Abstract
The hepatic glucose fasting response is gaining traction as a therapeutic pathway to enhance hepatic and whole-host metabolism. However, the mechanisms underlying these metabolic effects remain unclear. Here, we demonstrate the epidermal-type lipoxygenase, eLOX3 (encoded by its gene, Aloxe3), is a potentially novel effector of the therapeutic fasting response. We show that Aloxe3 is activated during fasting, glucose withdrawal, or trehalose/trehalose analogue treatment. Hepatocyte-specific Aloxe3 expression reduced weight gain and hepatic steatosis in diet-induced and genetically obese (db/db) mouse models. Aloxe3 expression, moreover, enhanced basal thermogenesis and abrogated insulin resistance in db/db diabetic mice. Targeted metabolomics demonstrated accumulation of the PPARγ ligand 12-KETE in hepatocytes overexpressing Aloxe3. Strikingly, PPARγ inhibition reversed hepatic Aloxe3–mediated insulin sensitization, suppression of hepatocellular ATP production and oxygen consumption, and gene induction of PPARγ coactivator-1α (PGC1α) expression. Moreover, hepatocyte-specific PPARγ deletion reversed the therapeutic effect of hepatic Aloxe3 expression on diet-induced insulin intolerance. Aloxe3 is, therefore, a potentially novel effector of the hepatocellular fasting response that leverages both PPARγ-mediated and pleiotropic effects to augment hepatic and whole-host metabolism, and it is, thus, a promising target to ameliorate metabolic disease.
Keywords: Hepatology, Metabolism
Keywords: Diabetes, Insulin signaling, Signal transduction
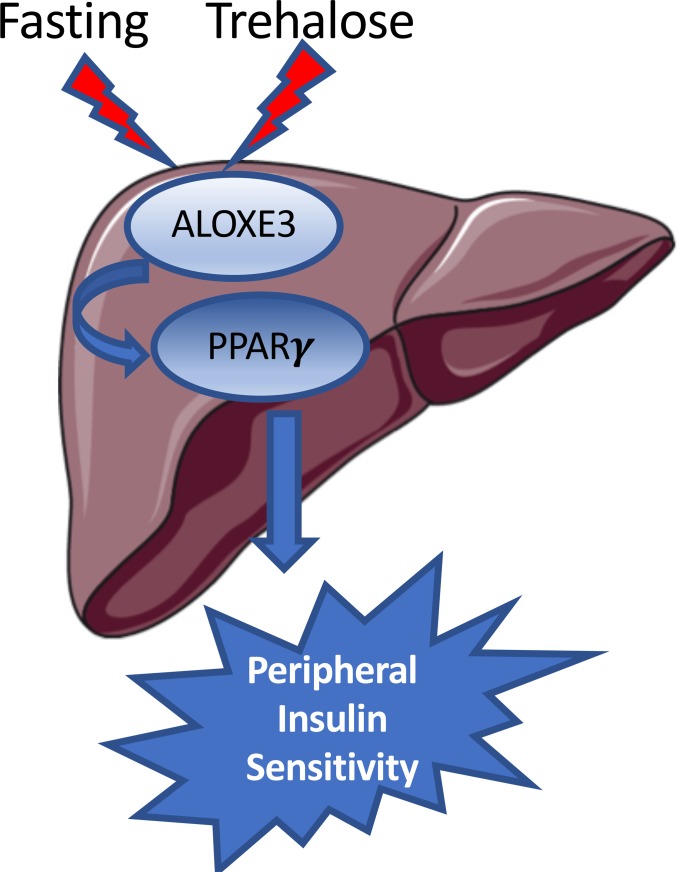
The lipoxygenase ALOXE3 is an effector of the hepatic fasting response that improves insulin sensitivity by activating hepatic PPARγ.
Introduction
Leveraging the generalized fasting and caloric restriction responses to mitigate metabolic disease is under intense investigation as a novel therapeutic approach against obesity, diabetes mellitus, and nonalcoholic fatty liver disease (NAFLD) (1, 2). The hepatocyte is uniquely positioned at the nexus of the portal circulation — where dietary macronutrients are sensed and triaged to their metabolic fate — and the peripheral circulation, from which macronutrients are taken up and utilized or stored. Its unique anatomic positioning also defines the hepatocyte’s role in coordinating the transition from fed to fasting states. Specifically, the hepatocyte senses fat, protein, and carbohydrate content and rapidly alters its metabolism and its endocrine functions to coordinate peripheral responses. However, the therapeutic promise of caloric restriction and generalized macronutrient fasting remains limited by its clinical impracticality and unsustainability. Our laboratory and others (1, 9, 11) have therefore examined specific pathways through which fasting and fasting-like mechanisms can be molecularly or pharmacologically leveraged to the benefit of the host.
Upon physiological macronutrient withdrawal, the canonical hepatic response is to shift to mobilize glycogen and oxidize fat from stores to produce glucose and ketone body fuel for the brain and heart. This begins with activation of proximal macronutrient sensing kinases, AMPK, and mTOR. These kinases act as primary regulators of macroautophagy as well as transcriptional fasting programming by hepatic transcription factors, including transcription factor EB (3); PPARγ coactivator-1α (PGC1α) (4); and transcriptional modifiers such as the sirtuin family of protein deacetylases (5). FGF21 is released from the liver of fasting rodents and humans to promote adipose “browning” (mitochondrial uncoupling to promote heat generation; ref. 6). The net effect of this signaling initiation is a networked cascade through which the liver provides energy substrate to the periphery during fasting and promotes peripheral insulin sensitivity for efficient absorption of the next meal.
We recently demonstrated that blocking hepatic glucose transporters genetically or pharmacologically closely recapitulates the therapeutic metabolic effects of generalized macronutrient fasting (7–11). Pharmacologic glucose transport blockade using the disaccharide glucose transporter inhibitor trehalose blocked mTOR and activated AMPK-dependent autophagic flux (9, 12). Moreover, trehalose blocked diet- and genetically induced steatosis via ATG16L1- and AMPK-dependent mechanisms, and it enhanced peripheral thermogenesis via hepatocyte TFEB- and FGF21-dependent mechanisms (11). Consistent with data from pharmacological glucose transporter (GLUT) blockade, genetic deletion of the hepatocyte GLUT8 similarly increased basal and fasting heat generation (10), enhanced hepatocyte fat oxidation, and inhibited hepatic de novo lipogenesis and triacylglycerol generation in mice fed a high-fructose diet (7, 8). Notably, in those experiments, GLUT8 blockade was associated with enhanced insulin tolerance, enhanced hepatic PPARγ expression, and concomitant activation of the PPARγ target UCP2 (8). Conversely, GLUT8 was upregulated in the livers of genetically diabetic mice (13). Together, prior data suggest that pharmacologic and genetic activation of hepatic glucose fasting–like responses are sufficient to mitigate metabolic disease. However, the mechanisms underlying this protection remain enigmatic.
Recent data illuminate a role for cellular lipoxygenases in the pathogenesis and protection from metabolic disease. In humans, a recent large cohort of adults with type 2 diabetes demonstrated a potentially protective role for 5-lipoxygenases ALOX5 and ALOX5AP against diabetes mellitus (14). Conversely, ALOX12/15 inhibition in rodent models inhibited oxidative stress, β cell deterioration, hepatic steatosis, and systemic inflammation (15–18). Together, these data suggest that intracellular lipid intermediary metabolism drives both tissue-level and whole-organism metabolic homeostasis. The concept that the fasting state might modulate lipoxygenase activity as part of the broadly adaptive effects of fasting on metabolism remained heretofore unexplored.
Here, we identified eLOXE3 (encoded by the Aloxe3 gene) as a fasting-induced epidermal-type lipoxygenase that acts as a potentially novel effector of the hepatic glucose fasting response in vitro and in vivo. Targeted Aloxe3 overexpression induced robust generation of the PPARγ ligand (19, 20) 12-KETE. In vivo, hepatic Aloxe3 expression enhanced insulin sensitivity, attenuated weight gain, and reduced hepatic steatosis in both diet- and genetically induced steatosis models. Moreover, hepatic Aloxe3 expression mitigated diet-induced dyslipidemia, and db/db mice expressing Aloxe3 exhibited attenuated weight gain, enhanced basal caloric expenditure, and PPARγ-dependent insulin sensitivity. Strikingly, we demonstrate that hepatocyte-specific PPARγ deletion is absolutely required for the insulin-sensitizing effect of hepatic Aloxe3 induction. Taken together, Aloxe3 is a fasting-responsive hepatocyte effector that is sufficient to attenuate insulin resistance, weight gain, and hepatic fat deposition in dietary and genetically obese models, in part by activating hepatic PPARγ. Aloxe3 is, thus, a novel therapeutic mechanism downstream of the fasting response, which can be leveraged against metabolic disease.
Results
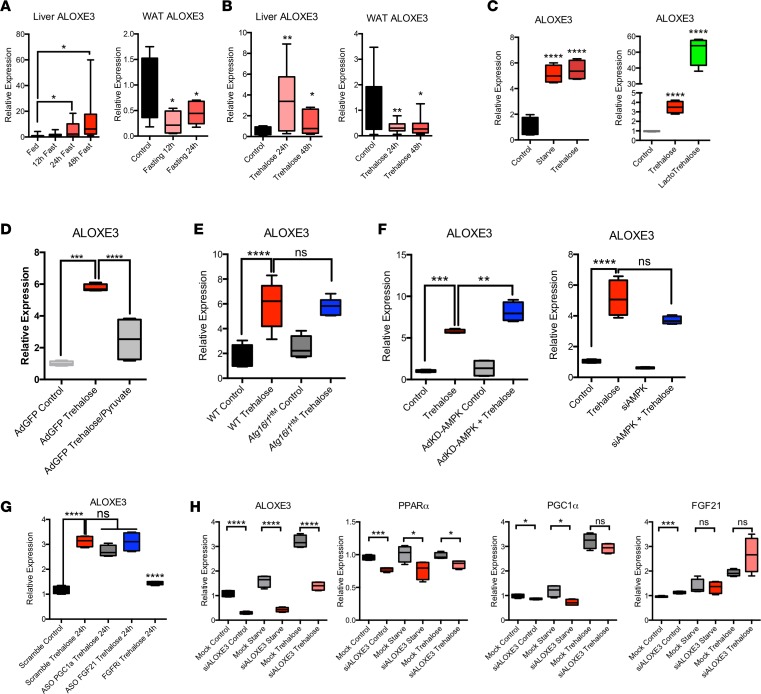
To test the hypothesis that Aloxe3 is induced by acute fasting or pseudo-fasting conditions, we subjected WT mice to 48-hour fasting. During acute fasting, hepatic Aloxe3 expression was increased and sustained at least through 48 hours (Figure 1A). Oral administration of the fasting-mimetic hepatic glucose transport inhibitor trehalose (3% PO, ad libitum), similarly induced Aloxe3 expression after 24-hour and 48-hour trehalose feeding (Figure 1B). The hepatic Aloxe3 response to fasting contrasted with fasting Aloxe3 expression in white adipose tissue (WAT), wherein Aloxe3 mediates fat differentiation and lipid storage. Accordingly, Aloxe3 was significantly suppressed during both fasting and trehalose feeding in epididymal WAT (Figure 1, A and B, respectively). We then modeled Aloxe3 expression in vitro to determine whether fasting-induced Aloxe3 expression is cell-autonomously regulated. Isolated primary murine hepatocytes were subjected to treatment by either low serum and glucose (0.5% serum, 1 g/l glucose) or trehalose (Figure 1C). In addition, we characterized fasting-mimetic effects of a trehalose analogue that resists enzymatic degradation by trehalases (21–23), called lactotrehalose (α-D-glucopyronosyl-(1,1)-α-D-galactopyranoside; Figure 1C and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.120794DS1). In each case, Aloxe3 expression was significantly increased, most potently by trehalose and lactotrehalose (Figure 1C and Supplemental Figure 1).
