Abstract
Chronic kidney disease (CKD) leads to decreased sensitivity to the metabolic effects of insulin, contributing to protein energy wasting and muscle atrophy. Targeted metabolomics profiling during hyperinsulinemic-euglycemic insulin clamp testing may help identify aberrant metabolic pathways contributing to insulin resistance in CKD. Using targeted metabolomics profiling, we examined the plasma metabolome in 95 adults without diabetes in the fasted state (58 with CKD, 37 with normal glomerular filtration rate [GFR]) who underwent hyperinsulinemic-euglycemic clamp. We assessed heterogeneity in fasting metabolites and the response to insulin to identify potential metabolic pathways linking CKD with insulin resistance. Baseline differences and effect modification by CKD status on changes with insulin clamp testing were adjusted for confounders. Mean GFR among participants with CKD was 37.3 compared with 89.3 ml/min per 1.73 m2 among controls. Fasted-state differences between CKD and controls included abnormalities in tryptophan metabolism, ubiquinone biosynthesis, and the TCA cycle. Insulin infusion markedly decreased metabolite levels, predominantly amino acids and their metabolites. CKD was associated with attenuated insulin-induced changes in nicotinamide, arachidonic acid, and glutamine/glutamate metabolic pathways. Metabolomics profiling suggests disruption in amino acid metabolism and mitochondrial function as putative manifestations or mechanisms of the impaired anabolic effects of insulin in CKD.
Keywords: Metabolism, Nephrology
Keywords: Amino acid metabolism, Insulin, Mitochondria
Targeted metabolomics analysis of the response to insulin clamp testing reveals underlying disruption in amino acid metabolism and mitochondrial function in chronic kidney disease patients.
Introduction
Insulin is a central hormone in the regulation of systemic metabolism. The metabolic effects of insulin include stimulation of cellular glucose uptake, amino acid transport, and glycogen formation, as well as suppression of hepatic gluconeogenesis and adipose tissue lipolysis.
Insulin also exerts additional anabolic effects, including stimulation of protein synthesis via activation of mTOR signaling. Defects in insulin sensitivity and secretion lead to diabetes, related vascular complications, and skeletal muscle wasting, which contribute to morbidity and disability (1–3).
Patients with chronic kidney disease (CKD) often exhibit decreased sensitivity to the metabolic actions of insulin, contributing to protein energy wasting (4, 5). Impaired insulin signaling in skeletal muscle after binding of insulin to its receptor is the best-described mechanism through which CKD promotes insulin resistance, and this may also contribute to muscle atrophy (6). Indeed, animal models of CKD demonstrate the postreceptor defect in insulin signaling leads to skeletal muscle protein breakdown, which provides amino acids to fuel cellular respiration and maintain energy homeostasis, consistent with the augmented skeletal muscle catabolism observed in patients with advanced CKD (7, 8).
The goal of this study was to extend our understanding of how CKD and insulin administration alter systemic metabolism. We evaluated nondiabetic human subjects with and without moderate-to-severe CKD using hyperinsulinemic-euglycemic clamps, and we applied a targeted metabolomics platform to interrogate the plasma metabolome before and after insulin infusion. We hypothesized that CKD and insulin infusion would each be associated with large differences in the plasma metabolome. We further hypothesized that CKD would modify the effects of insulin infusion on the plasma metabolome, thereby allowing identification of new CKD-related effects on insulin sensitivity.
Results
Study participants.
We studied 58 participants with CKD, defined by estimated glomerular filtration rate (eGFR) <60 ml/min per 1.73 m2, and 37 control subjects, defined by eGFR ≥60 ml/min per 1.73 m2. Adequate fasting plasma was available for 93 participants, and of these, 58 (40 participants with CKD) had both pre- and during-hyperinsulinemic–euglycemic clamp plasma samples (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122219DS1). The mean age was 62.3 years; 49% were women, and 76% were European descent (Table 1). Compared with controls, participants with CKD had more prevalent cardiovascular disease, were older, and had greater body weight, greater fat mass, lower fat-free mass, lower levels of serum bicarbonate, and greater levels of inflammatory markers. Mean eGFR among participants with CKD was 37.3 ml/min per 1.73 m2 compared with 89.3 ml/min per 1.73 m2 among controls.
