkers can supplement the mammographic method in cases where cancer cells cannot yet be observed. However, the method presented in this investigation was developed using a retrospective study design. A prospective study using the above estrogen quinone-derived protein adducts would help validate these as biomarkers for early detection of breast cancer.
Taken together, this evidence lends further support to the idea that the cumulative concentration of estrogen quinone-protein adducts is a significant predictor of the risk of developing breast cancer. Further, the methodology developed in this study may also be applicable to other epidemiological studies and clinical trials in the prevention and early detection of breast cancer.
Supporting information
(XLSX)
Abbreviations
- Alb
Albumin
- AUC
Area under curve
- BMA
Bayesian modeling averaging
- BCP
Breast cancer patient
- DF
Data fusion
- DT
Decision tress
- E2-2,3-Q
Estrogen-2,3-quinone
- E2-3,4-Q
Estrogen-3,4-quinone
- GWAS
Genome-wide association study
- Hb
Hemoglobin
- HC
Healthy control
- KNN
K-nearest neighbors
- MLP
Multilayer perceptron
- MSE
Mean square error
- NB
Niäve Bayes
- PLSR
Partial least square regression
- SNP
Single nucleotide polymorphism
- SVM
Support vector machine
Data Availability
All relevant data are provided within the supporting information file accompanying the manuscript.
Funding Statement
This work was supported in part by the National Science Council, Taiwan, through Grants NSC98-2314-B-371-004, NSC99-2314-B-005-001, and MOST-102-2221-E-005-084. However, the National Science Council, Taiwan, had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Besides, there was no additional external funding received for this study.
References
- 1.Egan RL. MAmmography, an aid to diagnosis of breast carcinoma. JAMA. 1962;182(8):839–43. 10.1001/jama.1962.03050470017004 [DOI] [Google Scholar]
- 2.Wolberg WH, Mangasarian OL. Multisurface method of pattern separation for medical diagnosis applied to breast cytology. Proc Natl Acad Sci USA. 1990;87(23):9193–6. 10.1073/pnas.87.23.9193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangasarian OL, Street WN, Wolberg WH. Breast cancer diagnosis and prognosis via linear programming. Oper Res. 1995;43(4):570–7. 10.1287/opre.43.4.570 [DOI] [Google Scholar]
- 4.Listgarten J, Damaraju S, Poulin B, Cook L, Dufour J, Driga A, et al. Predictive models for breast cancer susceptibility from multiple single nucleotide polymorphisms. Clin Cancer Res. 2004;10(8):2725–37. 10.1158/1078-0432.ccr-1115-03 [DOI] [PubMed] [Google Scholar]
- 5.Upstill-Goddard R, Eccles D, Ennis S, Rafiq S, Tapper W, Fliege J, et al. Support vector machine classifier for estrogen receptor positive and negative early-onset breast cancer. PLoS ONE. 2013;8(7):e68606 10.1371/journal.pone.0068606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aaroe J, Lindahl T, Dumeaux V, Saebo S, Tobin D, Hagen N, et al. Gene expression profiling of peripheral blood cells for early detection of breast cancer. Breast Cancer Res. 2010;12(1):R7 10.1186/bcr2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jerez-Aragones JM, Gomez-Ruiz JA, Ramos-Jimenez G, Munoz-Perez J, Alba-Conejo E. A combined neural network and decision trees model for prognosis of breast cancer relapse. Artif Intell Med. 2003;27(1):45–63. 10.1016/S0933-3657(02)00086-6 [DOI] [PubMed] [Google Scholar]
- 8.Rogan EG, Cavalieri EL. Estrogen metabolites, conjugates, and DNA adducts: possible biomarkers for risk of breast, prostate, and other human cancers. Adv Clin Chem. 2004;38:135–49. 10.1016/S0065-2423(04)38005-4 [DOI] [PubMed] [Google Scholar]
- 9.Jesneck JL, Mukherjee S, Nolte LW, Lokshin AE, Marks JR, Lo J, editors. Decision fusion of circulating markers for breast cancer detection in premenopausal women. Bioinformatics and Bioengineering, 2007 BIBE 2007 Proceedings of the 7th IEEE International Conference on; 2007 Oct. 14–17, 2007.