Figure 1. Aloxe3 is induced in response to fasting and pseudofasting.
(A) Aloxe3 expression in liver and white adipose tissue (WAT) from fasting WT mice (0–48 hours [h]). n = 6 each of fed, 12 h fasting, and 24 h fasting mice. For 48 h fasting mice, n = 7. (B) Aloxe3 expression in WT mice fed oral trehalose (3% in sterile water fed ad libitum, 0–48 h). For control and trehalose (24 h) mice, n = 5 and 6. For 48 h trehalose-treated mice, n = 6. (C) Aloxe3 expression in isolated primary murine hepatocytes treated with 0.5% FCS/1 g/l glucose media, 100 mM trehalose, or 100 mM lactotrehalose (24 h). n = 4 per group. (D) Aloxe3 expression in trehalose-treated isolated primary hepatocytes pretreated with or without 5 mM pyruvate. n = 4 per group. (E) Aloxe3 expression in trehalose-treated WT and Atg16l1-mutant mice in vivo (24 h). n = 6 per group. (F) Aloxe3 expression in response to trehalose in the presence or absence of kinase-dead AMPK overexpression or siRNA-mediated AMPK knockdown. n = 4 per group. (G) Aloxe3 expression in response to trehalose in the presence or absence of antisense oligonucleotide (ASO) directed against PGC1, or FGF21. FGF receptor 1–4 inhibitor (LY2874455) was included as a control to demonstrate Aloxe3 blockade in context. n = 4 per group. (H) Fasting-responsive marker gene expression in the presence or absence of trehalose following treatment with or without Aloxe3-directed siRNA. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by 2-tailed t test with Bonferroni-Dunn post hoc correction versus untreated controls group or versus the bracketed comparison group where indicated.
In light of evidence that Aloxe3 is a potentially novel hepatocyte fasting-responsive factor in mice and in isolated murine hepatocytes, we evaluated whether this factor is expressed in isolated human hepatocytes and in Huh7 and HepG2 human hepatoma cell lines. Consistent with immunoblot and immunohistochemical eLOX3 protein detection in human liver tissue (24), quantitative PCR (qPCR) analysis demonstrated ALOXE3 expression in both HepG2 and in primary human hepatocytes (Supplemental Figure 2).
To determine whether Aloxe3 induction depended upon energy substrate deficit, we treated primary hepatocytes with trehalose in the presence or absence of pyruvate. This provided energy substrate for the cell, independently of glucose transporter blockade. Pyruvate reversed trehalose-induced Aloxe3 induction by ~60% (P < 0.0001), suggesting that Aloxe3 expression is at least partly energy substrate dependent (Figure 1D). In light of our prior data in which we demonstrated that the autophagy complex protein ATG16L1 was required for autophagic and antisteatotic effects of trehalose in hepatocytes (9, 25), we examined whether ATG16L1 is required for Aloxe3 induction. We treated primary hepatocytes from WT littermates or from mice with homozygous hypomorphic Atg16l1 alleles (Atg16l1HM) with or without trehalose (24 hours). Aloxe3 mRNA quantification revealed robust Aloxe3 induction in WT hepatocytes, which was not suppressed in Atg16l1HM hepatocytes (Figure 1E). Similarly, AMPK inhibition (by kinase-dead AMPK overexpression or by Aloxe3 siRNA transfection; Figure 1F) and PPARα, PGC1α, and FGF21 knockdown each failed to reverse Aloxe3 induction after trehalose treatment or glucose and serum withdrawal (Figures 1G and 1H). Accordingly, moderate hepatocyte-specific overexpression of the fasting-induced transcription factor SIRT1 did not induce Aloxe3 expression (Supplemental Figure 3). In contrast, siRNA-based Aloxe3 knockdown attenuated PPARα and PGC1α at baseline and during glucose and serum withdrawal from hepatocytes in vitro, without effects on FGF21 expression (Figure 1H). Together, our data demonstrate that Aloxe3 is a hepatocyte lipoxygenase that is induced by glucose transporter blockade and energetic deficit via a mechanism that does not require canonical ATG16L1-dependent and AMPK-PGC1α/PPARα-FGF21 fasting mechanisms.
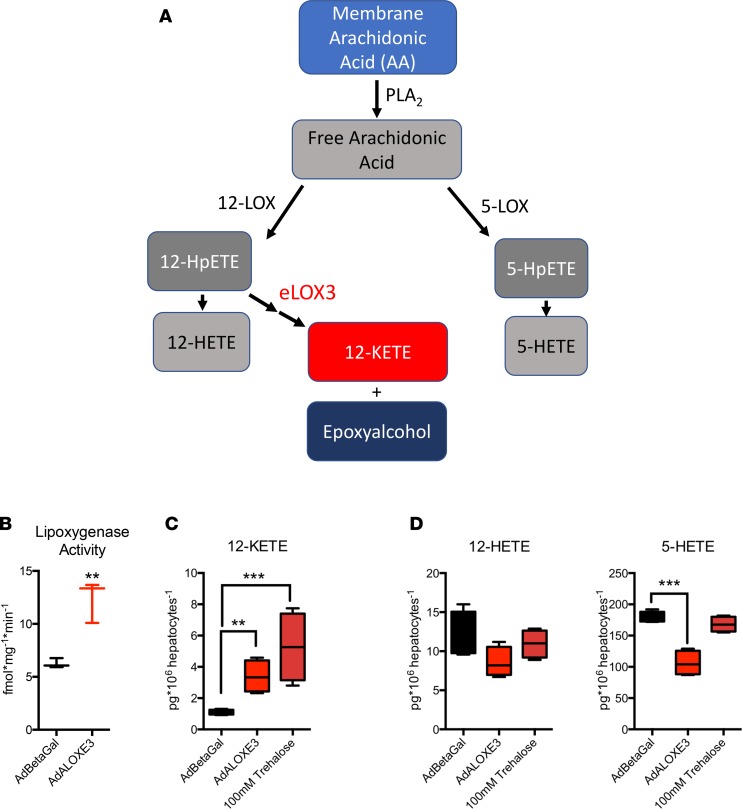
eLOX3 generates the PPARγ ligand 12-KETE and an epoxyalcohol from 12-HpETE in the metabolism of plasma membrane arachidonic acid (Figure 2A) (26–30). We tested whether Aloxe3 mediates similar lipid metabolism in hepatocytes upon forced Aloxe3 expression. We first evaluated the effect of Aloxe3 overexpression on hepatocyte lipoxygenase activity by measuring LOX substrate oxidation in cell extracts from hepatocytes overexpressing β galactosidase (β-gal) or Aloxe3. Consistent with prior data demonstrating intrinsic, latent dioxygenase activity of eLOX3 (31), Aloxe3 overexpression significantly increased total oxidized LOX substrate accumulation (Figure 2B) and 12-KETE accumulation in primary hepatocytes (Figure 2C). This effect of Aloxe3 overexpression on 12-KETE accumulation was indeed phenocopied by trehalose treatment (Figure 2C). Moreover, Aloxe3 overexpression decreased the abundance of alternate arachidonic acid metabolic pathway products 5-HETE and 12-HETE (Figure 2D).
Figure 2. Aloxe3 and trehalose induce the PPARγ ligand 12-KETE in murine hepatocytes.
(A) Arachidonic acid metabolism mediated by lipoxygenases 12-LOX, 5-LOX, and eLOX3 (adapted from refs. 26, 29, 58, 59). (B) Lipoxygenase activity in primary murine hepatocytes upon overexpression of β galactosidase or Aloxe3. n = 3 from 1 representative experiment of 2 experiments with similar results. (C and D) Quantitative GC-MS analysis of the stable lipoxygenase reaction products 12-KETE, 5-HETE, and 12-HETE. Data represent 4 independent cultures per group from 1 experiment, representing 2 independent experiments with similar results. **P < 0.01 and ***P < 0.001 by 2-tailed t test with Bonferroni-Dunn post hoc correction versus untreated controls group or versus the bracketed comparison group where indicated.