Table 1. Demographic characteristics by CKD status of analytic population (n = 95).

CKD is associated with differences in plasma amino acid and TCA cycle metabolism in the fasting state.
Substantial differences in the plasma fasting metabolome were evident among the CKD group compared with the controls (Figure 1). With adjustment, 48% (59 of 124) of detected plasma metabolites were significantly different between the 2 groups. Amino acids and their metabolites composed 41% (25 of 59) of these significant differences. The most pronounced reductions were seen in the glucogenic and ketogenic amino acids tryptophan and tyrosine, as well as the glucogenic amino acid serine (Figure 2A and Supplemental Table 1). Glycine and proline (glucogenic amino acids) were the only amino acids significantly higher in CKD patients compared with controls (Figure 2A and Supplemental Table 1).
Figure 1. Fold changes in fasting metabolites comparing CKD and controls (n = 93), adjusted for age, sex, race/ethnicity (European descent versus non–European descent), and weight.
Figure 2. Comparison of fasting metabolomic profiles and pathway differences between CKD and non-CKD participants (n = 93).
(A) Percent changes in TCA cycle metabolites and amino acid precursors to TCA cycle metabolites, comparing CKD (n = 58) and non-CKD (n = 35) participants. Fasting metabolomic pathways significantly different between CKD and controls. Blue represents glucogenic and ketogenic amino acids and green represents glucogenic amino acids. (B) Results from pathway enrichment analysis. Asterisks denote significance after adjustment for age, sex, race/ethnicity, body weight, and batch. PDH, pyruvate dehydrogenase; PC, pyruvate carboxylase; SDH, succinate dehydrogenase.
Concentrations of individual metabolites in the tryptophan metabolism and kynurenine pathway differed among participants with CKD compared with controls. Altered concentrations of metabolites within the kynurenine pathway included increased kynurenine (43% higher in CKD) with concomitant reductions in xanthurinate (13% lower in CKD) (Table 2 and Supplemental Table 1).
Table 2. Comparison of fasting metabolites between participants with and without CKD.
Several carbohydrate-related and TCA cycle metabolites were significantly altered in CKD, compared with those without CKD, including a marked elevation in fructose and a reduction in sorbitol. No differences were noted in the level of glycolytic intermediates (glucose 1-phosphate, glucose 6-phosphate, fructose 6-phosphate, and fructose 1-phosphate). The most markedly elevated metabolites in CKD were the TCA cycle intermediates fumarate (270% higher) and oxaloacetate (104% higher) (Figure 2A and Table 2). In contrast, there were no significant changes in metabolites downstream from oxaloacetate in the TCA cycle.
Fasting metabolomics reflects alterations in mitochondrial metabolic pathways in CKD.
Pathway analysis using Metaboanalyst demonstrated significant aberration in ubiquinone biosynthesis (P = 8.72 × 10–48) and the TCA cycle (P = 2.74 × 10–12) (Figure 2B) among CKD subjects compared with controls. Disruption in the ubiquinone pathway was characterized by lower tyrosine and higher hydroxyphenylpyruvate and homogentisate concentrations (Table 2). Many metabolic pathways differing in CKD involved amino acid metabolic pathways, including tyrosine, arginine, proline, phenylalanine, and tryptophan metabolism (Figure 2B) (Supplemental Table 2).
Insulin markedly influences the plasma metabolome.
