Abstract
Introduction
Exosomes, nanosized extracellular vesicles, are known to circulate through the blood stream to transfer molecular signals from tissue to tissue.
Methods
To determine whether exosomes affect aging in animals, we primarily identified the changes in exosomal miRNA contents during the aging process. In exosomes from 12-month-old mice, mmu-miR-126-5p and mmu-miR-466c-5p levels were decreased and mmu-miR-184-3p and mmu-miR-200b-5p levels were increased significantly compared with those of 3-month-old mice. Their levels in exosomes were partially correlated with those in tissues: levels of only mmu-miR-126-5p and mmu-miR-466c-5p in lungs and/or liver were decreased, but those of mmu-miR-184-3p and mmu-miR-200b-5p in tissues did not coincide with those of exosomes.
Results and discussion
In the aged tissues injected with young exosomes isolated from serum, mmu-miR-126b-5p levels were reversed in the lungs and liver. Expression changes in aging-associated molecules in young exosome-injected mice were obvious: p16Ink4A, MTOR, and IGF1R were significantly downregulated in the lungs and/or liver of old mice. In addition, telomerase-related genes such as Men1, Mre11a, Tep1, Terf2, Tert, and Tnks were significantly upregulated in the liver of old mice after injection of young exosomes.
Conclusion
These results indicate that exosomes from young mice could reverse the expression pattern of aging-associated molecules in aged mice. Eventually, exosomes may be used as a novel approach for the treatment and diagnosis of aging animals.
Keywords: exosome, injection, reverse aging, telomerase, biomarker, molecular therapy
Introduction
Understanding the regulatory mechanisms and the involved molecules underlying aging has aroused interest to prevent or delay aging or aging-associated diseases.1,2 It has been reported that the upregulated or downregulated miRNAs induce cellular senescence. Downregulated miR-24 in diploid fibroblasts,3 upregulated miR-203 in melanoma cells,4 and downregulated miRNA group, miR-26b, miR-181a, miR-201, and miR-424, in mammary epithelial cells5 are the examples of miRNAs inducing senescence of cells.
In cell-to-cell signaling in systemic aging, miRNAs are reported to be released in circulation and transferred to remote tissues. The released miRNAs can affect their levels in circulation in aged individuals,6 and in a recent study, they served as regulatory molecules to control aging speed.7 Therefore, they are strongly considered as aging-associated biomarkers, possibly determined by minimally invasive or noninvasive methods.8,9 So far, several studies comparing miRNA expression profiles from the blood of young and old animals have revealed differences in the expression levels of several miRNAs with aging: miR-17-5p, miR-16-5p, miR-21a-5p, and miR-34a in mice9,10 and let-7 and miR-34a in humans.11,12
One of the ways by which miRNAs are released in circulation is via vesicles blebbed out from cellular membranes. A representative type of these vesicles is exosomes, which are tens to hundreds of nanometers in diameter. The exosomes released from parent cells enter systemic circulation, which thus explains the signaling process among remote tissues.13–15 Cells under stress would release more exosomes in vitro to dispose unnecessary molecules or communicate their signals to the surrounding cells.16 Actually, aging is a type of cellular stress; thus, exosomes are secreted at higher levels from senescent cells than from normal cells.
It has been reported on cell-to-cell signaling via exosomes to explain systemic aging as a whole-body phenomenon.16–18 miRNAs are also contained in exosomes and released to various body fluids such as the saliva, synovial fluid, and blood; therefore, these miRNAs displaying changes in expression levels can be considered aging or aging-associated disease biomarkers. Indeed, upregulated miR-24-3p in exosomes was proposed as an aging biomarker in human saliva.19 In patients with osteoarthritis, expression levels of miR-16-2-3p, miR-6821-5p, miR-26a-5p, and miR-146a-5p in women and miR-201-5p and miR-6878-3p in men were altered in synovial exosomes in a sex-specific manner.20 In Alzheimer’s disease, induced miR-342-3p and miR-139b expression was considered a probable biomarker in plasma exosomes.21,22
However, limited information is available on changes in the miRNA contents of exosomes in naturally aged individuals and their effects in the aging process. Therefore, the identification of miRNA molecules deregulated in exosomes in the aging process would be required to understand the mechanisms underlying aging and may have potential applications in evaluating or reversing the aging status of an individual.
In this study, we primarily identified differentially expressed miRNAs (DERs) in exosomes from aged mice and compared them with those from young mice. In addition, we determined whether the exosomal level of DERs coincided with their levels in tissues, particularly in the lungs and liver. If the miRNAs in exosomes have regulatory capability in systemic aging, their increased levels in young exosomes were expected to exert a reversing effect on tissues of old mice. Therefore, after intravenously injecting exosomes from young mice to aged mice, changes in aging-associated molecule levels were analyzed in aged mice. To support the direct/indirect relationship of reverse-aging with injected young exosomes, we determined the expression level of previously reported molecular biomarkers for aging, such as p16Ink4A and TOR.23,24 IGF1R is involved in metabolic changes in the aging process, and upregulation of IGF1R was reported;25,26 thus, we also analyzed its expression level following the injection of young exosomes to old mice.
As another biomarker of aging, the expression of telomerase-associated genes could be considered based on previous studies. The absence of telomerase activity in most human somatic cells resulted in telomere shortening during aging.27 In addition, the telomerase expression in human bone marrow stromal cells prevented senescence-associated impairments,28 and transfection of a telomerase gene to old mice delayed aging and increased longevity.29 In this study, we analyzed the expression of telomerase gene expression as a feasible indicator of reverse-aging. The aim of this study is to identify aging-associated molecules in exosomes and whether they have an ability to induce reverse-aging in animals.
