Abstract
Although a subset of clear cell renal cell carcinoma (ccRCC) patients respond to immune checkpoint blockade (ICB), predictors of response remain uncertain. We investigated whether abnormal expression of endogenous retroviruses (ERVs) in tumors is associated with local immune checkpoint activation (ICA) and response to ICB. Twenty potentially immunogenic ERVs (πERVs) were identified in ccRCC in The Cancer Genome Atlas data set, and tumors were stratified into 3 groups based on their expression levels. πERV-high ccRCC tumors showed increased immune infiltration, checkpoint pathway upregulation, and higher CD8+ T cell fraction in infiltrating leukocytes compared with πERV-low ccRCC tumors. Similar results were observed in ER+/HER2− breast, colon, and head and neck squamous cell cancers. ERV expression correlated with expression of genes associated with histone methylation and chromatin regulation, and πERV-high ccRCC was enriched in BAP1 mutant tumors. ERV3-2 expression correlated with ICA in 11 solid cancers, including the 4 named above. In a small retrospective cohort of 24 metastatic ccRCC patients treated with single-agent PD-1/PD-L1 blockade, ERV3-2 expression in tumors was significantly higher in responders compared with nonresponders. Thus, abnormal expression of πERVs is associated with ICA in several solid cancers, including ccRCC, and ERV3-2 expression is associated with response to ICB in ccRCC.
Keywords: Immunology, Oncology
Keywords: Immunotherapy
Human endogenous retrovirus expression was identified as a feature of many cancers and correlated with exceptional response to immune checkpoint blockade in renal cell carcinoma.
Introduction
Immune checkpoint blockade (ICB) leads to durable objective responses in several cancer types (1). A high mutation burden, from exposure to exogenous carcinogens (2, 3) or intrinsic defects in DNA repair and replication (4, 5), predicts response to ICB in some cancer types (6), presumably because of somatic neoantigens. Further, expression of certain exogenous viruses in tumors, such as Epstein-Barr virus in gastric cancer (7) and NK/T cell lymphoma (8) and Merkel cell polyomavirus in Merkel cell cancer (9, 10), is also associated with response to ICB, presumably because of viral antigens. However, some cancers, such as clear cell renal cell carcinoma (ccRCC), have clinically significant and durable responses to ICB (11), despite low mutation burden and absence of known exogenous viral infection.
A recent study (12) reported that loss of the chromatin-modifying gene PBRM1 correlates with response to ICB in pretreated ccRCC patients. Intriguingly, in multiple cohorts of ccRCC patients, tumors with PBRM1 loss have lower levels of the CD8+ T cell marker (CD8A), IFN-γ (IFNG), and immune checkpoint genes compared with tumors with intact PBRM1 (12), although the mechanism underlying these correlations is unknown. Recent studies also show, that ccRCCs, while having low overall mutation burden, are enriched in frameshift mutations, which may be more immunogenic (13). However, the relationship between levels of frameshift mutations and response to ICB remains unclear. Emerging data also suggest a role for the metabolic environment in balancing (14) or suppressing (15) antitumor immunity in ccRCC. In summary, the mechanisms of response to ICB in ccRCC are currently unknown.
A substantial fraction of the human genome contains endogenous retroviruses (ERVs) (16), the expression of which is normally silenced in most somatic tissues. However, 66 ERVs are known to be transcribed in humans (17), and their expression has been reported in multiple cancers (18). Some recently integrated (19) and well-preserved (19) ERVs, such as those in the ERVK family, are known to retain a functional gag gene (20) and an open-reading frame in pol and env genes (20), and their simian equivalents are known to induce immune response in Indian rhesus macaques (21). Thus, abnormal expression of some potentially immunogenic ERVs (πERVs) in tumors may elicit an antitumor immune response spearheaded by CD8+ T cells. Tumors may progress by blocking this immune response through upregulation of immune checkpoint pathways, making them sensitive to ICB.
In this study, using RNA-sequencing (RNA-seq) data and previously published (18) ERV expression data of (mostly primary) tumors (n = 472 for ccRCC, n = 4,438 for 20 other cancers) from The Cancer Genome Atlas (TCGA), and a cohort of metastatic ccRCC patients treated with single-agent PD-1/PD-L1 blockade (n = 24) at two institutions, we evaluated the possibility that expression of πERVs in tumors induces local immune checkpoint activation (ICA) in a subset of tumors and associates with responsiveness to ICB.
Results
πERVs are abundant in 4 solid cancers from TCGA.