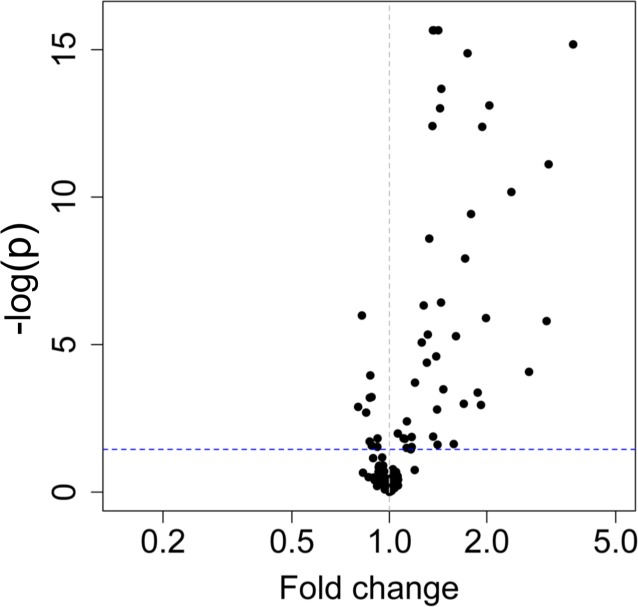
During the hyperinsulinemic-euglycemic clamp in the whole cohort, there were large and highly significant changes in many plasma metabolite concentrations, mostly characterized by reductions in plasma concentrations of amino acids and their associated metabolites (Supplemental Table 3). The largest reductions in individual metabolite concentrations were observed for kynurenine, isoleucine, α linolenic acid, and niacinamide (Figure 3A). The pathways most impacted by insulin infusion were tryptophan metabolism (P = 2.08 × 10–52) and the TCA cycle (P = 4.71 × 10–48) (Figure 3B). With adjustment, the largest differences in individual metabolite concentrations were noted for niacinamide, taurine, prostaglandin E, and linolenic acid (Table 3 and Supplemental Table 3). The greatest reductions observed for subgroups of metabolites involved arachidonic acid/lipid metabolism, fatty acid metabolism, and nicotinamide metabolism (Figure 4).
Figure 3. Comparison of metabolite concentrations before and during the insulin infusion as part of the hyperinsulinemic-euglycemic clamp procedure (n = 60).
(A) Volcano plot of fold change (FC) in metabolites with insulin infusion in entire cohort (n = 60). (B) Pathway analysis of changes with insulin infusion in the overall cohort (n = 60). (C) Pathway analysis of changes with insulin infusion comparing CKD (n = 40) with controls (n = 20). ALA, α linolenic acid; Ile, Isoleucine; L-Kyn, L-kynurenine; AA, arachidonic acid.
Table 3. Differences in plasma metabolites during the hyperinsulinemic-euglycemic clamp, compared with the fasting state.
Figure 4. Comparison of adjusted mean percent change in metabolite subgroups from fasting to during the insulin infusion at euglycemia (n = 60).
Adjusted for age, sex, race/ethnicity, weight, and batch.
CKD modifies the effect of insulin on pathways involving energy metabolism and arachidonic acid metabolism.
CKD was associated with significant differences in insulin response for 17 of 74 metabolites (23%). Most often, CKD was associated with attenuated reductions in plasma metabolite concentrations in response to insulin infusion. Amino acids and their metabolites composed 10 (59%) of the significant changes. With adjustment for baseline concentrations and other covariates, nicotinamide, prostaglandin E, taurine, glutamine, serine, arginine, phenylalanine, and valine were significantly higher in CKD during insulin infusion compared with controls (Table 4 and Supplemental Table 4). In contrast, the plasma glutamate concentration declined to a greater degree with insulin infusion among participants with CKD (Table 4). The principal metabolic pathways for which response to insulin varied among those with CKD compared with controls included nicotinamide metabolism (P = 0.001), arachidonic acid metabolism (P = 0.001), and glutamine and glutamate metabolism (P = 0.002) (Figure 3C) (Supplemental Table 5).
Table 4. Heterogeneity in the plasma metabolomics response to insulin infusion by chronic kidney disease.
The change in metabolites during insulin infusion was weakly correlated with glucose disposal during insulin clamp.