Materials and methods
Animals
Male C57BL/6 mice aged 3 weeks to 23 months were purchased from Central Lab Animal Inc. (Seoul, Korea). The animals were housed after purchase at the DGIST Animal Laboratory in accordance with the Institutional Animal Care Guidelines. The Animal Care and Use Committee of DGIST approved all experimental protocols to be applied to the mice. After 1 month of acclimatization, animals were either euthanized or used for exosome injection. In all cases, tissues (blood, liver, and lung) were sampled immediately.
Isolation of exosomes
Whole blood was sampled by retro-orbital bleeding, according to the previous protocols.30 Blood samples were collected in commercially available tubes with EDTA (BD Microtainer, BD, Franklin Lakes, NJ, USA). After collection of the whole blood samples, they were allowed to clot by leaving them at room temperature for 30 minutes. The clots were removed by centrifugation for 10 minutes at 1,000×g using a refrigerated centrifuge. The samples were maintained at −20°C until the exosome isolation was performed.
Exosomes were isolated by ExoQuick™ Exosome Precipitation Solution (System Biosciences, Inc., Palo Alto, CA, USA) in accordance with the manufacturer’s instructions. Briefly, 200 µL of serum was collected and centrifuged at 3,000×g for 15 minutes to eliminate cells and cell debris. The supernatant was transferred to a microtube, and an appropriate volume of precipitation solution was added. The mixture was incubated at 4°C for 30 minutes, and then centrifuged at 1,500×g for 30 minutes. The exosome pellet was resuspended in 100–500 µL of PBS.
Characterization of exosomes
The isolated exosomes were observed through transmission electron microscopy (TEM, Tecnai™ G2 Spirit, FEI Company, Hillsboro, OR, USA) to identify their morphology and the extent of dispersion. Prior to TEM observation, the samples were stained with methanolic uranyl acetate and lead citrate.31 The size distribution of exosomes was analyzed through dynamic light scattering (DLS; ZetaSizer Nano ZS, Malvern Instruments, Malvern, UK). The enrichment of exosomes was confirmed by the Western blotting analysis of exosome markers, such as CD63, CD9, and Flotillin-1.
Screening of aging-associated miRNAs in exosomes
For high-throughput screening of DERs in exosomes of aged mice, next-generation sequencing (NGS) was performed with small RNAs isolated from the exosomes of 3-, 8-, and 12-month-old mice (n=3). Total RNA was isolated from exosomes using the Hybride-R RNA Kit (GeneAll®, Seoul, Korea), in accordance with the manufacturer’s instructions. RNA integrity was confirmed with a bioanalyzer, using an Agilent RNA 6000 Pico Kit (Agilent Technologies, Santa Clara, CA, USA) with an RNA integrity number value >8 as a cutoff value. The isolated total RNA (50 ng) was processed for preparing a small RNA sequencing library using the TruSeq Small RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) in accordance with the manufacturer’s instruction. The total RNA sample ligated RNA 3′ and 5′ adapter with T4 RNA ligase (Promega, Madison, WI, USA) in the ATP-free buffer. The ligated RNAs were reverse transcribed to single-stranded cDNAs, using SuperScript II Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) with reverse transcription primers recommended by Illumina. The cDNAs were amplified in 11 cycles of PCR, using Illumina’s Primer Set. PCR products were run on a 12% tris-borate-EDTA (TBE) polyacrylamide gel, and a slice of gel containing fragments of 145–160 nt was excised. This fraction was eluted and the recovered cDNAs were precipitated, and quantification of library was performed through RT-PCR analysis, using a CFX96 real-time system (Bio-Rad Laboratories Inc., Hercules, CA, USA). The libraries were pooled in equimolar amounts and loaded on the flow cell of a HiSeq 2000 sequencing system (Illumina). Sequencing was performed to generate 1×50 bp length read.
Quantification of miRNA expression in exosomes
The amount of miRNAs in exosomes is usually not high; thus, miRNA levels in exosomes were validated with QX200 droplet digital PCR (ddPCR, Bio-Rad Laboratories Inc.). Total RNA was isolated from tissues using the Hybride-R RNA Kit (GeneAll), according to the manufacturer’s instruction. cDNA for miRNA analysis was synthesized from 5 ng of total RNA using the Universal cDNA Synthesis Kit II (Exiqon, Vedbaek, Denmark). PCR was performed in a 20 µL volume containing 10 µL 1× ddPCR EvaGreen Supermix, 2 µL cDNA, 350 nM primer sets, and deionized water to fill the remaining volume. miRNA LNA™ PCR primer sets (Qiagen, Hilden, Germany) were used for the detection of target miRNAs. 5S rRNA was used as an internal standard to determine the relative expression of target miRNAs. The prepared ddPCR assay mixture was loaded into a QX200 disposable droplet generator cartridge according to the manufacturer’s instruction. Briefly, 70 µL of Droplet Generation Oil (Bio-Rad Laboratories Inc.) for probes was loaded into each well. The cartridge was then placed inside the QX200 droplet generator. When droplet generation was complete, the droplets were transferred to a 96-well PCR plate by using a multichannel pipet (INTEGRA Biosciences, Zizers, Switzerland). The plate was heat-sealed with foil and placed in a conventional thermal cycler. Thermal cycling conditions for EvaGreen assays were as follows: 95°C for 10 minutes, 40 cycles of 95°C for 30 seconds, and 58°C for 1 minute (ramping rate to 2°C/s), and three final steps at 4°C for 5 minutes, 90°C for 5 minutes, and a 4°C indefinite hold to enhance stabilization.
Quantification of miRNA expression in tissues
The amount of miRNAs in tissues was determined by quantitative real-time PCR. Total RNA was isolated from tissues using the Hybride-R RNA Kit (GeneAll), according to the manufacturer’s instruction. cDNA for miRNA analysis was synthesized from 5 ng of total RNA using the Universal cDNA Synthesis Kit II (Exiqon). For the determination of miRNA expression by quantitative real-time PCR, the miRCURY LNA Universal RT micro RNA PCR LNA PCR primer sets for mmu-miR-126-5p, mmu-miR-466c-5p, mmu-miR-184-3p, and mmu-miR-200b-5p (Exiqon) and the SYBR Green PCR Kit (Exiqon) in an ABI7900HT Real-Time PCR System (Thermo Fisher Scientific) was used. The expression of miRNAs was determined by comparison of 5S rRNA expression in each sample.