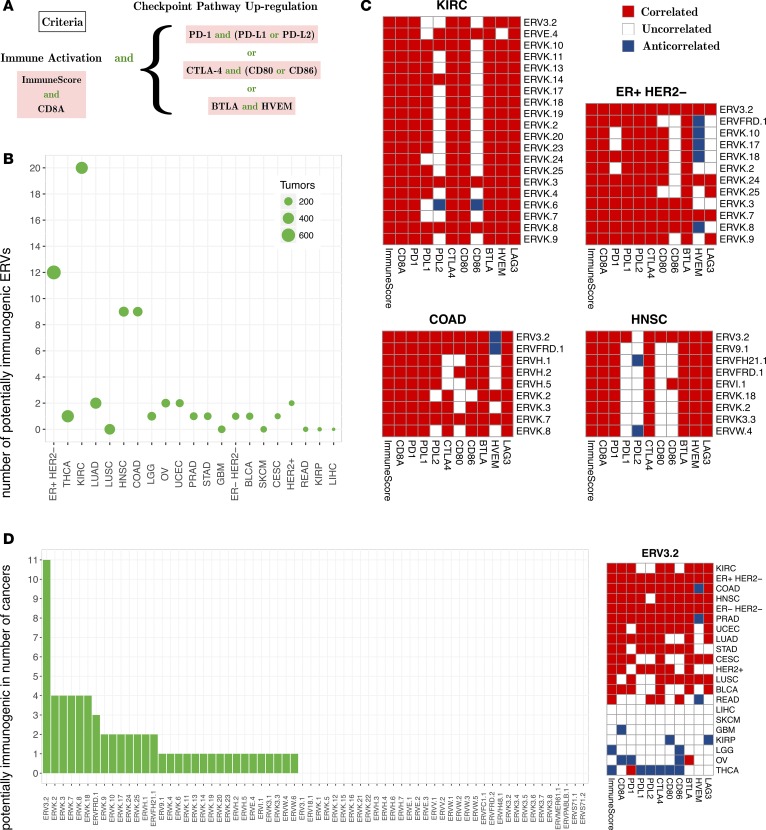
To identify πERVs (see the Methods for details), we evaluated 21 solid cancers from TCGA for correlation between expression levels (18) of 66 transcribed ERVs (17) and RNA-seq–based evidence of local ICA. As shown in Figure 1A, ICA criteria included markers of immune activation, namely overall immune infiltration (“ImmuneScore” from ESTIMATE, ref. 22) and expression of the cytotoxic T cell marker CD8A, and markers of checkpoint pathway upregulation, namely expression of genes in the PD-1, CTLA-4, and BTLA/HVEM pathways.
Figure 1. Potentially immunogenic ERVs are abundant in 4 solid cancers from TCGA.
(A) Immune checkpoint activation criteria used to identify potentially immunogenic ERVs (πERVs) in each solid cancer type. (B) The number of πERVs in each solid cancer type identified 4 cancer types with an unusually high number of πERVs. (C) Correlation (Spearman) between expression of πERVs (rows) and levels of immunological variables (columns) in the 4 cancer types. (D) ERV3-2 was identified as a πERV in 11 different cancer types. Cancer type acronyms are standard TCGA abbreviations (https://tcga-data.nci.nih.gov/).
Results of this analysis (summarized in Figure 1B) showed that πERVs are abundant in 4 cancers, namely ccRCC (from TCGA KIRC database, ER+/HER2− breast cancer (ER+ HER2−), colon cancer (COAD), and head and neck squamous cell cancer (HNSC). In these cancers, expression of 9–20 πERVs correlated with ICA, while expression of only 0 to 2 πERVs correlated with ICA in other cancers (Figure 1B). πERVs were most abundant in ccRCC (KIRC), where expression of 20 πERVs correlated with ICA, most of which (18 of 20) were members of the ERVK family (Figure 1C). While most ERVs were identified as πERVs in 0 or 1 cancer type, and several were identified across multiple diseases, ERV3-2 was identified as a πERV in 11 different solid cancers (Figure 1D), including the 4 cancers named above.
Expression of πERVs defines subtypes with differential ICA in ccRCC.
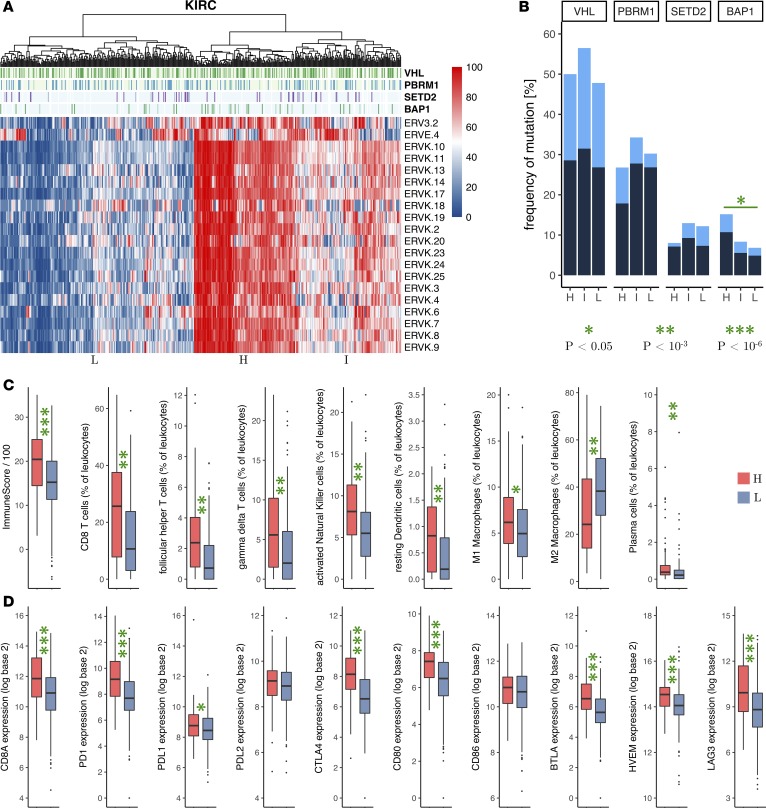
In TCGA ccRCC (KIRC) cohort, the 20 πERVs were mostly coexpressed. Hierarchical clustering of tumors by percentile expression of these 20 πERVs identified 3 distinct subtypes corresponding to high, intermediate, and low expression of πERVs (Figure 2A). Since loss-of-function mutations in chromatin regulatory genes (including PBRM1, SETD2, and BAP1) are frequently observed in ccRCC (23), and a recent study (12) reported an association between PBRM1 loss and response to non-first-line ICB in pretreated ccRCC, we compared the frequency of mutation of these genes in the 3 πERV expression–based subtypes in KIRC. As shown in Figure 2B, although there was no significant enrichment of VHL, PBRM1, or SETD2 mutations in the ccRCC tumors with a high expression of πERVs (πERV-high ccRCC tumors), there was a statistically significant enrichment of BAP1 mutations in the πERV-high tumors compared with ccRCC tumors with a low expression of πERVs (πERV-low tumors) (15.2% vs. 6.8%, odds ratio 2.44 [95% CI: 1.15–5.16], P = 0.028).