For metabolites demonstrating the largest differential effects of insulin by CKD status, correlations of change in metabolite with insulin sensitivity by insulin clamp (M/ΔI, where M represents the steady-state glucose infusion and ΔI represents the difference between basal and steady-state plasma insulin concentrations) were generally weak, particularly among participants with CKD (Supplemental Table 6). In comparison, correlations of changes in metabolites during insulin infusion with suppression of lipolysis and glucose disposal measured during an oral glucose tolerance test (OGTT) (performed on a separate day) were consistently stronger (Supplemental Table 7)
Discussion
We observed large differences in the fasting plasma metabolome comparing participants with and without CKD, large changes in the plasma metabolome during the hyperinsulinemic-euglycemic clamp in the whole cohort, and significant heterogeneity in the metabolomics response to insulin infusion as a function of CKD status. In the fasting state, notable differences comparing participants with CKD versus without CKD were observed in the TCA cycle, tryptophan metabolism, and metabolites related to biosynthesis of ubiquinone (coenzyme Q), a critical component of mitochondrial electron transport chain (ETC) function. During the hyperinsulinemic-euglycemic clamp, large differences in a broad array of metabolites were observed — largely, reductions in plasma concentrations in amino acids and associated metabolites, as well as profound changes in tryptophan metabolism and TCA cycle metabolic intermediates. We observed significant heterogeneity in the response to insulin by CKD status, with the most marked differences occurring in the metabolism of nicotinamide, a critical metabolite for energy metabolism and redox homeostasis, as well as glutamine and glutamate metabolism. Furthermore, we noted a weak correlation of the changes in these metabolites during insulin infusion versus fasting with glucose disposal rate (M/ΔI) that appeared to be more pronounced in CKD. Together, these findings point to abnormal mitochondrial function as a central mechanism or manifestation of the impaired anabolic response to insulin in CKD.
CKD promotes well-described catabolic and proinflammatory processes, including changes in lipid (9), amino acid (10), and protein metabolism (7, 11) that are linked to energy metabolism (12). CKD results in metabolic acidosis, which has been associated with increased protein degradation through glucocorticoid-dependent mechanisms (13) and augmentation of ATP-dependent proteolytic pathways (8). Indeed, we found marked differences in fasting metabolites between CKD and controls in amino acids and metabolites involved in mitochondrial energy metabolic pathways, which may have implications for both impaired amino acid metabolism and mitochondrial capacity for ATP generation. Anabolism is dependent on mitochondrial ATP production for protein translation, which in turn relies on respiratory fuel availability and efficiency of oxidative phosphorylation. In particular, a decrease in fasting plasma tryptophan levels and evidence for disruption of kynurenine metabolism in CKD compared with controls may reflect impaired de novo synthesis of nicotinamide adenine dinucleotide (NAD) necessary for mitochondrial biogenesis, biosynthesis, and redox homeostasis (via NADP+). This finding is consistent with a prior study demonstrating increased catabolism of tryptophan metabolites and disruption of kynurenine metabolites due to alterations in indoleamine 2,3 dioxygenase in CKD (10). Tryptophan and tyrosine are both glucogenic and ketogenic amino acids and serve as anaplerotic metabolites to replenish TCA cycle intermediates. In addition to its role as an anapleurotic agent, tyrosine is a central precursor for ubiquinone (coenzyme Q) biosynthesis critical for oxidative phosphorylation. Recently, coenzyme Q deficiency has been identified as an underlying mechanism driving oxidative stress and insulin resistance in mice and humans (14). Observed pathway differences in ubiquinone biosynthesis among patients with CKD compared with controls may indicate alteration in the integrity or efficiency of the ETC of the mitochondria, predisposing to mitochondrial oxidant production demonstrated to result in insulin resistance (15, 16). This may also help explain the previously observed uncoupling of muscle mitochondrial oxidative phosphorylation and oxidative stress in CKD (17–19).
Elevated fasting fumarate and oxaloacetate levels among those with CKD may be a hallmark of ETC and TCA cycle inhibition, contributing to mitochondrial oxidative stress associated with CKD (18, 20). Elevated levels of fumarate, a product of complex II (succinate dehydrogenase; SDH) of the ETC, which relies on flavin adenine dinucleotide instead of NAD, leads to increased oxidative stress via succinylation of glutathione (21). SDH is also the only complex that does not maintain the proton gradient (or proton motive force) across the inner mitochondrial membrane that is critical to chemiosmotic coupling powering ATP synthesis via oxidative phosphorylation (22). The high levels of fumarate combined with altered ubiquinone (coenzyme Q) biosynthesis, TCA cycle, and tryptophan/nicotinamide metabolic pathways in CKD suggest heightened activity of complex II, which is known to generate reactive oxygen species at a high rate (23, 24), especially under conditions of ischemia/reperfusion (25) and coenzyme Q depletion. Loss of mitochondrial coenzyme Q has been shown to drive complex II– and complex I–dependent mitochondrial oxidant production (14, 26).