Injection of young exosomes into old mice
Before investigating the effect of exosome injection from young to old mice, exosomes isolated from whole serum of a 3-month-old mouse was stained with exosome-specific dye (ExoGlow™-RNA, System Biosciences, Inc.) according to the manufacturer’s instruction, and then the exosome distribution in whole body was observed following the injection of the stained exosomes intravenously using an in vivo imaging system (IVIS® Spectrum, PerkinElmer Inc., Waltham, MA, USA; excitation/emission wavelength = 485 nm/538 nm). After 72 hours from exosome injection, lungs and liver tissues were dissected and observed by confocal microscopy (FV1200, Olympus Corporation, Tokyo, Japan).
To investigate the effect of young exosomes to old mice, exosomes from 4-week-old C57BL/6N male mice were injected intravenously in 18-month-old male mice (n=3). Exosomes for injecting into an aged mouse were prepared from 400 µL serum each (almost all serum from a 4-week-old mouse was used to inject into an aged mouse). As a control experiment (n=3), PBS was injected with the same volume of the exosome sample injected (averaging 100 µL per injection). After 24 hours from exosome injection, animals were euthanized, and the lungs and liver were sampled.
Western blotting analysis for aging-related molecules
Proteins were extracted from the lungs and liver of mice using extraction buffer (50 mM Tris–HCl, pH 8, 150 mM NaCl, 1% NP-40, 0.1% SDS, and 1 mM phenylmethylsulfonyl fluoride). After incubation on ice for 15 minutes, tissue extracts were centrifuged at 10,000×g for 30 minutes, and the supernatant was collected for analysis. Protein concentrations were determined using a BCA Protein Assay Kit (Bio-Rad Laboratories Inc.). Proteins (30 µg) were separated via SDS-PAGE on a 10% resolving gel and electroblotted onto nitrocellulose membranes using a transfer apparatus (Trans-Blot SD semi-dry transfer cell, Bio-Rad Laboratories Inc.) in accordance with the manufacturer’s instructions. Anti-MTOR, -p16Ink4A, and -IGF1R antibodies were obtained from Abcam, Inc. (Cambridge, MA, USA). Secondary antibodies conjugated with horseradish peroxidase were used, and the substrate for enhanced chemiluminescence (ECL) was purchased from Thermo Fisher Scientific. The intensity of protein bands was quantified by ImageJ 1.X developed by the National Institutes of Health.32 The expression level of proteins was determined in comparison with that of ACTB and GAPDH.
Analysis of telomerase-related gene expression
Total RNA from tissues was isolated using TaKaRa MiniBEST Universal RNA Extraction Kit (Takara Bio Inc., Shiga, Japan) in accordance with the manufacturer’s instructions. Single-strand cDNA was synthesized from 1 µg total RNA, using PrimeScript™ first strand cDNA Synthesis Kit (Takara Bio Inc.). For the determination of mRNA expression, RT-PCR was performed using gene-specific primer pairs specified in previous report10 and the Roche SYBR-Green® master mix in an ABI7900HT Real-Time PCR System (Thermo Fisher Scientific). The relative expression level of genes was determined in comparison with that of β-actin.
Statistical analysis
For all experiments, data from three independent experiments were analyzed using Student’s t-test and are reported as mean±SD. SigmaPlot version 12.3 was used (Systat Software, Inc., Chicago, IL, USA) to determine the P-values. A P-value <0.05 was considered statistically significant.
Results
Characterization of exosomes from mouse serum
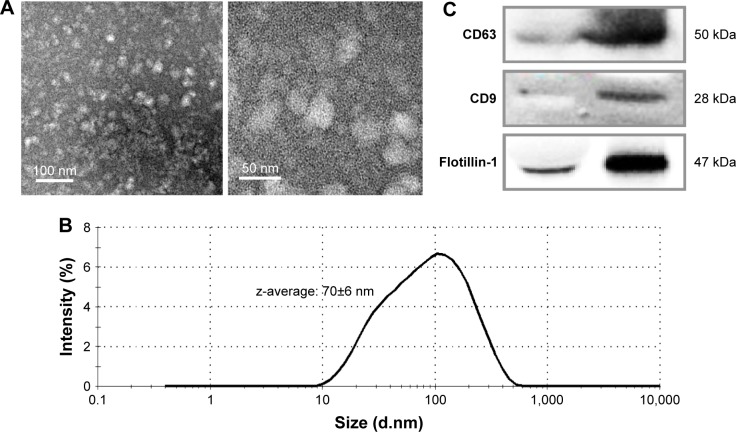
The isolated exosomes were visualized through TEM, after negative staining. Figure 1A shows that vesicles <100 nm in diameter were isolated by the methods used in this study. The size distribution was determined through DLS, which indicates that the average exosome size (z-average) in the main peak was 70±6 nm (average±SD, Figure 1B).
Figure 1.
Characterization of the isolated exosomes by transmission electron microscopy (A), dynamic light scattering (B), and Western blotting for exosome-specific biomarkers (C).
To confirm the enrichment of exosomes by the isolation procedure, the representative proteins expressed on exosomes were analyzed by Western blotting (Figure 1C). The enrichment of CD63, CD9, and Flotillin-1 was clearly enhanced in the isolated exosomes in comparison with those in the same amount of serum proteins.