Figure 2. Expression of πERVs defines subtypes with differential immune checkpoint activation in ccRCC (KIRC).
(A) Hierarchical clustering of tumors from TCGA (columns) by expression (percentile) of πERVs (rows) stratifies tumors into 3 subtypes (high [H], intermediate [I], and low [L]). (B) Frequency of VHL, PBRM1, SETD2, and BAP1 mutations (dark, truncating mutations; light, other nonsynonymous mutations) in the 3 subtypes. Comparison of (C) overall immune infiltration in tumors (“ImmuneScore”) and fractional composition of tumor-infiltrating leukocytes and (D) mRNA expression of CD8A (cytotoxic T cell marker) and immune checkpoint genes between πERV-high and πERV-low subtypes. Number of samples: (C) ImmuneScore (119 H, 228 L), all other categories (90 H, 134 L), (D) 119 H and 228 L. P values reported in bar plots and box plots are from Fisher’s exact test and Wilcoxon rank-sum test, respectively (all 2 sided). *P < 0.05, **P < 10–3, ***P < 10–6.
πERV-high tumors also had significantly higher immune infiltration (Figure 2C), a significantly higher fraction of CD8+ T cells among tumor-infiltrating leukocytes (Figure 2C), and significantly higher mRNA expression of the cytotoxic T cell marker CD8A (Figure 2D) compared with πERV-low tumors, indicating immune activation in the πERV-high tumors. In addition, follicular helper T cells, γδ T cells, activated NK cells, resting dendritic cells, and plasma cells constituted a significantly higher fraction of tumor-infiltrating leukocytes in the πERV-high tumors compared with that in πERV-low tumors (Figure 2C). Furthermore, M1 macrophages were more abundant in πERV-high tumors, whereas M2 macrophages were more abundant in πERV-low tumors (Figure 2C).
πERV-high tumors also had significantly higher mRNA expression of several checkpoint genes (PD-1, PD-L1, CTLA-4, CD80, BTLA, HVEM, LAG3) compared with πERV-low tumors (Figure 2D), indicating checkpoint pathway upregulation in πERV-high tumors. Interestingly, expression levels of most πERVs correlated with expression levels of PD-L1 but not PD-L2 and CD80 but not CD86 (Figure 1C). Consistently, in contrast to PD-L1 and CD80, PD-L2 and CD86 were not differentially expressed in πERV-high versus πERV-low tumors (Figure 2D).
Expression of ERV3-2 predicts response to ICB in ccRCC.
ccRCC (KIRC) showed the strongest evidence of πERV-associated ICA among the 21 solid cancers (Figure 1B), and ERV3-2 showed the most consistent correlation with ICA among the 66 transcribed ERVs (17) (Figure 1D). Consequently, we evaluated the expression of ERV3-2 in ccRCC tumors as a predictor of response to ICB.
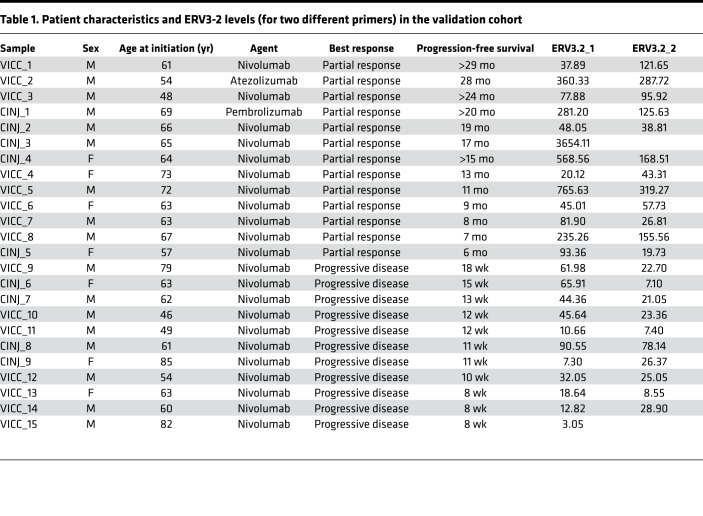
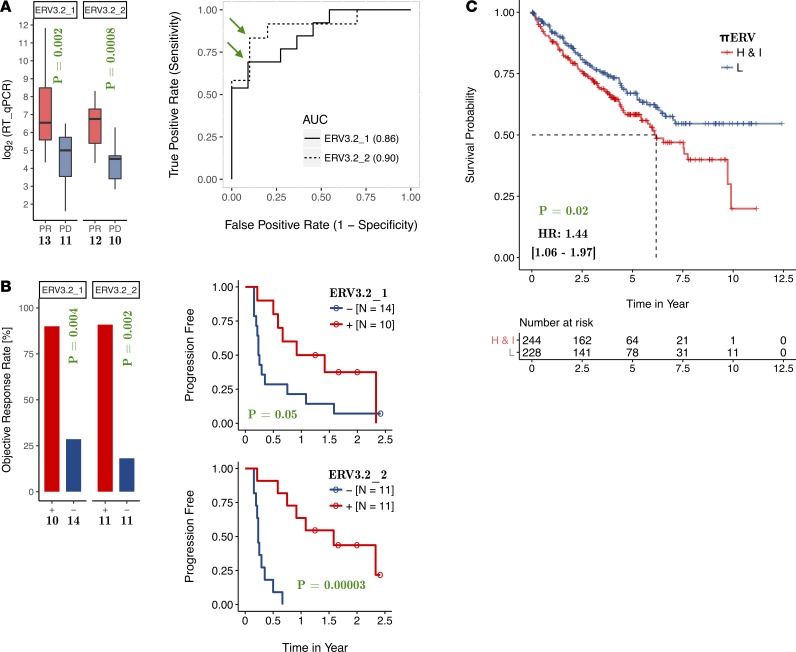
The RNA level of ERV3-2 was measured by real-time quantitative PCRs (RT-qPCRs) in tumors of 24 metastatic ccRCC patients treated with single-agent PD-1/PD-L1 antibody at the Vanderbilt-Ingram Cancer Center and the Rutgers Cancer Institute of New Jersey (Table 1). The cohort consisted of 13 patients with partial response, with progression-free survival of 6 months or longer, and 11 nonresponders who had immediate progressive disease. The RNA level of ERV3-2 was quantified using 2 different primers (see the Methods for details), referred to here as ERV3-2_1 and ERV3-2_2 (1 responder and 1 nonresponder failed in the case of ERV3-2_2). RNA levels of ERV3-2, as measured by either primer, were significantly higher (P = 0.002 for ERV3-2_1, P = 0.0008 for ERV3-2_2) in tumors from responders compared with tumors from nonresponders and were an excellent predictor of response to ICB (area under the receiver operating characteristic [ROC] curve: 0.86 for ERV3-2_1, 0.90 for ERV3-2_2) in the preliminary analysis, based on this collection of samples (Figure 3A).