The application of metabolomics to the rigorous hyperinsulinemic-euglycemic clamp method in our study provides more detailed insight into the dynamic metabolic response to insulin. Hyperinsulinemic-euglycemic clamp testing resulted in a pattern of changes in the plasma metabolome, suggesting an anabolic response and stimulation of mitochondrial energy metabolism to power biosynthesis. Insulin is known to activate the mTOR (27), leading to enhanced protein synthesis, mitochondrial activity and proliferation, and resistance to oxidative stress (28). The marked reductions in kynurenine, isoleucine, α linolenic acid, and niacinamide characterize the energetic dependence of the anabolic response and highlight the importance of branch chain amino acids and essential lipids in protein and lipid biosynthesis. Our findings of reductions in kynurenine and niacinamide indicate a role for tryptophan (de novo) and niacinamide (salvage pathway) metabolic pathways in NAD production requisite for the anabolic response to insulin (29). NAD synthesis is considered rate limiting for mitochondrial function. NAD is also essential for activation of the NAD-dependent protein deacetylases (sirtuins) important for activation of PGC-1α stimulation of mitochondrial biogenesis and for augmentation of metabolic and antioxidant activity (30, 31).
Using the hyperinsulinemic-euglycemic clamp as a provocative procedure, we observed significant heterogeneity by CKD status in the plasma metabolomics response to insulin, including abnormalities in pathways important for protein synthesis, inflammation, TCA cycle function, and antioxidant capacity. Interestingly, insulin-induced changes in these metabolites correlated poorly with glucose disposal measured during the clamp procedure, particularly among participants with CKD, suggesting that these changes reflect metabolic abnormalities distinct from insulin-mediated glucose uptake and not necessarily detected using common tests of glucose disposal. Specifically, marked abnormalities were observed in the pathways for glutamine/glutamate metabolism, arachidonic acid metabolism, and nicotinamide metabolism.
Glutamine is a conditionally essential amino acid, particularly important under catabolic and anabolic conditions, with a critical role in renal acid-base regulation via deamination and ammoniagenesis (32, 33), which is heightened with exposure to insulin (34). Glutamine is a precursor for α-ketoglutarate (a TCA cycle intermediate), supports cellular amino acid pools for protein and lipid synthesis, and regulates the mTOR pathway as well as redox homeostasis by serving as a substrate for glutathione (35). Impaired anabolic response to insulin in CKD is suggested by several lines of evidence. First, we observed disruption in glutamine and glutamate metabolic pathways with attenuation in the insulin-induced decline in numerous amino acids. Second, coinciding with attenuated insulin-induced changes in glutamine levels in CKD was a parallel lack of change in the anabolic amino acid metabolite leucic acid, derived from the branch chain amino acid leucine and shown to reverse immobilization induced muscle atrophy (36, 37) through mTOR activation (38). The attenuation of the insulin-induced decline was most marked in nicotinamide, prostaglandin E, and taurine in CKD. Third, CKD patients demonstrate alterations in protein metabolism reflected by weaker correlations of change in amino acid during insulin infusion with insulin sensitivity by (M/ΔI) compared with controls. These changes may indicate blunted insulin-mTOR signaling of mitochondrial biogenesis in the setting of persistent inflammation reflected in proportionately higher prostaglandin E2 levels (39) and abnormalities in arachidonic acid metabolism during insulin challenge. These findings motivate investigation of potentially reversible metabolic pathways linking the hypercatabolism associated with CKD with altered energy homeostasis, acid-base regulation, and loss of lean mass, increasing the risk of morbidity and mortality in CKD (40, 41).
Taken together, our results support 3 distinct metabolic disruptions to mitochondrial metabolism in CKD: altered coenzyme Q biosynthesis, disruption of NAD synthesis via impaired tryptophan metabolism and nicotinamide metabolism, and altered taurine metabolism.