Aging-associated miRNAs in exosomes
miRNA contents of exosomes isolated from 200 µL of serum of 3-, 8-, and 12-month-old mice (n=3) were analyzed using NGS analysis. The total number of miRNAs determined in exosomal fraction was 1,033. Among these miRNAs, the selection criteria for DERs were as follows: 1) continuously upregulated or downregulated in 8- and 12-month-old mice compared with that in 3- and 8-month-old mice, respectively, in >0.5 of log2 (Ratio); 2) statistically significant changes in expression, ie, P<0.05. Table 1 shows the list of DERs that satisfy both aforementioned criteria. In total, five miRNAs were selected as DERs: mmu-miR-126b-5p and mmu-miR-466c-5p were downregulated and mmu-miR-184-3p, mmu-miR-200b-5p, and mmu-miR-708-5p were upregulated depending on the age of the mice.
Table 1.
miRNA DERs in exosomes
| miRNA | 3 vs 8 months
|
8 vs 12 months
|
||
|---|---|---|---|---|
| Log2 (ratio) | P-value | Log2 (ratio) | P-value | |
| mmu-miR-126b-5p | −0.55 | 0.024 | −0.67 | 0.024 |
| mmu-miR-466c-5p | −0.58 | 0.045 | −1.92 | 0.0079 |
| mmu-miR-184-3p | 0.83 | 0.042 | 3.74 | 7.5×10−5 |
| mmu-miR-200b-5p | 0.86 | 0.038 | 1.49 | 0.030 |
| mmu-miR-708-5p | 4.78 | 0.028 | 3.20 | 0.018 |
Abbreviation: DERs, differentially expressed miRNAs.
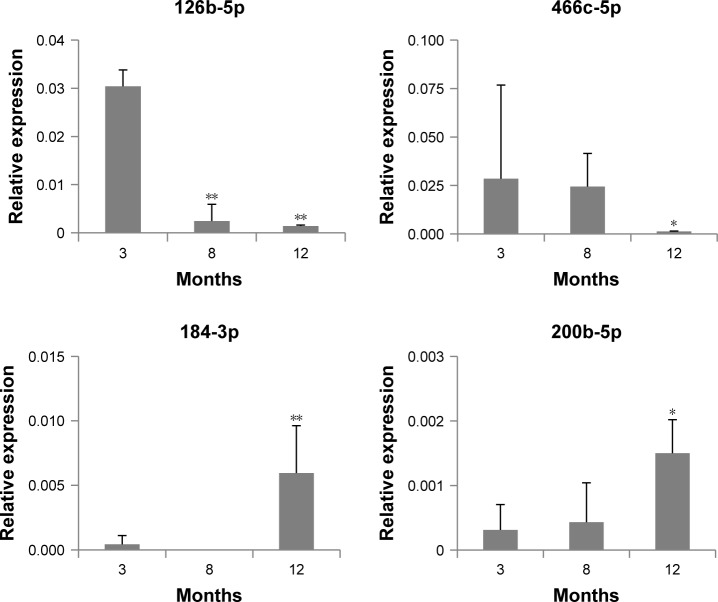
Expression changes in each miRNA in exosomes were confirmed by ddPCR analysis. Figure 2 shows the expression level of four DERs in 3-, 8-, and 12-month-old mice. In this figure, it is observed that mmu-miR-126b-5p was downregulated significantly at 8 and 12 months compared with that at 3 months. mmu-miR-466c-5p showed a trend for downregulation, and the change in 12-month-old mice was significant compared with that in 8-month-old mice. mmu-miR-184-3p and mmu-miR-200b-5p showed a strong trend for upregulation, and their expression in 12-month-old mice was significantly upregulated compared with that in 3-month-old mice. The results indicated that the expression level of selected DERs was closely related to the age of mice. In case of mmu-miR-708-5p, the expression did not change significantly in accordance with age (data not shown).
Figure 2.
Expression of miRNAs determined as differentially expressed miRNAs in exosomes, using droplet digital PCR.
Notes: The relative expression of miRNAs at each age was determined in comparison with that of 5S rRNA. The statistical significance of expression changes at 8 and 12 months of 126b-5p, 184-3p, and 200b-5p were determined by comparison with those of 3 months, but the significance for miR-466c-5p was compared to that of 8 months. *P<0.05; **P<0.01.
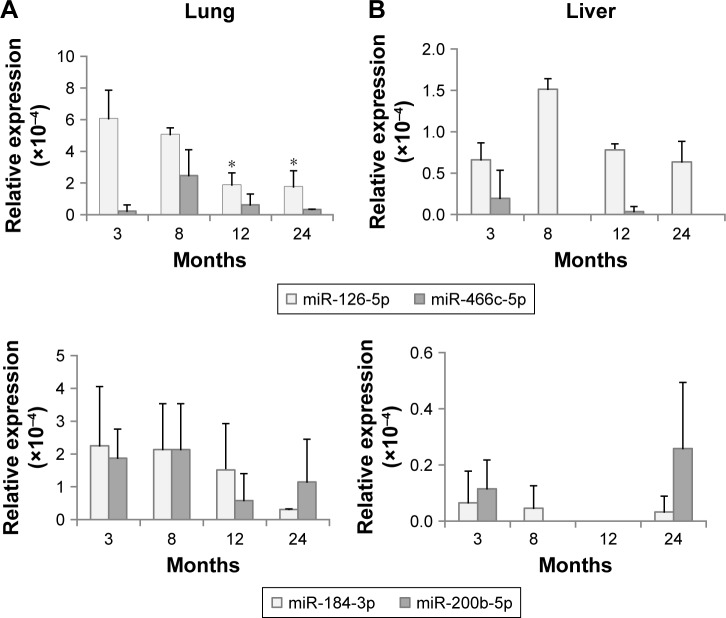
Tissue levels of aging-related miRNAs from exosomes
Until now, it is not clearly defined whether exosomal levels of miRNAs are closely linked to those of tissues. Figure 3 shows the expression of DERs in the lungs and liver. mmu-miR-126b-5p was downregulated in exosomes on aging (Figure 2) and also showed a decreasing trend in the lungs (Figure 3A). The expression of mmu-miR-126b-5p in 12- and 24-month-old mice was significantly decreased compared with that in 3-month-old mice (P<0.05). In liver, the decreased expression of mmu-miR-126b-5p was significantly observed in 12- and 24-month-old mice only when compared with that in 8-month-old mice (Figure 3B). The other downregulated miRNA in exosomes, mmu-miR-466c-5p, was not significantly downregulated in both lungs and liver (Figure 3A and B). However, downregulation of mmu-miR-466c-5p in lungs was obvious in 12- and 24-month-old mice compared with that in 8-month-old mice.