Table 1. Patient characteristics and ERV3-2 levels (for two different primers) in the validation cohort.
Figure 3. RNA expression of ERV3-2 predicts the response to immune checkpoint blockade in ccRCC.
(A) Expression of ERV3-2 is significantly higher in tumors from responders compared with tumors from nonresponders and is an excellent predictor of response to immune checkpoint blockade for both primers. Green arrows mark the optimal cutoffs that were subsequently used to stratify patients into ERV3-2+ or ERV3-2− groups for consistency checks. (B) The ERV3-2+ group has significantly higher objective response rates and longer progression-free survival compared with the ERV3-2− group for both primers. (C) In contrast, πERV-high/intermediate subtypes have shorter overall survival under standard therapy compared with πERV-low subtype in TCGA ccRCC (KIRC) cohort. The number of samples is specified in each panel. P values reported in bar plots, box plots, and Kaplan-Meier plots are from Fisher’s exact test, Wilcoxon rank-sum test, and log-rank tests, respectively (all 2 sided).
As a consistency check, patients were classified into ERV3-2+ and ERV3-2− groups, based on whether their tumors had higher or lower expression of ERV3-2 compared with the optimal cutoff inferred from the ROC curves (marked by green arrows in Figure 3A). As shown in Figure 3B, ERV3-2+ patients had significantly higher objective response rates compared with ERV3-2− patients for both ERV3-2_1 (90.0% vs. 28.6%, odds ratio 22.5 [95% CI: 2.1–240.5], P = 0.004) and ERV3-2_2 (90.9% vs. 18.2%, odds ratio 45.0 [95% CI: 3.5–584.3], P = 0.002). Consistently, ERV3-2+ patients had longer progression-free survival (Figure 3B) compared with ERV3-2− patients for both ERV3-2_1 (hazard ratio 0.53 [95% CI: 0.27–1.02], P = 0.05) and ERV3-2_2 (hazard ratio 0.15 [95% CI: 0.05–0.44], P = 0.00003).
These results suggest that ccRCC tumors with sufficiently high expression of ERV3-2 may be enriched in the pool of tumors sensitive to ICB. This is particularly interesting because patients with πERV-high and πERV-intermediate tumors have significantly shorter overall survival (hazard ratio 1.44 [95% CI: 1.06–1.97], P = 0.02) compared with patients with πERV-low tumors under standard therapy for ccRCC (Figure 3C). This is consistent with the enrichment of BAP1 mutations in the πERV-high subtype of ccRCC (Figure 2B), as BAP1 mutations are associated with poor prognosis in ccRCC (24).
Expression of πERVs defines subtypes with differential ICA in ER+ HER2−, COAD, and HNSC.
Like KIRC, πERV expression–based subtypes were also observed in ER+ HER2−, COAD, and HNSC (Supplemental Figure 1A, Supplemental Figure 2A, and Supplemental Figure 3A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121522DS1) in TCGA data. Thus, a clearly identifiable subset of tumors displays broad transcriptional activation of πERVs in these 4 cancers.