- 10.Jesneck JL, Mukherjee S, Yurkovetsky Z, Clyde M, Marks JR, Lokshin AE, et al. Do serum biomarkers really measure breast cancer? BMC Cancer. 2009;9:164 10.1186/1471-2407-9-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin C, Chen DR, Hsieh WC, Yu WF, Lin CC, Ko MH, et al. Investigation of the cumulative body burden of estrogen-3,4-quinone in breast cancer patients and controls using albumin adducts as biomarkers. Toxicol Lett. 2013;218(3):194–9. 10.1016/j.toxlet.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 12.Lin C, Hsieh W-C, Chen D-R, Kuo S-J, Yu W-F, Hu S-W, et al. Hemoglobin adducts as biomarkers of estrogen homeostasis: Elevation of estrogenquinones as a risk factor for developing breast cancer in Taiwanese Women. Toxicol Lett. 2014;225(3):386–91. 10.1016/j.toxlet.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 13.Colditz GA. Relationship between estrogen levels, use of hormone replacement therapy, and breast cancer. J Natl Cancer Inst. 1998;90(11):814–23. 10.1093/jnci/90.11.814 [DOI] [PubMed] [Google Scholar]
- 14.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354(3):270–82. 10.1056/NEJMra050776 [DOI] [PubMed] [Google Scholar]
- 15.Feigelson HS, Henderson BE. Estrogens and breast cancer. Carcinogenesis. 1996;17(11):2279–84. 10.1093/carcin/17.11.2279 [DOI] [PubMed] [Google Scholar]
- 16.Yager JD. Chapter 3: Endogenous estrogens as carcinogens through metabolic activation. J Natl Cancer Inst Monogr. 2000;2000(27):67–73. 10.1093/oxfordjournals.jncimonographs.a024245 [DOI] [PubMed] [Google Scholar]
- 17.Lavigne JA, Goodman JE, Fonong T, Odwin S, He P, Roberts DW, et al. The effects of catechol-o-methyltransferase inhibition on estrogen metabolite and oxidative DNA damage levels in estradiol-treated MCF-7 Cells. Cancer Res. 2001;61(20):7488–94. [PubMed] [Google Scholar]
- 18.Bolton JL, Thatcher GRJ. Potential mechanisms of estrogen quinone carcinogenesis. Chem Res Toxicol. 2008;21(1):93–101. 10.1021/tx700191p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavalieri E, Frenkel K, Liehr JG, Rogan E, Roy D. Chapter 4: Estrogens as Endogenous Genotoxic Agents—DNA Adducts and Mutations. J Natl Cancer Inst Monogr. 2000;2000(27):75–94. 10.1093/oxfordjournals.jncimonographs.a024247 [DOI] [PubMed] [Google Scholar]
- 20.Cavalieri EL, Stack DE, Devanesan PD, Todorovic R, Dwivedy I, Higginbotham S, et al. Molecular origin of cancer: catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc Natl Acad Sci USA. 1997;94(20):10937–42. 10.1073/pnas.94.20.10937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavalieri EL, Devanesan P, Bosland MC, Badawi AF, Rogan EG. Catechol estrogen metabolites and conjugates in different regions of the prostate of Noble rats treated with 4-hydroxyestradiol: implications for estrogen-induced initiation of prostate cancer. Carcinogenesis. 2002;23(2):329–33. 10.1093/carcin/23.2.329 [DOI] [PubMed] [Google Scholar]
- 22.Convert O, Van Aerden C, Debrauwer L, Rathahao E, Molines H, Fournier F, et al. Reactions of estradiol-2,3-quinone with deoxyribonucleosides: possible insights in the reactivity of estrogen quinones with DNA. Chem Res Toxicol. 2002;15(5):754–64. 10.1021/tx015561y [DOI] [PubMed] [Google Scholar]
- 23.Zahid M, Kohli E, Saeed M, Rogan E, Cavalieri E. The greater reactivity of estradiol-3,4-quinone vs estradiol-2,3-quinone with DNA in the formation of depurinating adducts: implications for tumor-initiating activity. Chem Res Toxicol. 2006;19(1):164–72. 10.1021/tx050229y [DOI] [PubMed] [Google Scholar]