Figure 3.
Expression of differentially expressed miRNAs in tissues, determined using droplet digital PCR.
Notes: (A) Expression in the lungs; (B) Expression in the liver. The statistical significance was determined by comparison with the expression at 3 months. *P<0.05.
mmu-miR-184-3p and mmu-miR-200b-5p were DERs that were upregulated with age in exosomes; however, their levels in the lungs and liver were not significantly changed. These results implied that the exosomal levels of miRNAs did not stringently match with those of individual tissues. The correlation of the miRNA expression levels in exosomes and tissues should be determined by separate tests for each miRNA.
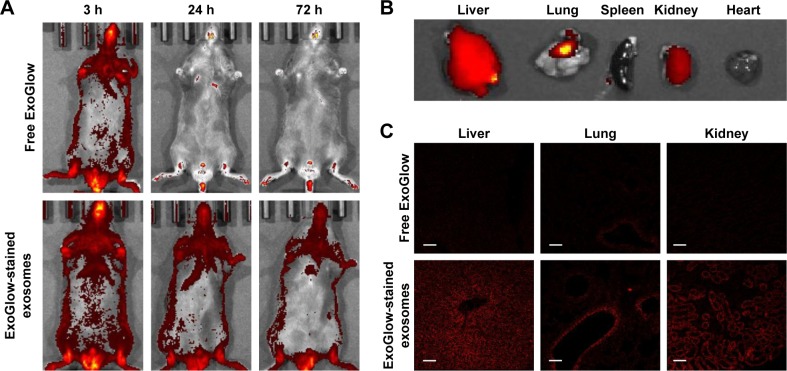
miRNA levels changed by injection of exosomes
Figure 4A shows that the isolated exosomes stained with RNA-specific dye ExoGlow were clearly observed by fluorescent emission at a wavelength of 538 nm, after intravenously injecting them into another mouse. The amount of exosomes stained with ExoGlow was time-dependently reduced but remained until 72 hours postinjection. However, the free ExoGlow dye almost disappeared at 24 hours as observed in the whole-body imaging using IVIS system (Figure 4A). The ex vivo image (Figure 4B) shows that exosomes were transferred to liver and lungs much higher than spleen, kidney, and heart. In the tissue sections of the sacrificed mice at 72 hours, the exosomes were also observed clearly in the liver, lung, and kidney, but free ExoGlow dye was not observed in those tissues (Figure 4C). These results indicated that exosomes isolated from the serum of a mouse can be successfully transferred to tissues of another mouse by intravenous injection.
Figure 4.
Distribution of exosomes after injection of exogenous exosomes labeled with an exosome-specific dye (ExoGlow™-RNA, System Biosciences, Inc.).
Notes: (A) Whole-body imaging for 3 days after exosome administration; (B) ex vivo imaging of exosome distribution in mouse tissues; (C) distribution of exosomes in the section of liver, lungs, and kidney observed using a confocal microscope.
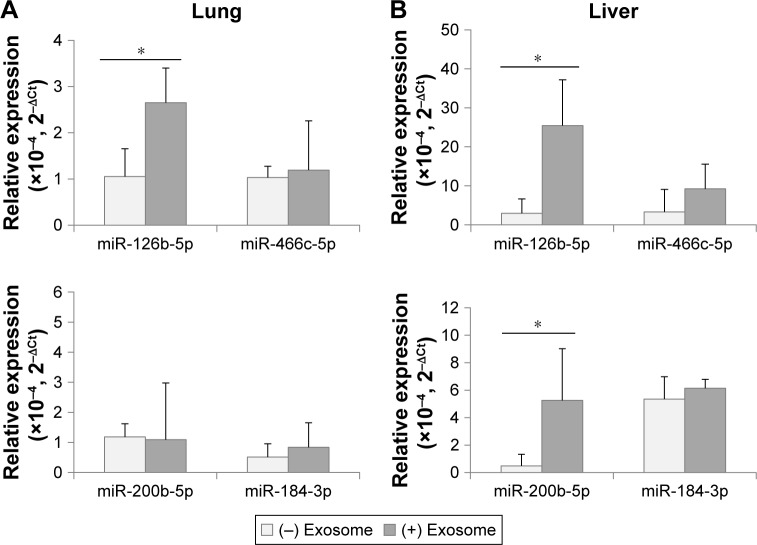
Following the injection of exosomes from 3-month-old mice, the DER levels were determined through ddPCR analysis in 18-month-old recipient mice. In Figure 5, mmu-miR-126-5p, which was determined to be downregulated with aging in exosomes and tissues, was significantly upregulated in the lungs and liver of young exosome-injected old mice. However, another downregulated DER, mmu-miR-466c-5p, did not significantly change after exosome injection. This result interpreted that the amount of mmu-miR-466c-5p in young exosome was not high enough to elevate its tissue level in aged mice or there was another regulatory process to determine it.
Figure 5.
Expression of differentially expressed miRNAs in lungs (A) and liver (B) after the injection of exosomes from 3-month-old to 18-month-old mice.
Note: *P<0.05.