Similar to KIRC, the πERV-high tumors had significantly higher immune infiltration, a significantly higher fraction of CD8+ T cells in infiltrating leukocytes, and significantly higher mRNA expression of the cytotoxic T cell marker CD8A compared with πERV-low tumors in these 3 cancers (Supplemental Figure 1, B–D; Supplemental Figure 2, B–D; and Supplemental Figure 3, B–D), indicating immune activation in the πERV-high subtype in all 4 cancers. Furthermore, M1 macrophages were more abundant in the πERV-high subtype of ER+ HER2− and COAD (but not HNSC), whereas M2 or M0 macrophages were more abundant in the πERV-low subtype of all 3 cancers (Supplemental Figure 1C, Supplemental Figure 2C, and Supplemental Figure 3C). Additionally, activated memory CD4+ T cells in COAD and HNSC and follicular helper T cells in HNSC comprised a significantly higher fraction of infiltrating leukocytes and regulatory T cells in COAD comprised a significantly lower fraction of T cells in the πERV-high tumors compared with the πERV-low tumors (Supplemental Figure 1C, Supplemental Figure 2C, and Supplemental Figure 3C).
As in KIRC, πERV-high tumors had significantly higher mRNA expression of checkpoint genes in the PD-1 and CTLA-4 pathways compared with πERV-low tumors in ER+ HER2− and COAD (Supplemental Figure 1D and Supplemental Figure 2D), indicating checkpoint pathway upregulation in the πERV-high tumors in these cancers. Interestingly, unlike that in ER+ HER2− and COAD, expression of most πERVs in HNSC did not correlate with the expression of the ligands of PD-1 and CTLA-4 (Figure 1C). Consistently, PD-L1 and PD-L2 were not differentially expressed in πERV-high versus πERV-low tumors in HNSC (Supplemental Figure 3D). Instead, the BTLA/HVEM pathway and LAG3 were upregulated in πERV-high tumors in HNSC (Supplemental Figure 3D).
πERV-high tumors in these cancers were also enriched in tumors with known potential predictors of ICA, namely in tumors with APOBEC mutagenesis (25) in ER+ HER2−, microsatellite instability–high (MSI-H) tumors in COAD, and HPV+ tumors in HNSC (Supplemental Figure 1E, Supplemental Figure 2E, and Supplemental Figure 3E). Although hypermutation due to APOBEC and MSI-H etiologies is known to be associated with ICA in ER+ HER2− and COAD, respectively (6), a detailed analysis (Supplemental Figure 4) demonstrated that πERV-high tumors showed evidence of immune activation and checkpoint pathway upregulation compared with πERV-low tumors in ER+ HER2−, both with and without APOBEC mutagenesis (25), as well as in both MSI-H and MSI-L/MSS in COAD, and in both HPV+ and HPV− in HNSC. These results demonstrate that πERV expression associates with ICA independently of APOBEC mutagenesis (25) status in ER+ HER2−, MSI-H status in COAD, and HPV status in HNSC.
Transcriptomic correlates of ERV expression point to chromatin alterations.
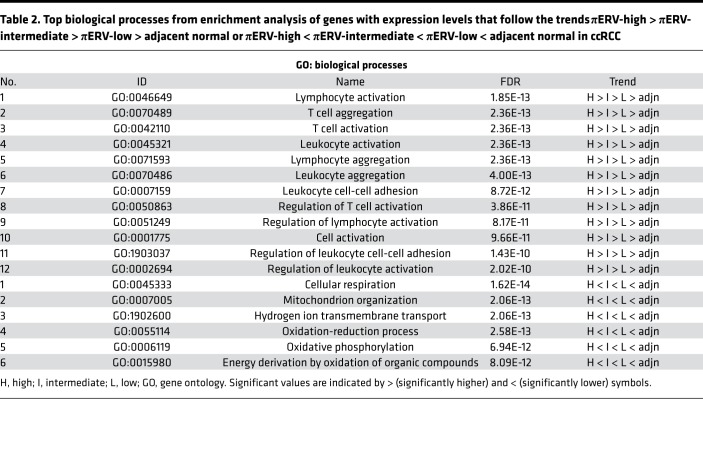
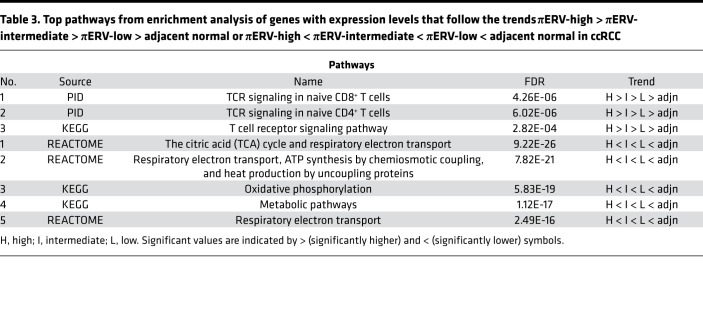
For further insight, we compared the gene expression profiles of the 3 πERV subtypes of ccRCC tumors, and tumor-adjacent normal tissues in TCGA data set. Expression levels of 1,048 genes followed the following trend: πERV-high > πERV-intermediate > πERV-low > adjacent normal; whereas 1,103 genes followed the following trend: πERV-high < πERV-intermediate < πERV-low < adjacent normal (where > and < represent statistically significant at P < 0.05 in 2-sided Wilcoxon rank-sum test, higher and lower, respectively). Enrichment analysis (Tables 2 and 3) of these two sets of genes using the ToppGene suite (26) showed that the former set was enriched in immune activation genes, whereas the latter set was enriched in genes associated with mitochondrial respiration, which often plays a role in determining disease behavior.
Table 2. Top biological processes from enrichment analysis of genes with expression levels that follow the trends πERV-high > πERV-intermediate > πERV-low > adjacent normal or πERV-high < πERV-intermediate < πERV-low < adjacent normal in ccRCC.