These abnormalities may explain suppression of mitochondrial biogenesis and mitochondrial function and heightened oxidative stress observed in prior studies of CKD (18, 42) and may lend support to a number of potential interventions designed to improve metabolic health and resistance to physiologic stress in CKD. Indeed, administration of coenzyme Q10 to advanced kidney disease patients in 1 randomized trial improved oxidative stress with reduction in isofuran levels (products of lipid peroxidation) (19). Furthermore, administration of nicotinamide riboside, a natural NAD precursor, has been shown to improve insulin sensitivity in high-fat diet–induced obese mice, with increased NAD leading to augmented mitochondrial sirtuin (SIRT3) activity, muscle mitochondrial content, and improved muscle endurance (43). SIRT3-5 has been shown to interact with proteins intimately involved with biologic processes of translation and energy metabolism critical to cellular stress resistance (44). Recent studies have uncovered a vital role of NAD and niacotinamide in tissue stress resistance using models of renal aging and renal ischemia/reperfusion (45, 46). Decreased NAD characteristic of aging and, in particular, renal aging may predispose renal tissue to damaging effects of cisplatin-induced renal injury (45). Kidney damage due to renal ischemia results in renal niacinamide depletion and fat accumulation reversed by niacinamide and PGC-1α–mediated NAD biosynthesis (46). The renoprotective effects of niacinamide-induced NAD production countering ischemia/reperfusion injury in PGC-1α–/– mice and cisplatin toxicity in aged mice are consistent with its role as a key effector of the protective metabolic benefits of mitochondrial biogenesis (45, 46). More recently, an alternative route of stimulating mitochondrial biogenesis has been reported in a diabetic kidney disease mouse model; taurine upregulated gene-1 (tug-1) overexpression improved the biochemical and histological features of diabetic kidney disease by interacting with PGC-1α to stimulate mitochondrial biogenesis and bioenergetics (47). Further evidence of the clinical importance of mitochondrial biogenesis stems from investigations of insulin sensitizers such as metformin and thiazolidinediones in models of obesity and diabetes. These agents increase PGC-1α in association with enhanced fatty acid oxidation, reduced lipid accumulation, and enhanced mitochondrial biogenesis (48–50) in skeletal muscle. Furthermore, insulin sensitizers result in reduction in amino acids metabolites (51, 52) and associate with attenuated loss of lean mass in older adults with diabetes (53). It is unclear if these effects translate to patients with CKD who demonstrate attenuated metabolome response to insulin. Future studies are needed to investigate the effects of insulin sensitizers on skeletal muscle metabolism and plasma metabolomics in nondiabetic CKD.
The main innovation of our study is the application of plasma metabolomics to provocative insulin administration to evaluate the broad metabolic effects of insulin and identify the mechanisms through which insulin action may be disturbed in CKD. Additional strengths of this investigation include the relatively large population evaluated with detailed methods and the focus on participants with advanced CKD and appropriate controls. We also employed a refined and broad-based targeted liquid chromatography–mass spectrometry metabolomics approach. Nonetheless, limitations include our inability to determine the mechanisms leading to changes in the plasma metabolome without simultaneous flux analysis or coinciding proteomic and transcriptomic analysis of tissues or organs. We were also unable to fully account for residual confounding by differences in unmeasured characteristics between those with CKD and controls.
In conclusion, our results suggest that abnormalities in ubiquinone biosynthesis, nicotinamide metabolism, and glutamate/glutamine metabolism are mechanisms or manifestations of insulin resistance in CKD that may be candidate targets for treatment.
Methods
Study population.
The Study of Glucose and Insulin in Renal Disease (SUGAR) is a cross-sectional study of glucose and insulin metabolism in moderate-to-severe nondiabetic CKD. From 2011–2014, we recruited 98 participants from Nephrology and Primary Care clinics associated with the University of Washington and neighboring institutions in Seattle (Washington, USA). Among the 98 recruited participants, 95 had adequate plasma samples collected for metabolomics, and of these, 60 had plasma samples before and after hyperinsulinemic-euglycemic clamp testing (40 CKD and 20 controls) (Supplemental Figure 1). We enrolled 58 participants with moderate-to-severe CKD, defined as eGFR <60 ml/min/1.73 m2. We then recruited 40 control subjects with eGFR ≥60 ml/min/1.73 m2 and spot urine albumin-creatinine ratio <30 mg/g, targeting the distributions of age, sex, and race of enrolled participants with CKD. Exclusion criteria for both groups included age <18 years, a clinical diagnosis of diabetes, maintenance dialysis or fistula in place, history of kidney transplantation, use of medications known to reduce insulin sensitivity (including corticosteroids and immunosuppressants), fasting serum glucose ≥126 mg/dl, and hemoglobin <10 g/dl.