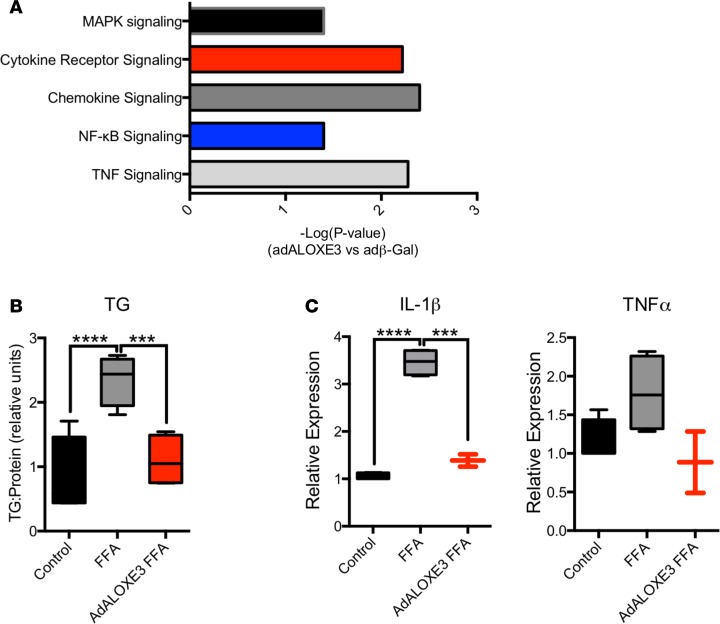
To ascertain transcriptome-wide effects of forced Aloxe3 overexpression, we next treated primary hepatocytes with adenovirus encoding β-gal or Aloxe3 prior to RNA sequencing (RNA-seq) analysis. Pathway analysis revealed that 5 of the 10 most downregulated processes in Aloxe3-overexpressing cells were devoted to inflammatory signaling, including TNFα, NF-κB, chemokine signaling, cytokine receptor signaling, and MAPK signaling (Figure 3A). Given these findings, and in light of the fact that Aloxe3 upregulation by trehalose correlates with reduced diet- and genetically induced steatosis (9, 25), we examined whether Aloxe3 attenuates fat-induced inflammatory signaling and triglyceride (TG) accumulation. Primary hepatocytes treated with BSA-conjugated fatty acids (FA) induced Il1b and Tnfa gene expression, as well as TG accumulation (Figure 3, B and C). These FA-induced effects on TG accumulation and Il1b and Tnfa gene expression effects were reversed in FA-treated cultures overexpressing Aloxe3.
Figure 3. Aloxe3 reduces inflammatory signaling and steatosis in hepatocytes.
(A) RNA-seq analysis of the top 9 downregulated genes in primary hepatocytes expressing either β galactosidase or Aloxe3. n = 3 from a single RNA-seq run performed once. The P value for significantly downregulated pathways is demonstrated as (–Log[P value]). (B) Hepatic TG accumulation and (C) IL-1β or TNFα expression in hepatocytes treated with BSA-conjugated fatty acids with or without Aloxe3 expression. n = 4 in B and n = 4 in C, single representative experiments, which were repeated twice with similar results. ***P < 0.001 and ****P < 0.0001 by 2-tailed t testing with Bonferroni-Dunn post hoc correction versus the bracketed comparison group as indicated.
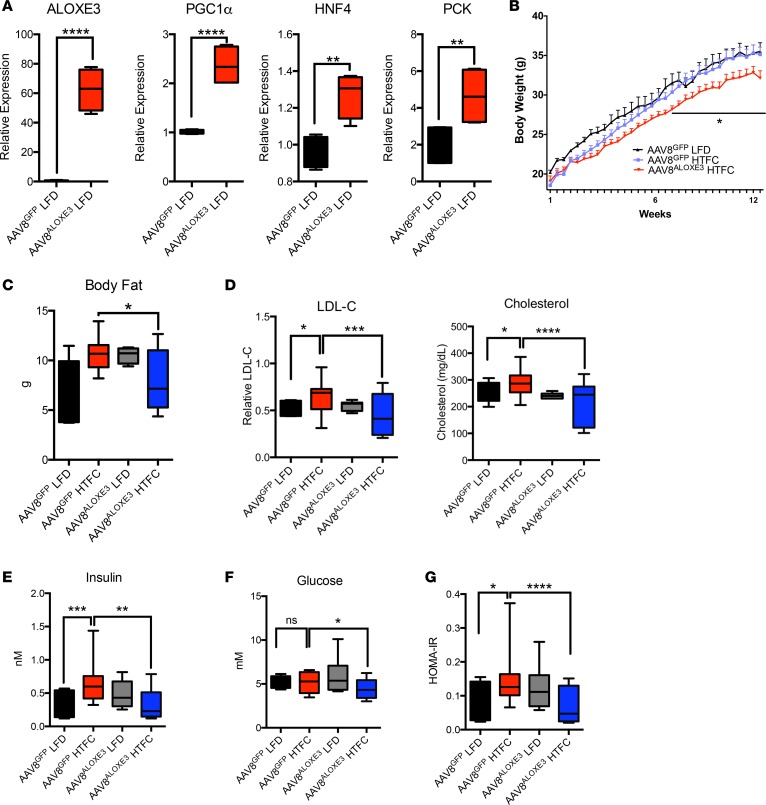
Aloxe3-deficient mice are not amenable to in vivo metabolic studies because germline Aloxe3 deficiency results in postnatal mortality secondary to massive skin permeability and water loss (32). To evaluate the in vivo metabolic consequences of hepatocyte Aloxe3 activation, we tested the in vivo effects of hepatocyte-directed Aloxe3 expression. We first confirmed overexpression of the Aloxe3 transgene in liver (Figure 4A) without changes in Aloxe3 expression in skeletal muscle, WAT, or brown adipose tissue (not shown). This correlated with upregulation of fasting and oxidative genes in unperturbed Aloxe3-overexpressing mice, including Aloxe3 (Figure 4A), Ppargc1a (PGC1), hepatocyte nuclear factor 4α (Hnf4A/HNF4α), and phosphoenolpyruvate carboxykinase 1 (Pck1, Figure 4A.).
Figure 4. Enhanced whole-body metabolism in mice ALOXE3-overexpressing mice.
(A) qPCR quantification of expression for oxidative and fasting-response genes in unperturbed mice expressing hepatocyte GFP or Aloxe3. (B) Body weight over time in low-fat or high–trans fat/cholesterol diet–fed mice expressing hepatocyte GFP or Aloxe3. (C) Body fat content in mice fed HTFC or LFD with or without hepatic Aloxe3 overexpression. (D) LDL-C and total cholesterol in mice fed HTFC or LFD with or without hepatic Aloxe3 overexpression. (E–G) Circulating insulin, glucose, and calculated HOMA-IR in LFD- and HTFC-fed mice overexpressing hepatocyte GFP or Aloxe3. Number of mice in each group is: 5, AAV8GFP LFD; 10, AAV8GFP HTFC; 5, AAV8ALOXE3 LFD; and 10, AAV8ALOXE3 HTFC. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by 2-tailed t test with Bonferroni-Dunn post hoc correction versus the bracketed comparison group as indicated.
We next evaluated the effect of hepatic Aloxe3 expression in mice fed low-fat diet (LFD) or steatogenic, high trans-fat/cholesterol diet (HTFC) (12 weeks). Mice overexpressing Aloxe3 fed HTFC gained significantly less weight, had lower total body mass (Figure 4B), and had lower body fat mass (Figure 4C) without changes in lean mass (not shown) when compared with Gfp-overexpressing mice fed HTFC. Accordingly, low density lipoprotein-cholesterol (LDL-C) and total cholesterol were significantly lowered in HTFC-fed mice overexpressing Aloxe3 (Figure 4D). Indices of glucose homeostasis were also improved by Aloxe3 overexpression in HTFC-fed mice (Figure 4, E–G). Specifically, HTFC feeding increased circulating insulin and homeostatic model assessment– insulin resistance (HOMA-IR) in GFP-overexpressing mice (Figure 4, E and G), without altering fasting plasma glucose (Figure 4F). In contrast, HTFC-fed mice overexpressing hepatic Aloxe3 were protected from HTFC-induced hyperinsulinemia and insulin resistance (Figure 4, E and G). Indeed, fasting glucose was also lowered in Aloxe3-overexpressing, HTFC-fed mice when compared with GFP-expressing, HTFC-fed mice (Figure 4F). Together, hepatocyte Aloxe3 expression was sufficient to reduce diet-induced weight gain, body fat accumulation, dyslipidemia, and insulin resistance.
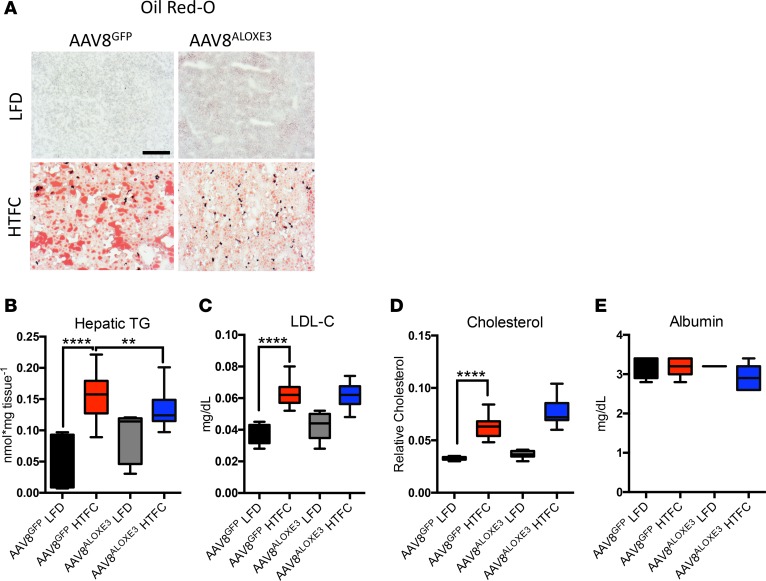
We next examined hepatic lipid metabolism in GFP- and Aloxe3-overexpressing mice fed LFD or HTFC. Frozen liver sections from HTFC-fed mice exhibited increased Oil Red O staining and macrosteatosis when compared with LFD-fed mice. Consistent with prior models of PPARγ activation (33–35), Aloxe3 expression in LFD-fed mice resulted in a mild basal TG accumulation (Figure 5, A and B). However, we did not observe evidence of hepatic inflammation or fibrosis by histological analysis of H&E- or trichrome-stained liver sections in LFD-fed mice after 12 weeks Aloxe3 overexpression (Supplemental Figure 4). Also consistent with prior reports on hepatic PPARγ activation, Aloxe3 overexpression modestly but significantly protected from HTFC-induced TG accumulation that took on a microsteatotic staining pattern (Figure 5, A and B). No changes in hepatic LDL-C or total cholesterol were observed (Figure 5, C and D) upon hepatic Aloxe3 overexpression in LFD- or HTFC-fed mice. None of our genetic or dietary manipulations had any effect on hepatic synthetic function, as ascertained by quantitative circulating albumin levels (Figure 5E).
Figure 5. Reduced hepatic steatosis correlates with Aloxe3-induced fasting responses.