Table 3. Top pathways from enrichment analysis of genes with expression levels that follow the trends πERV-high > πERV-intermediate > πERV-low > adjacent normal or πERV-high < πERV-intermediate < πERV-low < adjacent normal in ccRCC.
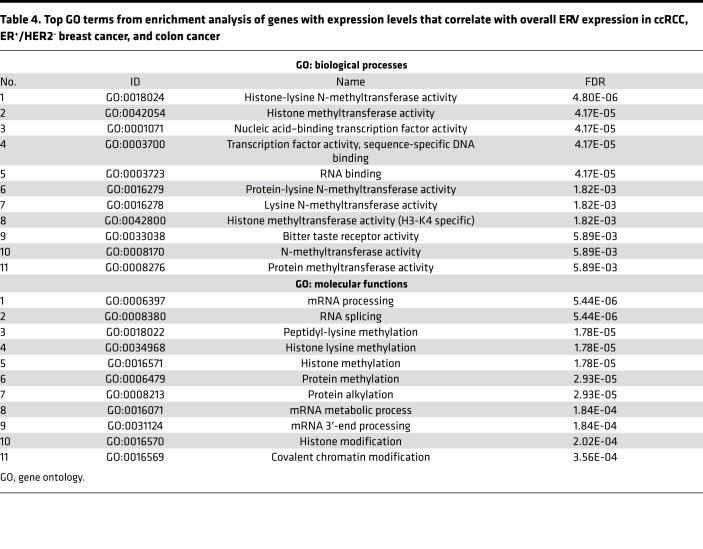
To identify the potential cause of ERV expression, we looked for genes with expression that was correlated with overall ERV expression in each cancer type (see the Methods for details). Six hundred fifty-seven genes satisfied this criterion in KIRC, ER+ HER2−, and COAD. Enrichment analysis of these genes using ToppGene (26) showed that “methyl” (methyltransferase and methylation) and “histone” pathways were significantly associated with overall ERV expression (Table 4). This suggests that epigenetic alterations, specifically those involving modification of histone methylation–based control of gene expression, may be a functional mechanism of ERV expression in these cancers.
Table 4. Top GO terms from enrichment analysis of genes with expression levels that correlate with overall ERV expression in ccRCC, ER+/HER2− breast cancer, and colon cancer.
Discussion
These results demonstrate that elevated expression of a set of πERVs is associated with gene expression-based evidence of ICA in several solid cancers and particularly strikingly in ccRCC. This finding suggests that broad transcriptional activation of πERVs, present in a subset of tumors, may induce antitumor immune response and subsequent upregulation of immune checkpoint pathways, making such tumors sensitive to ICB. Expression of 7 members of the ERVK family has previously been reported to be correlated with the expression of cytotoxins GZMA and PRF1 in ccRCC (18), and we found an association between ERV3-2 expression and response to ICB in a small cohort of 24 metastatic ccRCC patients. Although πERV-high tumors in ccRCC have poor prognosis under standard therapy, such tumors may have significantly improved outcomes with ICB.
In addition to CD8+ T cells, M1 macrophages were more abundant and M2 macrophages were less abundant in πERV-high ccRCC tumors compared with πERV-low tumors in TCGA. Since M1 macrophages have tumor-inhibitory properties and M2 macrophages may be tumor promoting (27), this suggests that πERV-high tumors have a relatively favorable immune microenvironment. Consistently, such enrichment of M1 polarization in macrophages has been previously reported in both mutation burden–associated (5, 6) and exogenous virus–associated (7) immunogenicity in other cancers.
Overall ERV expression in tumors correlated with the expression of genes involved in histone methylation and chromatin regulation in multiple cancers, including ccRCC. This suggests that ERV expression may be induced by some underlying dysfunction in chromatin organization, which plays a key role in the normal epigenetic silencing of ERVs. Of note, there was an enrichment of BAP1 mutations in the πERV-high subtype of KIRC, suggesting that BAP1 dysfunction may lead to some chromatin abnormalities resulting in πERV expression. BAP1 is a deubiquitinase and is known to functionally associate with chromatin regulating complexes (28), and its role in regulation of πERV expression warrants further investigation. A recent study (12) reported an association between loss of PBRM1 and response to non-first-line ICB in pretreated ccRCC. Although we observed no enrichment of PBRM1 mutations in the πERV-high subtype of KIRC, our results also suggest a link between changes in chromatin and an immune phenotype. It is important to consider that these chromatin modifiers exist with obligated loss of heterozygosity in ccRCC, due to the characteristic chromosome 3p deletion, such that extrapolation to other tumor types, such as papillary RCC or bladder cancers harboring these mutations, may be complex. Prior studies have also shown that ccRCC tumors are relatively enriched in potentially immunogenic frameshift mutations caused by small insertion/deletions (13). The relative association among frameshift mutation burden, presence of PBRM1 mutations, πERV expression, and clinical response to ICB needs to be explored in larger prospective studies.
In addition to ccRCC, overexpression of πERVs was also associated with ICA in ER+ HER2−, COAD, and HNSC. πERV overexpression overlapped somewhat with known genomic features potentially associated with ICA in these cancers, such as APOBEC mutagenesis (25) in ER+ HER2−, microsatellite instability in COAD, and HPV infection in HNSC, raising the possibility that πERV expression may be biologically related to these genomic features. However, a closer inspection revealed that πERV overexpression was associated with ICA independently of these features. Moreover, expression of ERV3-2 in particular was correlated with ICA in 11 solid cancers, including the 4 named above. Thus, like hypermutation or exogenous viral expression in tumors, πERV expression in tumors is another major mechanism of ICA operative in multiple solid cancers. Our results suggest that overexpression of πERVs, especially ERV3-2, should be investigated further as a feature of immune response in ccRCC and other solid cancers.