Classification of CKD.
CKD was defined as eGFR <60 ml/min per 1.73 m2. Serum creatinine and cystatin C (Gentian) were measured in fasting serum collected immediately prior to the hyperinsulinemic-euglycemic clamp using a Beckman DxC automated chemistry analyzer. Creatinine and cystatin C concentrations are traceable to isotope dilution mass spectrometry and ERM-DA471/IFCC, respectively. Interassay coefficients of variation were 1.5%–3.0%. GFR was estimated from creatinine and cystatin C concentrations using the CKD-EPI formula (54).
Hyperinsulinemic-euglycemic clamp.
Plasma for metabolomics was available for 60 participants who had pre- and during-hyperinsulinemic-euglycemic clamp studies. For the clamps studies, each participant was admitted to the University of Washington Clinical Research Center after an overnight fast. I.v. catheters were placed in peripheral veins in each arm, with 1 arm being heated to allow for the sampling of arterialized blood. After collection of fasting plasma, a short i.v. GTT (not utilized for for this analysis) was performed with an infusion of 20% dextrose (11.4 g/m2 over 60 seconds) and frequent plasma sampling for 30 minutes. Immediately afterward, an insulin infusion was initiated as a prime (160 mU/m2/min for 5 minutes) followed by a constant rate (80 mU/m2/min). A variable rate infusion of unlabeled 20% dextrose was administered to maintain blood glucose (measured every 5 minutes) at approximately 90 mg/dl. Beginning 120–150 minutes after initiation of the insulin infusion, the dextrose infusion rate was held constant for 30 minutes, over which 3 steady-state plasma samples were obtained 15 minutes apart. Plasma samples from fasting blood drawn prior to the clamp and from the middle of the 3 steady-state clamp blood draws were used for the metabolomics assays.
Measurement of glucose and insulin homeostasis.
Calculation of standard parameters of insulin sensitivity measured from hyperinsulinemic-euglycemic clamp technique and OGTT were determined as previously reported (4). The insulin sensitivity (M/ΔI) was calculated as (glucose infusion rate × concentration of infused glucose)/(insulin concentration at steady state – fasting insulin concentration). The glucose infusion rate was calculated as the glucose infusion rate during the last 30 minutes of the clamp adjusted for the drift in plasma glucose concentration using Steels’ non–steady-state equation. The inclusion of insulin concentration in the denominator accounts for variability in the achieved insulin concentration and is supported by the linear relationship between insulin concentration and glucose disposal rate within individuals in a prior dose-range study.
A standard 75-g OGTT was performed 1 week after the clamp. Insulin and glucose values obtained before and during the OGTT were use to derive the Matsuda index, a common index of insulin sensitivity, as previously reported (55). We further measured nonesterified fatty acid insulin sensitivity index (NEFA index) (56, 57) as a marker of suppression of lipolysis. NEFA levels were measured at the Northwest Lipids Research Laboratories (Seattle, Washington, USA).
Metabolomics.