(A) Oil Red O staining in livers from low-fat or high–trans fat/cholesterol–fed mice overexpressing GFP or Aloxe3. (B–D) Hepatic tissue quantification of triglycerides, LDL-C, total cholesterol in low-fat or high–trans fat/cholesterol–fed mice overexpressing empty vector or Aloxe3. (E) Serum albumin measurements in mice analyzed in A–D Number of mice in each group is: 5, AAV8GFP LFD; 10, AAV8GFP HTFC; 5, AAV8ALOXE3 LFD; and 10, AAV8ALOXE3 HTFC. Scale bar: 100 μm. **P < 0.01 and ****P < 0.0001 by 2-tailed t test with Bonferroni-Dunn post hoc correction versus untreated controls group or versus the bracketed comparison group where indicated.
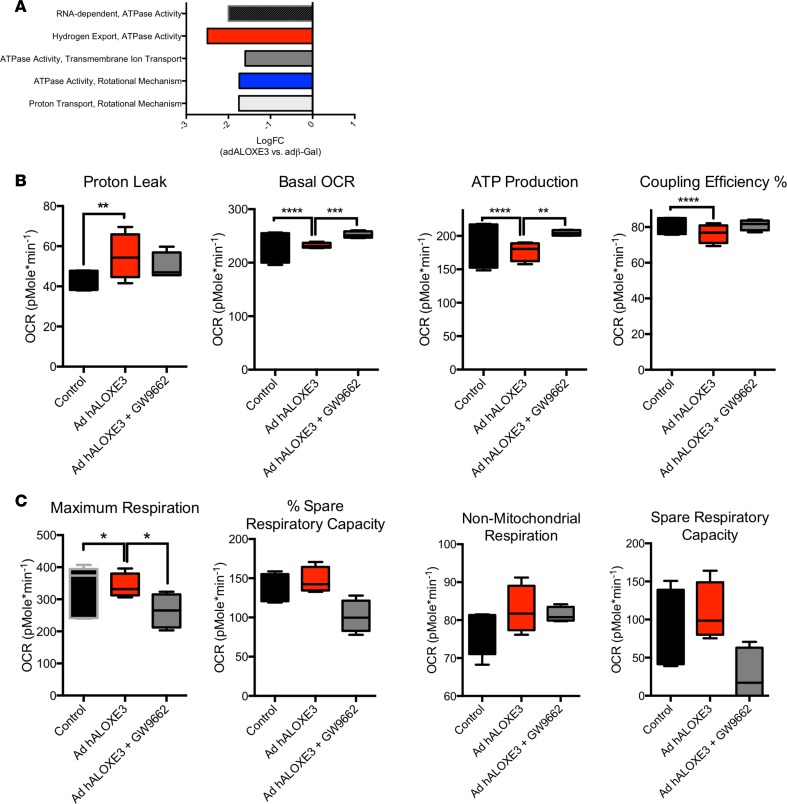
To examine mechanistically how Aloxe3 might activate hepatocyte-starvation–like responses, we evaluated the effects of Aloxe3 overexpression on mitochondrial respiratory function at the cellular and molecular levels. RNA-seq analysis of primary hepatocytes overexpressing Aloxe3 revealed that 6 of the 10 most downregulated molecular processes encompassed mitochondrial electron transport function, including proton transport, ATPase activity, transmembrane ion transport, and hydrogen export (Figure 6A). We therefore tested functionally the hypothesis that Aloxe3 mitigates ATP production by inducing hepatocyte mitochondrial uncoupling. Seahorse analysis of hepatocytes overexpressing Aloxe3 exhibited elevated proton leak, ATP production, and coupling efficiency, concomitant with suppressed basal oxygen consumption rate and enhanced glycolytic rate when compared with hepatocytes expressing β-gal (Figure 6B). Each of these parameters was partly or fully reversed in the presence of the PPARγ inhibitor GW9662 (Figure 6B). No changes in nonmitochondrial oxygen consumption were observed (Figure 6C), suggesting that Aloxe3 specifically affected mitochondrial energy metabolism.
Figure 6. Hepatic PPARγ is required for Aloxe3 to induce hepatic energetic inefficiency.
(A) RNA-seq analysis of the top 5 downregulated molecular processes in hepatocytes overexpressing Aloxe3. Highlighted are ATPase-related and mitochondrial coupling processes. Graphed is logFC relative to cultures expressing β galactosidase. n = 3 from a single RNA sequencing run performed once. (B and C) Seahorse XF96 analysis of proton leak, basal OCR, ATP production, coupling efficiency, and nonmitochondrial oxygen consumption in AML12 cells overexpressing Aloxe3 with or without GW9662 (PPARγ inhibitor) treatment. n = 8 independent cultures combined from 2 distinct experimental runs. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by 2-tailed t testing with Bonferroni-Dunn post hoc correction versus the bracketed comparison group as indicated.
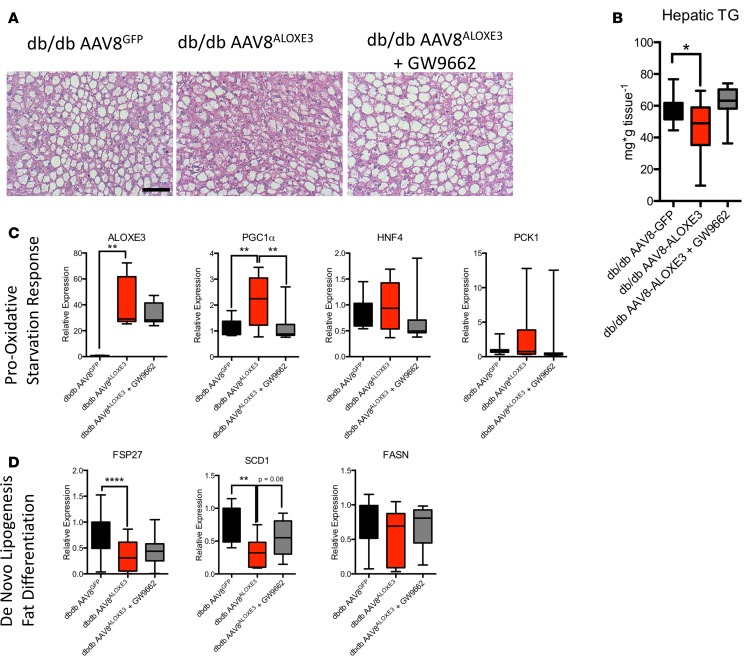
Mechanistic interrogation in vivo was executed to determine whether hepatocyte Aloxe3 mediated peripheral insulin and glucose homeostasis in a leptin-dependent manner. To that end, we overexpressed Aloxe3 or GFP in db/db mice, which lack the leptin receptor. Again, in the liver, we observed modest, statistically significant attenuation of hepatic TG accumulation in db/db mice overexpressing Aloxe3 when compared with db/db controls (Figure 7, A and B). We confirmed Aloxe3 overexpression correlated with increases in oxidative and fasting-response genes Ppargc1a, Hnf4a, and Pc1 (Figure 7C). Moreover, Aloxe3 overexpression reduced genetic markers of de novo lipogenesis in db/db mice, including Fsp27, Scd1, and Fasn (Figure 7D). Concomitant PPARγ inhibition by the inhibitor GW9662 produced either statistically significant or trends toward significant reversal in each of these Aloxe3-altered marker genes (Figure 7, C and D).
Figure 7. Reduced hepatic steatosis correlates with Aloxe3-induced fasting and reduction of de novo lipogenesis.
(A) H&E staining in livers from low-fat or high–trans fat/cholesterol–fed mice overexpressing empty vector or Aloxe3. (B) Hepatic tissue triglyceride quantification in db/db mice overexpressing GFP or Aloxe3 in the presence or absence of GW9662. (C and D) qPCR quantification of expression for oxidative and fasting-response genes (C) and de novo lipogenic genes (D) in db/db mice expressing GFP or Aloxe3 with or without GW9662 administration. Number of mice in each group is: 10, db/db AAV8GFP; 10, db/db AAV8ALOXE3; 10, AAV8ALOXE3 + GW9662. Scale bar: 100 μm. **P < 0.01, or ****P < 0.0001 by 2-tailed t testing with Bonferroni-Dunn post hoc correction versus the bracketed comparison group as indicated.
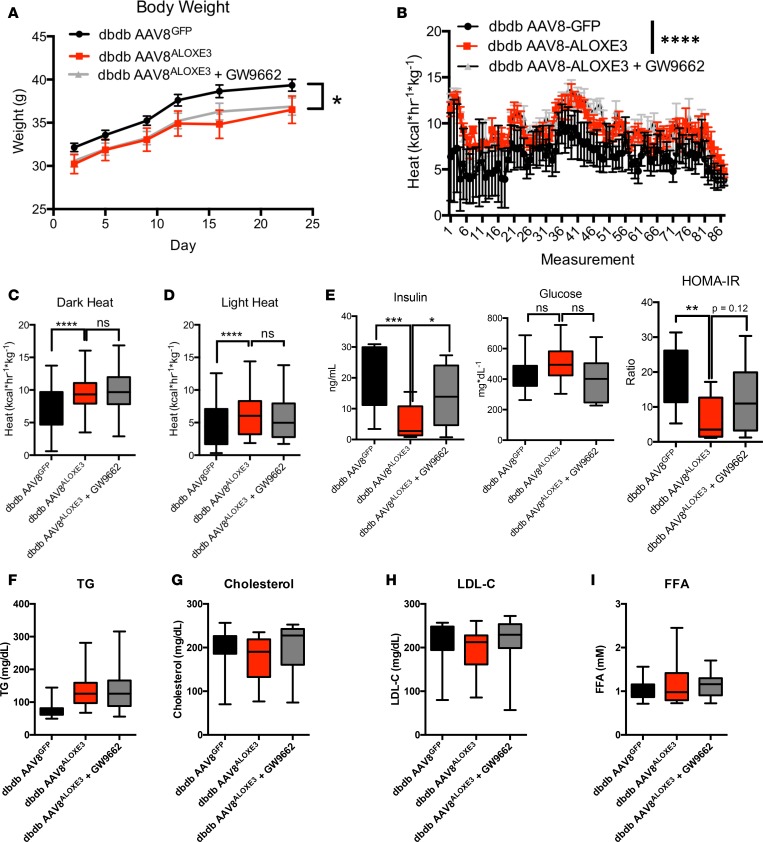
Although baseline weights were not statistically different, db/db mice overexpressing Aloxe3 gained significantly less weight and had a lower end-of-trial weight than db/db mice over the 28-day trial (Figure 8A). Indirect calorimetry revealed that the attenuated weight gain in aloxe3 db/db mice was associated with enhanced light- and dark-cycle heat generation and O2-CO2 exchange (Figure 8, B-D). Neither heat generation nor O2-CO2 exchange in Aloxe3db/db was affected by GW9662 coadministration (Figure 8, B–D).