Because the expression level of mmu-miR-200b-5p and mmu-miR-184-3p in tissues did not coincide with that of exosomes in the aging process (Figures 3 and 4), it was not expected that their tissue levels in age mice could be affected by the injection of young exosomes. As expected, mmu-miR-200b-5p in the lungs and mmu-miR-184-3p in both the lungs and liver did not significantly change by injection of young exosomes. The exception was mmu-miR-200b-5p in the liver, which significantly changed.
Reverse expression of aging-related molecules in the lungs and liver
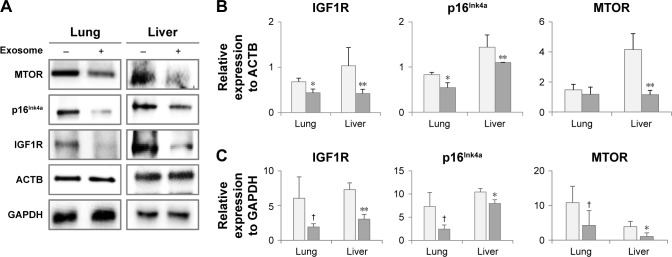
The injection of young exosomes in old mice was performed to examine the possibility of the ability of exosomes to alter the expression of aging-associated signaling molecules in tissues. As aging-related signaling molecules, the expression of p16Ink4A, MTOR, and IGF1R was analyzed by Western blotting and compared with that of ACTB and GAPDH as shown in Figure 6A. Compared with ACTB expression (Figure 6B), IGF1R and p16Ink4A were significantly down-regulated in both the lungs and liver of aged mice after injection of exosomes from young mice, while MTOR was significantly downregulated only in the liver. In lungs, the average levels of MTOR were decreased, but not significant. However, IGF1R, p16Ink4A, and MTOR in liver were significantly downregulated compared with GAPDH as an internal standard (Figure 6C). In lungs, the expression changes of three molecules were not significant, but showed strong trend of downregulation (P<0.10).
Figure 6.
Expression of aging-associated biomarkers following the injection of exosomes from 3-month-old to 18-month-old mice.
Notes: Following the analysis of protein expression by Western blotting (A), relative quantity of protein expression was determined by comparison with ACTB expression (B) and GAPDH expression (C). †P<0.1; *P<0.05; **P<0.01.
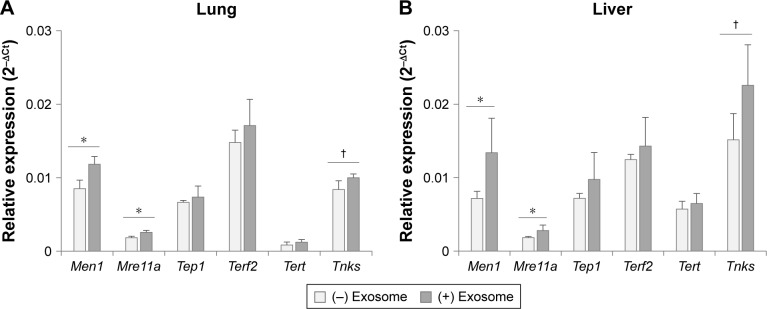
As other aging biomarkers, telomerase-related proteins were also analyzed in the tissues of injected mice. As shown in Figure 7, among the six genes involved in the telomerase complex (Men1, Mre11a, Tep1, Terf2, Tert, and Tnks), two genes (Men1 and Mre11a) were reversed significantly in the lungs and liver of aged mice by injection of young exosomes. In case of Tnks, the expression was statistically nonsignificant but showed a distinct trend toward significance (P=0.051 and 0.078 in lungs and liver, respectively).
Figure 7.
Expression of telomerase-related genes in lungs (A) and liver (B) after the injection of exosomes from 3-month-old to 18-month-old mice.
Notes: †P<0.1; *P<0.05.
Overall, the reversed expression of aging-related molecules was observed in aged mice after injection of young exosomes, although there were some differences based on the type of tissues and molecules.
Discussion
Because exosomes are endogenous vesicles, they have unique advantages as a source of miRNA biomarkers: lengthening the half-life of miRNAs in circulation by protection from a vesicular membrane and harboring enhanced miRNAs as signaling molecules compared with the parent cells.10,33,34 The target organs that analyze the effect of young exosomes in this study were liver and lungs because they were supposed to respond primarily for the changes in blood circulation. It was supported by the transfection of ExoGlow-stained exosomes, which showed stronger fluorescence intensity in liver and lung than other tissues such as spleen, kidney, and heart (Figure 4B).
Consequently, downregulated mmu-miR-126b-5p and mmu-miR-466c-5p and upregulated mmu-miR-184-3p, mmu-miR-200b-5p, and mmu-miR-708-5p were considered candidate exosomal biomarkers in aged mice (Table 1). Expression changes in those miRNAs, observed on NGS analysis, were validated through ddPCR analysis, but four of them, except mmu-miR-708-5p, were clearly confirmed to be regulated in an age-dependent manner (Figure 2).
Downregulation of mmu-miR-126a-5p in mice is reported to be associated with experimental autoimmune encephalomyelitis,35 and hsa-miR-126-5p in human (same sequence to mmu-miR-126a-5p, CAUUAUUACUUUUG GUACGCG) is known to be a tumor suppressor.36–38 However, the sequence of mmu-miR-126b-5p (AUUAUUACU CACGGUACGAGUU) has been determined only in mice;39 however, its function has not been reported. Regarding other DERs, mmu-miR-466c-5p was downregulated and mmu-miR-200b-5p and mmu-miR-184-3p were upregulated in exosomes, and their sequences were found to be unique in mice.40
The role of mmu-miR-466c-5p in aging has not been reported; however, mmu-miR-466c-5p was reported to be regulated by IGF1R and IR in a mouse preadipocyte cell line.41 In the previous study, it was reported that mutant mouse Igf1r and Insr genes (knockout) downregulated mmu-miR-466c-5p mature miRNA. In our study, the IGF1R was downregulated in 18-month-old mice after injection of the exosomes from 3-month-old mice (Figure 6), but no significant changes were observed in mmu-miR-466c-5p expression levels in the tissues of aged mice (Figure 5). There was a possibility that although mmu-miR-466c-5p was increased in aged tissue by the injection young exosomes with upregu-lated mmu-miR-466c-5p, it was compensated owing to the downregulated IGF1R in aged tissues simultaneously.