- 24.Martucci CP, Fishman J. P450 enzymes of estrogen metabolism. Pharmacol Ther. 1993;57(2–3):237–57. 10.1016/0163-7258(93)90057-K [DOI] [PubMed] [Google Scholar]
- 25.Hayes CL, Spink DC, Spink BC, Cao JQ, Walker NJ, Sutter TR. 17 beta-estradiol hydroxylation catalyzed by human cytochrome P450 1B1. Proc Natl Acad Sci USA. 1996;93(18):9776–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spink DC, Spink BC, Cao JQ, Gierthy JF, Hayes CL, Li Y, et al. Induction of cytochrome P450 1B1 and catechol estrogen metabolism in ACHN human renal adenocarcinoma cells. J Steroid Biochem Mol Biol. 1997;62(2–3):223–32. 10.1016/S0960-0760(97)00024-1 [DOI] [PubMed] [Google Scholar]
- 27.Butterworth M, Lau SS, Monks TJ. 17β-estradiol metabolism by hamster hepatic microsomes: Comparison of catechol estrogen o-methylation with catechol estrogen oxidation and glutathione conjugation. Chem Res Toxicol. 1996;9(4):793–9. 10.1021/tx9501952 [DOI] [PubMed] [Google Scholar]
- 28.Cao K, Stack DE, Ramanathan R, Gross ML, Rogan EG, Cavalieri EL. Synthesis and structure elucidation of estrogen quinones conjugated with cysteine, N-acetylcysteine, and glutathione. Chem Res Toxicol. 1998;11(8):909–16. 10.1021/tx9702291 [DOI] [PubMed] [Google Scholar]
- 29.Parl FF, Dawling S, Roodi N, Crooke PS. Estrogen metabolism and breast cancer: a risk model. Ann N Y Acad Sci. 2009;1155:68–75. 10.1111/j.1749-6632.2008.03676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng SH, Tsou MH, Liu MC, Jian JJ, Cheng JC, Leu SY, et al. Unique features of breast cancer in Taiwan. Breast Cancer Res Treat. 2000;63(3):213–23. 10.1023/A:1006468514396 [DOI] [PubMed] [Google Scholar]
- 31.Huang C-S, Lin C-H, Lu Y-S, Shen C-Y. Unique features of breast cancer in Asian women—breast cancer in Taiwan as an example. The Journal of steroid biochemistry and molecular biology. 2010;118(4–5):300–3. 10.1016/j.jsbmb.2009.12.017 [DOI] [PubMed] [Google Scholar]
- 32.Hajiloo M, Damavandi B, Hooshsadat M, Sangi F, Mackey JR, Cass CE, et al. Breast cancer prediction using genome wide single nucleotide polymorphism data. BMC Bioinformatics. 2013;14(Suppl 13):S3 10.1186/1471-2105-14-s13-s3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F, Deng Y, Drabier R. Multiple biomarker panels for early detection of breast cancer in peripheral blood. Biomed Res Int. 2013;2013:781618 10.1155/2013/781618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galvan-Tejada CE, Zanella-Calzada LA, Galvan-Tejada JI, Celaya-Padilla JM, Gamboa-Rosales H, Garza-Veloz I, et al. Multivariate Feature Selection of Image Descriptors Data for Breast Cancer with Computer-Assisted Diagnosis. Diagnostics (Basel, Switzerland). 2017;7(1):9 10.3390/diagnostics7010009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polascik TJ, Oesterling JE, Partin AW. Prostate specific antigen: a decade of discovery—what we have learned and where we are going. J Urol. 1999;162(2):293–306. 10.1016/S0022-5347(05)68543-6 [DOI] [PubMed] [Google Scholar]
- 36.Gorelik E, Landsittel DP, Marrangoni AM, Modugno F, Velikokhatnaya L, Winans MT, et al. Multiplexed immunobead-based cytokine profiling for early detection of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(4):981–7. 10.1158/1055-9965.epi-04-0404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are provided within the supporting information file accompanying the manuscript.