Figure 8. Enhanced whole-body energy metabolism in db/db diabetic mice overexpressing Aloxe3.
(A) Body weight over time in db/db mice expressing empty vector or Aloxe3 in the presence or absence of GW9662. (B) Heat generation over time in db/db mice expressing GFP or Aloxe3. (C and D) Indirect calorimetric quantification of light- and dark-cycle heat generation in db/db mice expressing Aloxe3 or GFP treated with or without GW9662. (E) Fasting serum insulin determined by ELISA, serum glucose determined by colorimetric assay, and calculated HOMA-IR index based on glucose and insulin data. (F–I) Serum TG, cholesterol, LDL-C, and FFA content in db/db mice with or without hepatic Aloxe3 overexpression and with or without GW9662 treatment. Number of mice in each group is: 10, db/db AAV8GFP; 10, db/db AAV8ALOXE3; 10, AAV8ALOXE3 + GW9662. *P < 0.05, **P < 0.01, ***P < 0.001, and < 0.0001 by 2-tailed t test with Bonferroni-Dunn post hoc correction versus bracketed comparison groups.
Because hepatic Aloxe3 expression enhanced peripheral insulin sensitivity in HTFC-fed mice (Figure4), and because PPARγ agonism by thiazolidinediones (TZDs) enhances peripheral insulin sensitivity (36, 37), we evaluated whether Aloxe3 enhances peripheral insulin and glucose homeostasis dependent on PPARγ and yet independent of the leptin receptor. Fasting circulating insulin and HOMA-IR were significantly lower in Aloxe3db/db mice without changes in circulating glucose when compared with db/db mice expressing GFP. The reduction in circulating insulin and HOMA-IR were reversed in mice concomitantly treated with the PPARγ inhibitor GW9662, suggesting that Aloxe3 improves glucose and insulin homeostasis via a PPARγ-dependent mechanism. In contrast with our diet-induced model, however, targeted hepatic Aloxe3 expression did not reduce circulating lipids in a leptin receptor–deficient mice. Together, the data in our leptin-deficient model elucidate leptin- and PPARγ-dependent functions by Aloxe3.
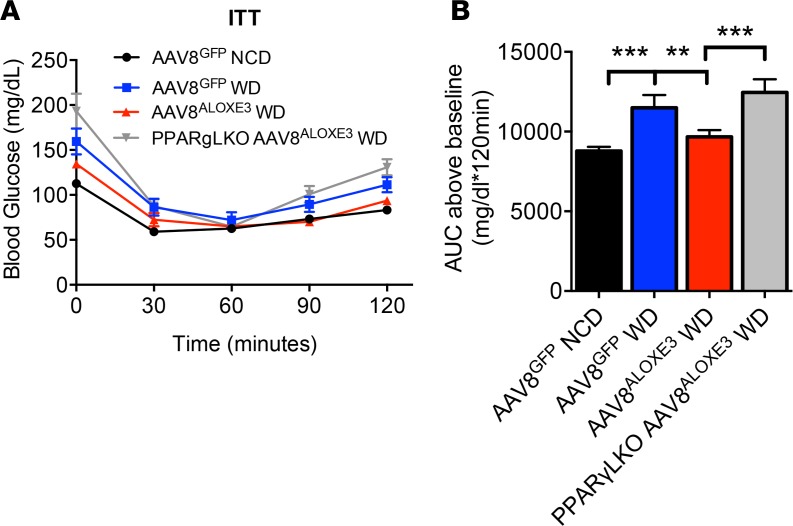
To gain further specificity regarding the role of hepatocyte PPARγ in Aloxe3-enhanced insulin sensitivity, we generated mice harboring a hepatocyte-specific PPARG deletion (hereafter referred to as PPARγ-LKO mice) by crossing mice with homozygous floxed Pparg alleles with mice expressing Cre recombinase driven by the albumin promoter. Mice were placed on a chow or Western diet (WD) to induce insulin resistance over 12 weeks; they were then subjected to insulin tolerance testing (ITT, Figure 9, A and B). Area under the ITT curve analysis revealed that WD feeding increased AUC in GFP-overexpressing animals, whereas AUC was reduced in Aloxe3-overexpressing animals. In striking contrast, WD-fed Aloxe3-overexpressing PPARγ-LKO mice had a significantly elevated AUC when compared with WD-fed Aloxe3-overexpressing mice harboring WT hepatic PPARγ.
Figure 9. Insulin sensitization by Aloxe3 requires hepatic PPARγ.
(A) Insulin tolerance testing in WT or PPARγ-LKO mice expressing hepatic GFP or Aloxe3 after chow or Western diet. (B) Quantification of total area under the glucose time curve. **P < 0.01 and ***P < 0.001 by 2-tailed t test with Bonferroni-Dunn post hoc correction between bracketed comparison groups as indicated. Number of mice in each group is: 4, AAV8GFP Chow; 5, AAV8GFP WD; 5, AAV8ALOXE3 WD; and 4, PPARγ-LKO AAV8ALOXE3 WD.
Discussion
The concept of the fasting and caloric restriction response as a therapeutic to be leveraged in longevity, healthspan, and metabolism from rodents to early primates to humans (1, 38–43) is gaining traction. Already in the clinical realm, caloric restriction improved multiple indices related to aging, cardiovascular disease, and diabetes mellitus (2, 44–46). However, strict and prolonged adherence to caloric restriction, intermittent fasting, or any intensive dietary restriction directed toward weight loss carries with it several practical barriers. Therefore, identifying the therapeutic determinants of the hepatic glucose fasting response opens the promise of novel, targeted, and sustainable treatments against metabolic and other diseases. Here, we show that Aloxe3 is activated by the adaptive hepatic fasting response and is itself sufficient to enhance insulin sensitivity, increase basal caloric expenditure, and reduce diet-induced weight gain and fat accumulation, hepatic steatosis, and dyslipidemia.
Fully defining the metabolic functions of Aloxe3 has been previously limited by its basal tissue expression and distribution and by the dramatic phenotype observed in animals and humans deficient for this enzyme. Aloxe3 was first identified as an epidermal-type lipoxygenase that mediates skin differentiation via hydroperoxide isomerase activity (27, 47). Humans born with a defect in this gene develop ichthyosiform disease, characterized by profound skin barrier dysfunction and transepidermal water loss (28, 30). This was confirmed in murine models of Aloxe3 deficiency, which succumb to massive dehydration without intervention shortly after birth (32). Subsequent work demonstrated a role for Aloxe3 in adipocyte differentiation via PPARγ agonism in cultured adipocytes, as well (29). This introduced the possibility that Aloxe3 has important functions that extend beyond epidermal tissues. However, a potential function in liver was largely ignored, perhaps in part because nonfasting mRNA is relatively low when compared with epidermis and tongue by Northern blot detection (32) and by reverse transcription PCR analysis of murine liver (48). However, we show here that serum and glucose withdrawal, trehalose and lactotrehalose (trehalose analog) treatment in vitro, trehalose feeding, and acute fasting in vivo each robustly induce Aloxe3 mRNA expression. This raised the prospect that Aloxe3 mediates part of the physiological hepatic fasting response. We postulate that the observed increased insulin sensitivity in hepatic Aloxe3–overexpressing mice reflects an adaptive response that enhances peripheral insulin sensitivity in preparation for a subsequent meal after prolonged fasting. This function would be particularly useful to maximize macronutrient absorption for animals in which meals are few and far between (e.g., intermittently fasting or hibernating animals). Indeed, this teleological rationale has been proposed for other physiological fasting-induced pathways, such as FGF21 signaling (49, 50).
The data herein demonstrate that Aloxe3 ameliorated the diet-induced obesity and db/db diabetic phenotypes, at least in part, via a PPARγ-dependent mechanism — most prominently with regard to the insulin-sensitizing effects of Aloxe3. Aloxe3 overexpression and trehalose treatment in hepatocytes induced the PPARγ ligand 12-KETE at cellular concentrations that appear to be physiologically relevant (19). Indeed, 12-KETE induced sebocyte PPARγ signaling at ~1,500 ng/ml (19), whereas our calculated 12-KETE concentrations in Aloxe3-overexpressing hepatocytes (accounting for a hepatocyte volume of roughly 3 fL, femtoliters; ref. 51) indicate concentration of 12-KETE is in the range of 1,000 ng/ml (Figure 2C). In addition, PPARγ blockade by GW9662 administration in isolated hepatocytes and in vivo reversed Aloxe3-mediated changes in hepatocyte oxygen consumption rate, insulin resistance, and PGC1α upregulation. Finally, liver-specific PPARγ deletion abrogated Aloxe3-enhanced insulin sensitivity. Together, these data introduce a potentially novel hepatic Aloxe3–PPARγ axis that improves whole-body insulin sensitivity. These findings are consistent with the previously demonstrated control of peripheral insulin sensitivity via hepatic PPARγ in ob/ob mice (34, 35). However, given that GW9662 did not significantly reverse Aloxe3 effects on hepatic fat accumulation and whole-body thermogenesis, our data indicate that Aloxe3 exerts both PPARγ-dependent and -independent functions to enhance hepatic and extrahepatic metabolic homeostasis.
It should be noted that genetic disruption of both 12-LOX and 15-LOX in multiple diabetic models largely recapitulates the Aloxe3-mediated enhancement of hepatic insulin sensitivity and reduced hepatic steatosis (15–17). Therefore, it is plausible that — in addition to (or perhaps in lieu of) generating the PPARγ ligand 12-KETE — eLOX3 either depletes 12-LOX products 12-HpETE and 12-HETE (Figure 2) to the host’s benefit, or 12-LOX and 15-LOX deletion shunts arachidonic acid down the eLOX3 enzymatic pathway to activate PPARγ. A third distinct possibility is that Aloxe3 mediates its PPARγ-activating effects independently of upstream lipoxygenases. Three key pieces of data support this possibility. First, hepatocytes are not considered overall to be a significant source of total lipoxygenase activity. This is confirmed by our own data suggesting that basal intrinsic hepatocyte lipoxygenase activity is relatively low — on the order of 1–10 fmol/mg/min (Figure 2B). Second, eLOX3 exhibits latent intrinsic dioxygenase activity, although this catalysis is relatively inefficient when compared with its hydroperoxide isomerase activity (31). Third, in adipose tissue, eLOX3 generated PPARγ agonists in the absence of identifiable upstream lipoxygenases (29). However, regardless of the origin and fate of the lipid intermediaries going through the Aloxe3 pathway, the net benefit of Aloxe3 expression involves both PPARγ-dependent and -independent pathways. The species, origin, and precise bioactivity of the lipid intermediaries that underlie these therapeutic effects now arise as a rich topic for further interrogation.