In summary, our results suggest that a set of πERVs is broadly overexpressed in a subset of tumors and is associated with ICA in ccRCC. Analysis of larger cohorts and different cancers, development of clinical-grade assays of πERV expression, and prospective clinical trials are needed to validate πERV expression as a predictor of response to ICB in ccRCC and other solid cancers.
Methods
Processing of TCGA RNA-seq data.
RNA-seqV2–scaled estimates were obtained from Broad Genome Data Analysis Centers (GDAC) (http://gdac.broadinstitute.org) and TCGA data portal (https://tcga-data.nci.nih.gov/docs/publications/tcga/). The data were median-adjusted so that the median-scaled estimate was unity in each sample and was then used as input for the ESTIMATE (22) and CIBERSORT (29) algorithms to quantify the level of immune infiltration in tumor (“ImmuneScore”) and the composition of infiltrating leukocytes. Only unambiguous (P < 0.05) CIBERSORT outputs were retained. For the remainder of the analysis, the median-adjusted data (x) were log transformed to y = log2(1 + 1,023 × x), so that the lowest possible expression was 0 units and the median expression was 10 units in each sample.
Source of the remaining TCGA data.
ERV expression data for a large subset of TCGA cohort were downloaded from a recent study (18) that quantified normalized expression levels of 66 transcribed ERVs (17) by direct remapping from the raw RNA-seq data. Only tumors for which both mRNA and ERV expression data were available were included in the analyses (n = 472 for ccRCC, n = 4,438 for 20 other cancers). ERBB2 focal copy number data and ESR1 mRNA expression data obtained from Broad GDAC were used to classify breast cancer into clinical subtypes (ER+/HER2−, ER−/HER2−, HER2+), and each subtype was analyzed separately. Clinical and mutation data were downloaded from cBioPortal (http://www.cbioportal.org), APOBEC enrichment data were compiled from P-MACD (25) files from Broad GDAC, and microsatellite status and HPV status were compiled from auxiliary files from TCGA Data Portal.
Search for πERVs.
An ERV was considered potentially immunogenic if its expression was correlated with both immune activation and checkpoint pathway upregulation (Figure 1A). The immune activation criterion was considered satisfied if the expression of an ERV correlated with immune infiltration (“ImmuneScore” from ESTIMATE, ref. 22) and mRNA expression of CD8A (marker of CD8+ T cell infiltration). The checkpoint pathway upregulation criterion was considered satisfied if the expression of an ERV correlated with the PD-1 pathway (i.e., PD-1 and at least one of its ligands) or the CTLA-4 pathway (i.e., CTLA-4 and at least one of its ligands) or the BTLA/HVEM pathway. In each cancer type, we tested whether any of the 66 transcribed ERVs (17) satisfied both immune activation and checkpoint pathway upregulation criteria.
Gene expression-based enrichment analyses.
By defining overall ERV expression as the total fraction of RNA-seq reads that mapped to the 66 transcribed ERVs (17), genes with expression that correlated (Spearman Rho > 0, P < 0.05) with overall ERV expression in each cancer type were identified. Six hundred fifty-seven genes satisfied this criterion in KIRC, ER+ HER2−, and COAD. An enrichment analysis of these genes was performed using the ToppGene (26) suite to identify enriched gene ontology terms.
Patient samples in the validation cohort.
Formalin-fixed, paraffin-embedded (FFPE) metastatic ccRCC samples from patients treated with single-agent PD-1/PD-L1 blockade were collected at the Vanderbilt-Ingram Cancer Center (using an IRB-exempt waiver of consent, approved by the Vanderbilt University Medical Center Human Research Protection Program) and the Rutgers Cancer Institute of New Jersey (with approval from the Rutgers Institutional Review Board). All tissues were evaluated by a pathologist using hematoxylin and eosin staining, and only the samples containing ≥70% tumor cells were included in this study. The cohort included 13 patients who experienced partial response, with progression-free survival of at least 6 months, and 11 patients who demonstrated immediate progressive disease.
Quantification of ERV3-2 expression in the validation cohort.
Total RNA isolation was performed using the RNAeasy FFPE Kit (Qiagen). DNAse treatment was performed during RNA isolation using RNase-free DNase I (Qiagen). RNA quality and concentration were assessed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). First-strand cDNA synthesis was performed using 250 ng total RNA, random hexamers, and the SuperScript IV Reverse Transcriptase Kit (Life Technologies). RT-qPCRs were performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using the SYBR green master mix reagent (Life Technologies). Primer sequences for ERV3-2 and HPRT1 (fwd: GACACTGGCAAAACAATGCAGAC, rev: TGGCTTATATCCAACACTTCGTGG) were designed using PrimerBank (30). ERV3-2 was quantified using two different primers: ERV3-2_1 (fwd: 5′-CAAGAGGCGGCATAGAAGCAA-3′, rev: 5′-GGAGAGTAGCTTGGGGTTTCA-3′) and ERV3-2_2 (fwd: 5′-AGCCATTTACAAAGAAAGGGGAC-3′, rev: 5′-CTATGCCGCCTCTTGTCTGAT-3′). All analyses were performed in triplicate, and relative RNA levels were determined using HPRT1 as an endogenous internal control. A HeLa control RNA sample was included for interplate calibration. ERV3-2 expression level was calculated using the ΔΔCt method.