Molecular changes in tissues of aged mice injected with exosomes from young mice were analyzed especially for aging-associated biomarkers, such as p16Ink4A and MTOR. p16Ink4A and MTOR are known to be involved in signaling in the aging process and are upregulated with aging.23,24 In our study, p16Ink4A and MTOR were downregulated in the lungs and liver (except for MTOR in the lungs) after injection of young exosomes. This result could be designated as a critical response to exosomal injection from young to old mice, which reversed the expression pattern of the representative aging-associated molecules.
IGF1R is another aging-associated molecule, which is involved in metabolic changes in the aging process, and upregulation of IGF1R is a part of aging-related signaling.25,26 In the current study, after the injection of young exosomes to old mice, IGF1R was significantly downregulated in the lungs and liver (Figure 6), which could be interpreted as the reversed expression of IGF1R by injection of young exosomes.
However, expression changes in mmu-miR-200b-5p and mmu-miR-184-3p in tissues after injection of exosomes were not clearly observed. This result implied that the addition of exogenous young exosomes with lower miRNA levels compared with those of existing exosomes in the recipient aged mice could not affect the tissue levels of miRNAs in them. The increased level of mmu-miR-200b-5p in the liver of injected mice seems to be the result of indirect regulation of its expression. However, it might be also related to the enhanced response of reversed expression of aging-associated molecules in liver, such as MTOR (Figure 6) and telomerase-related genes (Figure 7).
Taken together, these results supported the possible role of exosomes in cell-to-cell signaling in the aging process at the whole-body level. Obviously, the effect of exosome injection in old mice would be systemic response and not limited to a change in single miRNA expression. Therefore, mmu-miR-126b-5p was only a representative molecule in this study. The result on mmu-miR-126b-5p indicated that exosome injection could result in molecular transfer from exogenous exosomes to the recipient tissues, even though the other DER levels in exosomes did not accurately coincide with those of tissues. It was revealed from this study that mmu-miR-126b-5p and mmu-miR-466c-5, which were identified in tissues with extremely low expression levels, were more clearly detected in exosomes. Using the exosomes, induction of reverse-aging would be attainable via circulatory system, wherein young exosomes could be transported to each tissue in old mice.
We propose that young exosomes could cause reverse- aging of old mice, but it can be said that there are still limitations in this study. Even though we verify a functional importance of exosomes in aging process of animal body, the detailed molecular mechanism in exosome-to-cell or cell-to exosomes signaling could not be explained. Until now, it has not been clear whether the molecular expression levels are totally synchronous between tissues and exosomes in the same body. The expression level of 184-3p and 200b-5p in exosomes was increased with aging (Figure 2); however, there were no significant changes in their expression level in tissues (Figure 3). This result made it difficult to understand whether the controlled transfer of molecules between exo-some-to-tissue occurred in aging. Another limitation of this study is that the results are just an acute response of young exosomes injected to old mice; thus, the effect of chronic transfection by young exosomes should be investigated further to identify the physiological response in old mice.
Conclusion
The effect of exosome injection can be considered a systemic process caused by highly regulated contents of exosomes. To our knowledge, exosomal miRNAs as aging-associated molecules in mice are reported for the first time in this study. Further investigation on signaling pathways altered by exosome injection is required to explain the mechanism of reversed expression of aging biomarkers. The novel approach presented here involves the use of exosomes carrying exogenous genetic information as a potential clinical tool for more effective treatment of aging-associated diseases.
Acknowledgments
This study was supported in part by the Ministry of Science and ICT (2016R1A2B4014728) and DGIST Basic Research Fund (18-BT-01, 18-BT-02, 18-LC-01, and 18-BD-06). We would like to express our thanks to Dr Hasan Al Faruque for his support to confirm the exosome distribution in vivo.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Baker DJ, Wijshake T, Tchkonia T, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gladyshev TV, Gladyshev VN. A disease or not a disease? Aging as a pathology. Trends Mol Med. 2016;22(12):995–996. doi: 10.1016/j.molmed.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lal A, Kim HH, Abdelmohsen K, et al. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3(3):e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noguchi S, Mori T, Otsuka Y, et al. Anti-oncogenic microRNA-203 induces senescence by targeting E2F3 protein in human melanoma cells. J Biol Chem. 2012;287(15):11769–11777. doi: 10.1074/jbc.M111.325027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overhoff MG, Garbe JC, Koh J, Stampfer MR, Beach DH, Bishop CL. Cellular senescence mediated by p16INK4A-coupled miRNA pathways. Nucleic Acids Res. 2014;42(3):1606–1618. doi: 10.1093/nar/gkt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olivieri F, Rippo MR, Procopio AD, Fazioli F. Circulating inflamma-miRs in aging and age-related diseases. Front Genet. 2013;4:121. doi: 10.3389/fgene.2013.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Kim MS, Jia B, et al. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature. 2017;548(7665):52–57. doi: 10.1038/nature23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noren Hooten N, Fitzpatrick M, Wood WH, 3rd, et al. Age-related changes in microRNA levels in serum. Aging. 2013;5(10):725–740. doi: 10.18632/aging.100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Khanna A, Li N, Wang E. Circulatory miR34a as an RNAbased, noninvasive biomarker for brain aging. Aging. 2011;3(10):985–1002. doi: 10.18632/aging.100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JH, Lee BR, Choi ES, et al. Reverse expression of aging-associated molecules through transfection of miRNAs to aged mice. Mol Ther Nucleic Acids. 