The current study examines therapeutic effects of hepatic Aloxe3 induction. From a clinical therapeutic perspective, virus-based genetic therapy for many monogenic diseases has already reached clinical care (52). However, this therapeutic approach is not yet fully optimized for polygenic diseases, such as obesity, insulin resistance, and NAFLD (53). Although our physiological, morphometric, and liver histological data did not reveal problematic effects of Aloxe3 gene overexpression after 12 weeks (Figure 4, Figure 5, and Supplemental Figure 4), a genetic overexpression approach at present is likely to be a more distant clinical therapy when compared with precision small-molecule therapeutics. In addition, although the full extent of Aloxe3 regulation has yet to be elucidated as a means to leverage Aloxe3-enhancing pathways, we demonstrated here that Aloxe3 is potently upregulated by generalized stimuli such as fasting, trehalose feeding, serum withdrawal, and flavone-class GLUT inhibition. In contrast, canonical fasting intermediates AMPK, PGC1α, FGF21, and PPARα were dispensable for trehalose-induced Aloxe3 expression. Accordingly, SIRT1 overexpression was insufficient to upregulate Aloxe3 in vivo. It is therefore intriguing that we were able to achieve enhanced in vitro Aloxe3 upregulation comparable with viral Aloxe3 overexpression by treatment with the trehalase-resistant trehalose analogue lactotrehalose (21–23, 54, 55). Our characterization of this glucose-galactose trehalose analogue, and the finding that lactotrehalose exhibits enhanced fasting-mimetic potency, is a critical advance for at least 2 reasons. First, abundant trehalase expression in human gut, liver, brain, kidney, and reproductive tissues raises the possibility that trehalose-based therapies may be limited by trehalase-mediated trehalose degradation. By corollary, trehalase-resistant compounds such as lactotrehalose are postulated to confer enhanced efficacy. Secondly, it is well understood that GLUT8 and GLUT2 (the 2 most highly expressed hepatic glucose transporters; ref. 7) are competitively inhibited by galactose (56–58). In light of the fact that galactose is one of the saccharide moieties that comprises lactotrehalose, the data suggest that altering the saccharide moieties in trehalose modulates potency (and perhaps also selectivity) based on each GLUT’s substrate predilection.
Taken together, although greater detail regarding intermediary regulation of Aloxe3 is warranted, augmenting Aloxe3 expression and other components of the hepatic fasting response is now feasible through fasting itself, glucose withdrawal, or trehalose/analogue-class GLUT inhibition. The palatability, and heavy first-pass enterohepatic kinetics (which minimizes peripheral tissue GLUT side effects) indeed make trehalose and its analogues especially attractive candidate nutraceuticals to elicit this response. Future directions should interrogate the fasting response–inducing potential, kinetics, and efficacy of trehalose and its analogs in mitigating metabolic disease (21, 25, 59). In addition, it will be critical to understand the extent to which Aloxe3 regulation in other tissues is necessary or sufficient to augment tissue-specific adaptive fasting responses. Finally, understanding the full cadre of epoxyalcohols and other lipid products of eLOX3 enzymatic activity, and their specific roles in the mitigating metabolic disease, are of special pharmaceutical and clinical interest (15, 60–63).
In summary, we identified the lipoxygenase Aloxe3 as a potentially novel effector of the hepatic fasting response that is sufficient to augment basal caloric expenditure, and ameliorate insulin resistance, weight gain, and hepatosteatosis. (the meaning I hope to convey is that Aloxe3 does all of the above). The rapidly rising prevalence of each of these major public health problems throughout the Western and developing worlds mandates novel therapeutic pathway generation and leveraging thereof. We assert that further interrogation into how hepatic glucose transport mediates the networked adaptive hepatic fasting response will advance the field toward new and effective human therapy against metabolic disease.
Methods
Mouse models and treatment.
All mice were caged in specific pathogen-free barrier housing with 12-hour light-dark cycles and free access to water and rodent chow. For transgenic studies, WT C57B/6J mice and Leprdb/db mice were obtained directly from the Jackson Laboratory. Upon arrival, mice were randomized to experimental assignments and equilibrated for a minimum of 7 days in the specific pathogen-free vivarium with automated 12-hour light-dark cycles and free access to water and rodent chow. Prior to initiating metabolic measurements. ATG16L1HM mice were a provided by Herbert “Skip” W. Virgin’s laboratory (Washington University School of Medicine) (64). PPARγ-LKO mice were obtained directly from the Jackson Laboratory to the laboratory of David Rudnick (Washington University School of Medicine) and, after propagation, were provided for experimentation.
Adeno-associated viruses overexpressing either GFP or Aloxe3 under control of the thyroid binding globulin (TBG) promoter were obtained as ready-to-use viral stocks from Vector Biolabs. Viral particles (1 × 1011 per animal) were injected via tail vein in 6-week-old mice 10 days prior to dietary initiation or 38 days prior to sacrifice in the genetic obesity model (e.g., 10 days rest period + 28 day dietary stimulus) Aloxe3 expression was quantified in pilot experiments by qPCR analysis of hepatic tissue, skeletal muscle tissue, and brown and epididymal WAT 10 days after injection and confirmed selective hepatic Aloxe3 overexpression (not shown).
Antisense oligonucleotides (ASOs) were obtained from IONIS Pharmaceuticals as ready-to-inject oligomers, and they were used precisely as described previously (65). GW9662 was obtained from Cayman Chemicals (catalog 70785). WD is from Harlan Teklad (catalog TD88137); HTF-C diet is from Research Diets (catalog 09100301).
Lactotrehalose synthesis and purification.
Synthesis and purification of lactotrehalose was carried out precisely as described (66). We confirmed 98%–99% purity by 1H-NMR (not shown).
Lipoxygenase activity assay.
LOX activity was quantified using a lipoxygenase activity assay kit (Biovision, catalog 978-100) per manufacturer instructions.
Serum analyses.
Fasting blood glucose was measured via glucometer using tail vein blood. For all other serum analyses, submandibular blood collection was performed immediately prior to sacrifice, and serum was separated. Insulin ELISA (MilliporeSigma, catalog EZRMI-13K), TG (Thermo Fisher Scientific, catalog TR22421), cholesterol (Thermo Fisher Scientific, catalog TR13421), and free FA (FFA) (Wako Diagnostics, catalog 999-34691, 995-34791, 991-34891, and 993-35191) quantification was performed using commercially available reagents according to manufacturer’s directions. Albumin levels were quantified using an AMS LIASYS Chemistry Analyzer.
Hepatic lipids.
Lipids were extracted from ~100 mg hepatic tissue homogenized in 2:1 chloroform/methanol. Extract (0.25%–0.5% of each) was evaporated overnight prior to biochemical quantification of TGs, LDL-C, cholesterol, and FFA using reagents described above, according to manufacturer’s directions.
Oil Red O staining.
Methanol-fixed frozen sections from WT and transgenic mice were stained according to described protocols (7, 9, 12).
Body composition analysis and indirect calorimetry.
Body composition analysis was carried out in unanesthetized mice as described (7, 8, 67) using an EchoMRI 3-1 device (Echo Medical Systems) via the Washington University Diabetic Mouse Models Phenotyping Core Facility.
The first 4 hours in the cage were unmeasured in order to allow for acclimation time. Thereafter, the oxygen and CO2 consumption and production were quantified for a minimum of 1 light cycle (0601–1800 hours) and for 1 dark cycle (1801–0600 hours). Volume O2 (VO2), VCO2, heat, movement, and respiratory exchange ratio (RER) were automatically calculated using TSE Phenomaster software.
In vitro metabolism (Seahorse XF96) assays.
Seahorse assays were performed using a Seahorse XF96 analyzer Mito Stress Test Kit (Agilent Technologies) according to manufacturer specifications. Hepatocyte cultures were seeded at 20,000 cells per well prior to assay.
Immunoblotting.
Immunoblotting was performed as described (67). Anti-GAPDH (catalog 5174) was obtained from Cell Signaling Technologies. LC3B antiserum was obtained from Novus Biologicals (catalog NB-100-2200).
Indirect calorimetry.
Oxygen consumption, CO2 production, RER, and heat production were measured using the Phenomaster system (TSE) via the Washington University Diabetic Mouse Models Phenotyping Core Facility as described (8, 67). Metabolic parameters were documented every 13 minutes.
Cell cultures and treatment.
Primary murine hepatocytes obtained from WT mice were isolated as described (7, 9–11) and cultured and maintained in regular DMEM growth media (MilliporeSigma, catalog D5796) containing 10% FBS. For in vitro starvation experiments, starved media containing 1 g/l glucose and 0.5% FBS was used. Cultures were lysed in Trizol and subjected to downstream analysis. In vitro genetic knockdown was achieved via siRNA transfection using Lipofectamine 3000 from Invitrogen (catalog L3000015). Trehalose was obtained from MilliporeSigma and was >97% purity by HPLC. Trehalose water (3%, w/v) fed ad libitum was used in all in vivo experiments. Huh7 and HepG2 cell lines were obtained directly from the American Type Culture Collection (ATCC). Human primary hepatocyte cDNAs were obtained directly from Sciencell (catalog 5204).
qPCR.
qPCR was performed as previously reported (7, 12) with some modifications. Snap-frozen livers or cultured hepatocytes were homogenized in Trizol reagent (Invitrogen, catalog 15596026). RNA isolated according to the manufacturer’s protocol was reverse-transcribed using the Qiagen Quantitect reverse transcriptase kit (Qiagen, catalog 205310). cDNA was subjected to qPCR using the SYBR Green master mix reagent (Applied Biosystems, catalog 4309155). Expression data are depicted as a mean ratio of target gene expression relative to 36B4 (housekeeping gene) expression in each sample, ± SEM. Primer sequences for qPCR are shown in Supplemental Table 1.