Statistics.
P values reported in bar plots, box plots, and Kaplan-Meier plots are from Fisher’s exact test, Wilcoxon rank-sum test, and log-rank tests, respectively. Statistical significance was assessed at P < 0.05 in 2-sided tests. False discovery rates used the Benjamini-Hochberg procedure, and hazard ratios are from univariate cox regression. Two variables were considered correlated if they had Spearman Rho > 0 and P < 0.05, anticorrelated if they had Spearman Rho < 0 with P < 0.05, and uncorrelated if P ≥ 0.05.
Study approval.
Work in this study was performed with the approval of the Rutgers Institutional Review Board. Informed consent was deemed unnecessary by the Institutional Review Board of the Vanderbilt University Medical Center Human Research Protection Program.
Author contributions
The study was designed by AP, AADC, SG, GB, and WKR. Data collection was performed by AP and AADC. Samples and clinical data were analyzed by KEB, MS, GR, CCS, BGV, SG, and WKR. The literature search was performed by AP, AADC, SG, GB, and WKR. Figures were created and data analysis was performed by AP and AADC. Data interpretation was performed by AP, AADC, SG, GB, and WKR. Writing was performed by AP, AADC, KEB, SG, GB, and WKR.
Supplementary Material
Acknowledgments
The study was supported by NIH grants R01-CA198482 (to WKR), K24-CA172355 (to WKR), T32-CA009582-28 (to AADC), and P30-CA072720 (to AP, MS, GR, SG, and GB); a Department of Defense grant CA160728P1 (to WKR); the American Association for Cancer Research — Kure It Research Grant for Immunotherapy in Kidney Cancer (to WKR); a Cancer Research Institute Irvington post-doctoral fellowship (to KEB); a New Jersey Commission on Cancer Research post-doctoral fellowship (to AP); an anonymous gift for precision medicine (to SG); and the Val Skinner Foundation (to SG). Some of this work was performed at the Aspen Center for Physics, which is supported by National Science Foundation grant PHY-1607611 (to AP, AADC, and GB).
Version 1. 08/23/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2018;3(16):e121522. https://doi.org/10.1172/jci.insight.121522.
Contributor Information
Anshuman Panda, Email: ap1022@scarletmail.rutgers.edu.
Aguirre A. de Cubas, Email: a.decubas@vanderbilt.edu.
Joshua Kra, Email: jk1393@rwjms.rutgers.edu.
Tina Mayer, Email: mayertm@cinj.rutgers.edu.
Jonathan S. Serody, Email: jonathan_serody@med.unc.edu.
Shridar Ganesan, Email: ganesash@cinj.rutgers.edu.
Gyan Bhanot, Email: gbhanot@rci.rutgers.edu.
W. Kimryn Rathmell, Email: kimryn.rathmell@Vanderbilt.edu.
References
- 1.Kim JW, Eder JP. Prospects for targeting PD-1 and PD-L1 in various tumor types. Oncology (Williston Park, NY) 2014;28 Suppl 3:15–28. [PubMed] [Google Scholar]
- 2.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehnert JM, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest. 2016;126(6):2334–2340. doi: 10.1172/JCI84940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panda A, et al. Identifying a clinically applicable mutational burden threshold as a potential biomarker of response to immune checkpoint therapy in solid tumors. JCO Precis Oncol. 2017:2017. doi: 10.1200/PO.17.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panda A, et al. Immune activation and benefit From avelumab in EBV-positive gastric cancer. J Natl Cancer Inst. 2018;110(3):316–320. doi: 10.1093/jnci/djx213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwong YL, et al. PD1 blockade with pembrolizumab is highly effective in relapsed or refractory NK/T-cell lymphoma failing l-asparaginase. Blood. 2017;129(17):2437–2442. doi: 10.1182/blood-2016-12-756841. [DOI] [PubMed] [Google Scholar]
- 9.Nghiem PT, et al. PD-1 blockade with pembrolizumab in advanced merkel-cell carcinoma. N Engl J Med. 2016;374(26):2542–2552. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufman HL, et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016;17(10):1374–1385. doi: 10.1016/S1470-2045(16)30364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao D, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turajlic S, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18(8):1009–1021. doi: 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 14.Ascierto ML, et al. The intratumoral balance between metabolic and immunologic gene expression Is associated with anti-PD-1 response in patients with renal cell carcinoma. Cancer Immunol Res. 2016;4(9):726–733. doi: 10.1158/2326-6066.CIR-16-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siska PJ, et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight. 2017;2(12):93411. doi: 10.1172/jci.insight.93411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 17.Mayer J, Blomberg J, Seal RL. A revised nomenclature for transcribed human endogenous retroviral loci. Mob DNA. 2011;2(1):7. doi: 10.1186/1759-8753-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohn O, Hanke K, Bannert N. HERV-K(HML-2), the best preserved family of HERVs: endogenization, expression, and implications in health and disease. Front Oncol. 2013;3:246. doi: 10.3389/fonc.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löwer R, et al. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90(10):4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu HL, et al. Identification and spontaneous immune targeting of an endogenous retrovirus K envelope protein in the Indian rhesus macaque model of human disease. Retrovirology. 2016;13:6. doi: 10.1186/s12977-016-0238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapur P, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol. 2013;14(2):159–167. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45(9):970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan A, et al. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep. 2015;5:14273. doi: 10.1038/srep14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaFave LM, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21(11):1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40(Database issue):D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.