2017;6:106–115. doi: 10.1016/j.omtn.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai CY, Wu YT, Yu SL, et al. Modulated expression of human peripheral blood microRNAs from infancy to adulthood and its role in aging. Aging Cell. 2014;13(4):679–689. doi: 10.1111/acel.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Owczarz M, Budzinska M, Domaszewska-Szostek A, et al. miR-34a and miR-9 are overexpressed and SIRT genes are downregulated in peripheral blood mononuclear cells of aging humans. Exp Biol Med (Maywood) 2017;242(14):1453–1461. doi: 10.1177/1535370217720884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol. 2011;2011:842849. doi: 10.1155/2011/842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luga V, Zhang L, Viloria-Petit AM, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151(7):1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 15.Clayton A, Turkes A, Dewitt S, Steadman R, Mason MD, Hallett MB. Adhesion and signaling by B cell-derived exosomes: the role of integrins. FASEB J. 2004;18(9):977–979. doi: 10.1096/fj.03-1094fje. [DOI] [PubMed] [Google Scholar]
- 16.Urbanelli L, Buratta S, Sagini K, Tancini B, Emiliani C. Extracellular vesicles as new players in cellular senescence. Int J Mol Sci. 2016;17(9):1408. doi: 10.3390/ijms17091408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiels PG, Stenvinkel P, Kooman JP, McGuinness D. Circulating markers of ageing and allostatic load: A slow train coming. Pract Lab Med. 2017;7:49–54. doi: 10.1016/j.plabm.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21(12):1406–1415. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- 19.Machida T, Tomofuji T, Ekuni D, et al. MicroRNAs in salivary exosome as potential biomarkers of aging. Int J Mol Sci. 2015;16(9):21294–21309. doi: 10.3390/ijms160921294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolhe R, Hunter M, Liu S, et al. Gender-specific differential expression of exosomal miRNA in synovial fluid of patients with osteoarthritis. Sci Rep. 2017;7(1):2029. doi: 10.1038/s41598-017-01905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lugli G, Cohen AM, Bennett DA, et al. Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS One. 2015;10(10):e0139233. doi: 10.1371/journal.pone.0139233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu CG, Song J, Zhang YQ, Wang PC. MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol Med Rep. 2014;10(5):2395–2400. doi: 10.3892/mmr.2014.2484. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-menocal L, Pham SM, Mateu D, et al. Aging increases p16INK4a expression in vascular smooth-muscle cells. Biosci Rep. 2010;30(1):11–18. doi: 10.1042/BSR20080128. [DOI] [PubMed] [Google Scholar]
- 24.Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493(7432):338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Chiu JF, Gagne J, Fukagawa NK. Age-related differences in insulin-like growth factor-1 receptor signaling regulates Akt/FOXO3a and ERK/Fos pathways in vascular smooth muscle cells. J Cell Physiol. 2008;217(2):377–387. doi: 10.1002/jcp.21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Essers P, Tain LS, Nespital T, et al. Reduced insulin/insulin-like growth factor signaling decreases translation in Drosophila and mice. Sci Rep. 2016;6:30290. doi: 10.1038/srep30290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornsby PJ. Telomerase and the aging process. Exp Gerontol. 2007;42(7):575–581. doi: 10.1016/j.exger.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonsen JL, Rosada C, Serakinci N, et al. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol. 2002;20(6):592–596. doi: 10.1038/nbt0602-592. [DOI] [PubMed] [Google Scholar]
- 29.Bernardes de Jesus B, Vera E, Schneeberger K, et al. Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol Med. 2012;4(8):691–704. doi: 10.1002/emmm.201200245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenarczyk M, Lam V, Jensen E, et al. Cardiac injury after 10 gy total body irradiation: indirect role of effects on abdominal organs. Radiat Res. 2013;180(3):247–258. doi: 10.1667/RR3292.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao FQ, Craig R. Capturing time-resolved changes in molecular structure by negative staining. J Struct Biol. 2003;141(1):43–52. doi: 10.1016/s1047-8477(02)00546-4. [DOI] [PubMed] [Google Scholar]
- 32.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathiyalagan P, Sahoo S. Exosomes-based gene therapy for microRNA delivery. Methods Mol Biol. 2017;1521(1521):139–152. doi: 10.1007/978-1-4939-6588-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu M, Xing H, Xun Z, et al. Exosome-based small RNA delivery: Progress and prospects. Asian J Pharm Sci. 2018;13(1):1–11. doi: 10.1016/j.ajps.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewkowicz P, Cwiklińska H, Mycko MP, et al. Dysregulated RNA-induced silencing complex (RISC) assembly within CNS corresponds with abnormal miRNA expression during autoimmune demyelination. J Neurosci. 2015;35(19):7521–7537. doi: 10.1523/JNEUROSCI.4794-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seike M, Goto A, Okano T, et al. miR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106(29):12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mudduluru G, Abba M, Batliner J, et al. A systematic approach to defining the microRNA landscape in metastasis. Cancer Res. 2015;75(15):3010–3019. doi: 10.1158/0008-5472.CAN-15-0997. [DOI] [PubMed] [Google Scholar]
- 38.Bofill-De Ros X, Gironella M, Fillat C. miR-148a- and miR-216a-regulated oncolytic adenoviruses targeting pancreatic tumors attenuate tissue damage without perturbation of miRNA activity. Mol Ther. 2014;22(9):1665–1677. doi: 10.1038/mt.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castellano L, Stebbing J. Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues. Nucleic Acids Res. 2013;41(5):3339–3351. doi: 10.1093/nar/gks1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang HR, Schoenfeld LW, Ruby JG, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24(10):992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boucher J, Charalambous M, Zarse K, et al. Insulin and insulin-like growth factor 1 receptors are required for normal expression of imprinted genes. Proc Natl Acad Sci U S A. 2014;111(40):14512–14517. doi: 10.1073/pnas.1415475111. [DOI] [PMC free article] [PubMed] [Google Scholar]