RNA-seq.
RNA-seq was performed by the Washington University Genome Technology Access Center (GTAC). Primary data, processed data, and metadata for these experiments were deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo), with accession number GSE116516.
Library preparation was performed with 10 μg of total RNA with a Bioanalyzer RIN score greater than 8.0. Ribosomal RNA was removed by poly-A selection using Oligo-dT beads (mRNA Direct kit, Invitrogen). mRNA was then fragmented in buffer containing 40 mM tris acetate pH 8.2, 100 mM potassium acetate, and 30 mM magnesium acetate and heated to 94°C for 150 seconds. mRNA was reverse transcribed to yield cDNA using SuperScript III RT enzyme (Invitrogen, per manufacturer’s instructions) and random hexamers. A second strand reaction was performed to yield double-stranded cDNA (ds-cDNA). cDNA was blunt ended, had an A base added to the 3′ ends, and then had Illumina sequencing adapters ligated to the ends. Ligated fragments were then amplified for 12 cycles using primers incorporating unique index tags. Fragments were sequenced on an Illumina HiSeq-3000 using single reads extending 50 bases.
RNA-seq reads were aligned to the Ensembl release 76 top-level assembly with STAR version 2.0.4b. Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version 1.4.5. Transcript counts were produced by Sailfish version 0.6.3. Sequencing performance was assessed for total number of aligned reads, total number of uniquely aligned reads, genes and transcripts detected, ribosomal fraction known junction saturation, and read distribution over known gene models with RSeQC version 2.3.
To enhance the biological interpretation of the large set of transcripts, grouping of genes/transcripts based on functional similarity was achieved using the R/Bioconductor packages GAGE and Pathview. GAGE and Pathview were also used to generate pathway maps on known signaling and metabolism pathways curated by Kyoto Encyclopedia of Genes and Genomes (KEGG).
Liquid chromatography-tandem mass spectral analysis of eicosanoids.
Eicosanoids (5-HETE, 12-HETE, and 12-KETE) were extracted with 300 μl of 1:1 methanol/water, containing 2 ng deuterated 12-HETE-d8 (Cayman Chemical) as the internal standard. This extract (33% of total) was injected; therefore, 0.67 ng represents the total internal standard analyzed. The quantity of eicosanoids calculated based on the peak are the ratio between the analyte and internal standard. Eicosanoid and internal standard analyses were performed using a Shimadzu 20AD HPLC system, and a LeapPAL autosampler coupled to a tandem mass spectrometer (API 4000, API-Sciex). These were operated in negative ion MRM mode. The sample was injected on to a Thermo-Keystone betasil C-18 HPLC column (2 × 100 mm, 3 μm) with mobile phases (A, 5 mM ammonium fluoride in water; B, 100 % acetonitrile). The data processing was conducted with Analyst 1.6.3 (API-Sciex).
ITT.
ITT was performed precisely as reported previously (8). Briefly, mice were fasted 4 hours prior to i.p. administration of 0.75 U/kg insulin. Glucose was monitored by glucometer (One Touch Ultra, Lifescan) over the course of 2 hours following injection.
Statistics.
Data were analyzed using GraphPad Prism version 6.0 (RRID:SCR_015807). A P value less than 0.05 was defined as statistically significant. Data shown are as mean ± SEM. Unpaired 2-tailed homoscedastic t tests with Bonferroni post hoc correction for multiple comparisons were used for all analyses unless otherwise noted in the figure legends. Two-way ANOVA was also used for analyses with 2 independent variables.
Study approval.
All animal procedures were reviewed and approved by the Washington University School of Medicine Animal Studies Committee.
Author contributions
BJD conceived and coordinated the study and wrote the paper. CBH, YZ, ALM, HF, MJG, BMS, AIS, and BJD designed, performed, and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Medical Research Program under Award No. W81XWH-17-1-0133. Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. This work was also supported by grants from the NIH/National Center for Advancing Translational Sciences (NCATS) grant UL1TR002345, the Children’s Discovery Institute (MI-FR-2014-426) AGA-Gilead Sciences Research Scholar Award in Liver Disease, the Washington University Digestive Disease Research Core Center (BJD) (P30DK52574), the Washington University Diabetes Research Center (P30DK020579), Nutrition & Obesity Research Center (P30DK056341), and the Robert Wood Johnson Foundation. BJD is a Scholar of the Washington University Child Health Research Center (K12HD076224) and of the Children’s Discovery Institute (MI-FR-2014-426). BMS was supported by NIH (R15 AI117670) to BMS. ALM was supported by the Washington University Spencer T. Olin Fellowship, the Washington University NIGMS Institutional Training Grant in Cell and Molecular Biosciences (T32GM007067), and National Science Foundation Graduate Student Fellowship (DGE-1143954). We thank Herbert Virgin at Washington University School of Medicine for the ATG16L1HM mouse line and David Rudnick (Washington University) for the liver-specific PPARγ-deficient mice.
Version 1. 08/23/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reference information: JCI Insight. 2018;3(16):e120794. https://doi.org/10.1172/jci.insight.120794.
Contributor Information
Yiming Zhang, Email: yiming.zhang@wustl.edu.
Hideji Fujiwara, Email: hfujiwara@wustl.edu.
References
- 1.Longo VD, Mattson MP. Fasting: molecular mechanisms and clinical applications. Cell Metab. 2014;19(2):181–192. doi: 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnosky AR, Hoddy KK, Unterman TG, Varady KA. Intermittent fasting vs daily calorie restriction for type 2 diabetes prevention: a review of human findings. Transl Res. 2014;164(4):302–311. doi: 10.1016/j.trsl.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15(6):647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura MT, Yudell BE, Loor JJ. Regulation of energy metabolism by long-chain fatty acids. Prog Lipid Res. 2014;53:124–144. doi: 10.1016/j.plipres.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Chalkiadaki A, Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol. 2012;8(5):287–296. doi: 10.1038/nrendo.2011.225. [DOI] [PubMed] [Google Scholar]
- 6.Degirolamo C, Sabbà C, Moschetta A. Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nat Rev Drug Discov. 2016;15(1):51–69. doi: 10.1038/nrd.2015.9. [DOI] [PubMed] [Google Scholar]
- 7.Debosch BJ, Chen Z, Saben JL, Finck BN, Moley KH. Glucose transporter 8 (GLUT8) mediates fructose-induced de novo lipogenesis and macrosteatosis. J Biol Chem. 2014;289(16):10989–10998. doi: 10.1074/jbc.M113.527002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeBosch BJ, Chen Z, Finck BN, Chi M, Moley KH. Glucose transporter-8 (GLUT8) mediates glucose intolerance and dyslipidemia in high-fructose diet-fed male mice. Mol Endocrinol. 2013;27(11):1887–1896. doi: 10.1210/me.2013-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBosch BJ, et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci Signal. 2016;9(416):ra21. doi: 10.1126/scisignal.aac5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer AL, et al. Enhanced Hepatic PPARα Activity Links GLUT8 Deficiency to Augmented Peripheral Fasting Responses in Male Mice. Endocrinology. 2018;159(5):2110–2126. doi: 10.1210/en.2017-03150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.et al. Transcription Factor EB (TFEB)-dependent Induction of Thermogenesis by the Hepatocyte Solute Carrier 2A (SLC2A) Inhibitor Trehalose. Autophagy. doi: 10.1080/15548627.2018.1493044. doi: 10.1080/15548627.2018.1493044. [published online ahead of print Kuly 12, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayer AL, et al. SLC2A8 (GLUT8) is a mammalian trehalose transporter required for trehalose-induced autophagy. Sci Rep. 2016;6:38586. doi: 10.1038/srep38586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorovits N, Cui L, Busik JV, Ranalletta M, Hauguel de-Mouzon S, Charron MJ. Regulation of hepatic GLUT8 expression in normal and diabetic models. Endocrinology. 2003;144(5):1703–1711. doi: 10.1210/en.2002-220968. [DOI] [PubMed] [Google Scholar]
- 14.Tsekmekidou XA, et al. Assessment of association between lipoxygenase genes variants in elderly Greek population and type 2 diabetes mellitus. Diab Vasc Dis Res. 2018;15(4):340–343. doi: 10.1177/1479164118756241. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Clemente M, et al. Disruption of the 12/15-lipoxygenase gene (Alox15) protects hyperlipidemic mice from nonalcoholic fatty liver disease. Hepatology. 2010;52(6):1980–1991. doi: 10.1002/hep.23928. [DOI] [PubMed] [Google Scholar]
- 16.Samala N, Tersey SA, Chalasani N, Anderson RM, Mirmira RG. Molecular mechanisms of nonalcoholic fatty liver disease: Potential role for 12-lipoxygenase. J Diabetes Complicat. 2017;31(11):1630–1637. doi: 10.1016/j.jdiacomp.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez-Perez M, et al. Inhibition of 12/15-Lipoxygenase Protects Against β-Cell Oxidative Stress and Glycemic Deterioration in Mouse Models of Type 1 Diabetes. Diabetes. 2017;66(11):2875–2887. doi: 10.2337/db17-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole BK, Morris MA, Grzesik WJ, Leone KA, Nadler JL. Adipose tissue-specific deletion of 12/15-lipoxygenase protects mice from the consequences of a high-fat diet. Mediators Inflamm. 2012;2012:851798. doi: 10.1155/2012/851798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dozsa A, et al. PPARγ-mediated and arachidonic acid-dependent signaling is involved in differentiation and lipid production of human sebocytes. J Invest Dermatol. 2014;134(4):910–920. doi: 10.1038/jid.2013.413. [DOI] [PubMed] [Google Scholar]
- 20.Hammond VJ, et al. Novel keto-phospholipids are generated by monocytes and macrophages, detected in cystic fibrosis, and activate peroxisome proliferator-activated receptor-γ. J Biol Chem. 2012;287(50):41651–41666. doi: 10.1074/jbc.M112.405407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walmagh M, Zhao R, Desmet T. Trehalose Analogues: Latest Insights in Properties and Biocatalytic Production. Int J Mol Sci. 2015;16(6):13729–13745. doi: 10.3390/ijms160613729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.et al. Efficient Enzymatic Synthesis of Disaccharide, alpha-D-Galactosyl alpha-D-Glucoside, by Trehalose Phosphorylase from Thermoanaerobacter brockii. J Appl Glycosci. 2001;48(2):135–137. doi: 10.5458/jag.48.135. [DOI] [Google Scholar]
- 23.Kim HM, Chang YK, Ryu SI, Moon SG, Lee SB. Enzymatic synthesis of a galactose-containing trehalose analogue disaccharide by Pyrococcus horikoshii trehalose-synthesizing glycosyltransferase: Inhibitory effects on several disaccharidase activities. J Mol Catal B Enzym. 2007;49(1):98–103. [Google Scholar]
- 24.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 25.Mardones P, Rubinsztein DC, Hetz C. Mystery solved: Trehalose kickstarts autophagy by blocking glucose transport. Sci Signal. 2016;9(416):fs2. doi: 10.1126/scisignal.aaf1937. [DOI] [PubMed] [Google Scholar]
- 26.O’Donnell VB, Murphy RC. New families of bioactive oxidized phospholipids generated by immune cells: identification and signaling actions. Blood. 2012;120(10):1985–1992. doi: 10.1182/blood-2012-04-402826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Z, Schneider C, Boeglin WE, Marnett LJ, Brash AR. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc Natl Acad Sci USA. 2003;100(16):9162–9167. doi: 10.1073/pnas.1633612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fürstenberger G, et al. Role of epidermis-type lipoxygenases for skin barrier function and adipocyte differentiation. Prostaglandins Other Lipid Mediat. 2007;82(1-4):128–134. doi: 10.1016/j.prostaglandins.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Hallenborg P, et al. Epidermis-type lipoxygenase 3 regulates adipocyte differentiation and peroxisome proliferator-activated receptor gamma activity. Mol Cell Biol. 2010;30(16):4077–4091. doi: 10.1128/MCB.01806-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mashima R, Okuyama T. The role of lipoxygenases in pathophysiology; new insights and future perspectives. Redox Biol. 2015;6:297–310. doi: 10.1016/j.redox.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y, Brash AR. Dioxygenase activity of epidermal lipoxygenase-3 unveiled: typical and atypical features of its catalytic activity with natural and synthetic polyunsaturated fatty acids. J Biol Chem. 2010;285(51):39866–39875. doi: 10.1074/jbc.M110.155374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krieg P, et al. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J Invest Dermatol. 2013;133(1):172–180. doi: 10.1038/jid.2012.250. [DOI] [PubMed] [Google Scholar]
- 33.Yang SJ, et al. Activation of peroxisome proliferator-activated receptor gamma by rosiglitazone increases sirt6 expression and ameliorates hepatic steatosis in rats. PLoS One. 2011;6(2):e17057. doi: 10.1371/journal.pone.0017057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavrilova O, et al. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 2003;278(36):34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 35.Matsusue K, et al. Liver-specific disruption of PPARgamma in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J Clin Invest. 2003;111(5):737–747. doi: 10.1172/JCI17223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hauner H. The mode of action of thiazolidinediones. Diabetes Metab Res Rev. 2002;18 Suppl 2:S10–S15. doi: 10.1002/dmrr.249. [DOI] [PubMed] [Google Scholar]
- 37.He L, Liu X, Wang L, Yang Z. Thiazolidinediones for nonalcoholic steatohepatitis: A meta-analysis of randomized clinical trials. Medicine (Baltimore) 2016;95(42):e4947. doi: 10.1097/MD.0000000000004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webber J, Taylor J, Greathead H, Dawson J, Buttery PJ, Macdonald IA. The effects of fasting on the thermogenic, metabolic and cardiovascular responses to infused adrenaline. Br J Nutr. 1995;74(4):477–490. doi: 10.1079/BJN19950152. [DOI] [PubMed] [Google Scholar]
- 39.Ahmet I, Wan R, Mattson MP, Lakatta EG, Talan M. Cardioprotection by intermittent fasting in rats. Circulation. 2005;112(20):3115–3121. doi: 10.1161/CIRCULATIONAHA.105.563817. [DOI] [PubMed] [Google Scholar]
- 40.Wan R, Camandola S, Mattson MP. Intermittent fasting and dietary supplementation with 2-deoxy-D-glucose improve functional and metabolic cardiovascular risk factors in rats. FASEB J. 2003;17(9):1133–1134. doi: 10.1096/fj.02-0996fje. [DOI] [PubMed] [Google Scholar]
- 41.Colman RJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325(5937):201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmes D. Metabolism: Fasting induces FGF21 in humans. Nat Rev Endocrinol. 2016;12(1):3. doi: 10.1038/nrendo.2015.202. [DOI] [PubMed] [Google Scholar]
- 43.Fazeli PK, et al. FGF21 and the late adaptive response to starvation in humans. J Clin Invest. 2015;125(12):4601–4611. doi: 10.1172/JCI83349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Look AHEAD Research Group. et al. Cardiovascular effects of intensive lifestyle intervention in type 2 diabetes. N Engl J Med. 2013;369(2):145–154. doi: 10.1056/NEJMoa1212914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Redman LM, Ravussin E. Caloric restriction in humans: impact on physiological, psychological, and behavioral outcomes. Antioxid Redox Signal. 2011;14(2):275–287. doi: 10.1089/ars.2010.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sathananthan M, et al. Six and 12 Weeks of Caloric Restriction Increases β Cell Function and Lowers Fasting and Postprandial Glucose Concentrations in People with Type 2 Diabetes. J Nutr. 2015;145(9):2046–2051. doi: 10.3945/jn.115.210617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinzig A, Heidt M, Fürstenberger G, Marks F, Krieg P. cDNA cloning, genomic structure, and chromosomal localization of a novel murine epidermis-type lipoxygenase. Genomics. 1999;58(2):158–164. doi: 10.1006/geno.1999.5816. [DOI] [PubMed] [Google Scholar]
- 48.Heidt M, Fürstenberger G, Vogel S, Marks F, Krieg P. Diversity of mouse lipoxygenases: identification of a subfamily of epidermal isozymes exhibiting a differentiation-dependent mRNA expression pattern. Lipids. 2000;35(7):701–707. doi: 10.1007/s11745-000-0576-0. [DOI] [PubMed] [Google Scholar]
- 49.Lin Z, et al. Adiponectin mediates the metabolic effects of FGF21 on glucose homeostasis and insulin sensitivity in mice. Cell Metab. 2013;17(5):779–789. doi: 10.1016/j.cmet.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Markan KR, et al. Circulating FGF21 is liver derived and enhances glucose uptake during refeeding and overfeeding. Diabetes. 2014;63(12):4057–4063. doi: 10.2337/db14-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, et al. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat Protoc. 2012;7(6):1068–1085. doi: 10.1038/nprot.2012.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ribeil JA, et al. Gene Therapy in a Patient with Sickle Cell Disease. N Engl J Med. 2017;376(9):848–855. doi: 10.1056/NEJMoa1609677. [DOI] [PubMed] [Google Scholar]
- 53.Gao M, Liu D. Gene therapy for obesity: progress and prospects. Discov Med. 2014;17(96):319–328. [PubMed] [Google Scholar]
- 54.Urbanek BL, et al. Chemoenzymatic synthesis of trehalose analogues: rapid access to chemical probes for investigating mycobacteria. Chembiochem. 2014;15(14):2066–2070. doi: 10.1002/cbic.201402288. [DOI] [PubMed] [Google Scholar]
- 55.Meints LM, et al. Rapid One-step Enzymatic Synthesis and All-aqueous Purification of Trehalose Analogues. J Vis Exp. 2017;10(120):3791. doi: 10.3791/54485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt S, Joost HG, Schürmann A. GLUT8, the enigmatic intracellular hexose transporter. Am J Physiol Endocrinol Metab. 2009;296(4):E614–E618. doi: 10.1152/ajpendo.91019.2008. [DOI] [PubMed] [Google Scholar]
- 57.Leturque A, Brot-Laroche E, Le Gall M. GLUT2 mutations, translocation, and receptor function in diet sugar managing. Am J Physiol Endocrinol Metab. 2009;296(5):E985–E992. doi: 10.1152/ajpendo.00004.2009. [DOI] [PubMed] [Google Scholar]
- 58.Colville CA, Seatter MJ, Jess TJ, Gould GW, Thomas HM. Kinetic analysis of the liver-type (GLUT2) and brain-type (GLUT3) glucose transporters in Xenopus oocytes: substrate specificities and effects of transport inhibitors. Biochem J. 1993;290(Pt 3):701–706. doi: 10.1042/bj2900701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rundell SR, et al. Deoxyfluoro-d-trehalose (FDTre) analogues as potential PET probes for imaging mycobacterial infection. Org Biomol Chem. 2016;14(36):8598–8609. doi: 10.1039/C6OB01734G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab. 2012;23(7):351–363. doi: 10.1016/j.tem.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Wahli W, Braissant O, Desvergne B. Peroxisome proliferator activated receptors: transcriptional regulators of adipogenesis, lipid metabolism and more... Chem Biol. 1995;2(5):261–266. doi: 10.1016/1074-5521(95)90045-4. [DOI] [PubMed] [Google Scholar]
- 62.Yu Z, Schneider C, Boeglin WE, Brash AR. Epidermal lipoxygenase products of the hepoxilin pathway selectively activate the nuclear receptor PPARalpha. Lipids. 2007;42(6):491–497. doi: 10.1007/s11745-007-3054-4. [DOI] [PubMed] [Google Scholar]
- 63.Boeglin WE, Kim RB, Brash AR. A 12R-lipoxygenase in human skin: mechanistic evidence, molecular cloning, and expression. Proc Natl Acad Sci USA. 1998;95(12):6744–6749. doi: 10.1073/pnas.95.12.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soufi N, et al. Inhibiting monoacylglycerol acyltransferase 1 ameliorates hepatic metabolic abnormalities but not inflammation and injury in mice. J Biol Chem. 2014;289(43):30177–30188. doi: 10.1074/jbc.M114.595850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bassily RW, el-Sokkary RI, Silwanis BA, Nematalla AS, Nashed MA. An improved synthesis of 4-azido-4-deoxy- and 4-amino-4-deoxy-alpha,alpha-trehalose and their epimers. Carbohydr Res. 1993;239:197–207. doi: 10.1016/0008-6215(93)84215-R. [DOI] [PubMed] [Google Scholar]
- 67.DeBosch BJ, Kluth O, Fujiwara H, Schürmann A, Moley K. Early-onset metabolic syndrome in mice lacking the intestinal uric acid transporter SLC2A9. Nat Commun. 2014;5:4642. doi: 10.1038/ncomms5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.