The liquid chromatography–tandem mass spectrometry (LC-MS/MS) metabolite data were acquired at the Northwest Metabolomics Research Center using a targeted metabolic profiling LC-MS/MS assay that has been described previously (58). Briefly, prior to LC-MS/MS, blood samples were prepared by protein precipitation (using methanol), centrifugation (21,694 g for 10 minutes at 4°C), drying, and reconstitution. The LC-MS/MS studies were performed on an Agilent 1260 LC (Agilent Technologies) coupled to a AB Sciex QTrap 5500 MS (AB Sciex) system equipped with a standard electro-spray ionization (ESI) source. Each sample was injected twice (10 μl and 2 μl) for analysis in negative and positive ionization modes, respectively. Both chromatographic separations were performed using identical hydrophilic interaction chromatography (HILIC) methods on a SeQuant ZIC-cHILIC column (150 × 2.1 mm × particle size 3.0 μm, Merck KGaA). The flow rate was 0.3 ml/min. The mobile phase was composed of Solvents A (10 mM ammonium acetate in 90% H2O/10% acetonitrile + 0.2% acetic acid, Thermo Fisher Scientific) and Solvent B (10 mM ammonium acetate in 90% acetonitrile/10% H2O + 0.2% acetic acid, Thermo Fisher Scientific) . After the initial 2-minute isocratic elution at 90% Solvent B, the percentage of Solvent B was decreased to 50% at t = 5 minutes. The composition of Solvent B was maintained at 50% for 4 minutes (t = 9 minutes), and then the percentage of Solvent B was returned to 90% at 12 minutes and stayed at 90% Solvent B for the duration of chromatographic separation (total run time = 20 minutes). The metabolite identities were confirmed by spiking the pooled serum sample used for method development with mixtures of standard compounds. Quality control samples consisting of a pooled serum sample were run once for every 10 biological samples to allow instrument monitoring and signal normalization as needed. Four isotope-labeled internal standards were used to monitor sample preparation. The extracted multiple reaction montitoring (MRM) peaks were integrated using MultiQuant 2.1 software (AB Sciex). Pre- and during-hyperinsulinemic-euglycemic clamp samples testing 74 metabolites occurred over 2 separate days, but for each person, pre- and during-clamp samples were tested on the same day. Fasting samples were all tested together on a third day after additional metabolites were added to the assay. This accounted for the discrepancy of 124 fasting samples compared with 74 metabolites assayed from pre- and during-hyperinsulinemic-euglycemic clamp samples.
Statistics.
Since metabolite distributions were greatly skewed, all plasma metabolite concentrations were transformed by log(1 + x) prior to performing statistical analysis. To compare the fold changes in metabolite levels between CKD and non-CKD participants, we used linear regression with robust Huber-White standard errors of the log-transformed metabolite on CKD status, additionally adjusted for the potential confounders of age, sex, race/ethnicity (European descent versus non–European descent), and weight. To account for the correlation of measurements within participants, we examined the fold changes associated with the clamp procedure via a linear mixed model with random intercepts, regressing the log-transformed metabolite on the sample type (during clamp versus fasting sample), adjusted for the covariates listed above, as well as batch. Finally, to evaluate whether the effect of the hyperinsulinemic-euglycemic clamp procedure differed between CKD and non-CKD participants, we again used a linear mixed model approach with random intercepts, in which the log-transformed metabolite was regressed on sample type (clamp versus fasting), CKD status (CKD versus non-CKD), and the interaction of the 2, additionally adjusting for age, sex, race/ethnicity, weight, and batch. In each analysis, we used a Benjamini-Hochberg FDR of 10% to declare statistical significance to account for multiple comparisons (59). Under this approach, 10% of the metabolites identified as significant would be expected to be false positives. All of the statistical analyses described above were conducted using R, version 3.4.0 (60). Targeted metabolic pathway analysis was performed using Metaboanalyst version 3.5 (61). Pathway topology analysis was performed analyzing differences in fasting plasma metabolite between CKD and controls. Fold change analysis of discrete metabolites was performed using Metaboanalyst software to calculate the change between fasting and during insulin infusion plasma metabolites group averages for the entire cohort, which was used to construct a volcano plot. Using pathway topology analysis, we identified significant metabolic pathways affected by insulin infusion using log-transformed pre- and during-infusion metabolite levels. Determination of significant metabolic pathways modified by CKD status involved comparing the fold change of each metabolite calculated by subtracting log-transformed values of during-clamp from the preclamp time points. Data is presented for hyperinsulinemic-euglycemic clamp–induced plasma changes in the overall cohort and comparing CKD with non-CKD. Pearson correlations of changes in plasma metabolite concentrations (during insulin infusion versus fasting) with parameters of insulin sensitivity from the clamp (M/ΔI) and OGTT (Matsuda Index and NEFA index) were estimated for metabolites showing the greatest difference in insulin response between CKD and non-CKD. Data is available to the public (http://dx.doi.org/10.17632/9z7ncwvxnz.1).
Author contributions
Conceptualization was contributed by BR, LRZ, IHDB, KU, SEK, BK, JH, DR, and JLG. Methodology was contributed by DD, HG, DR, KU, and SEK. Formal analysis was contributed by BR and LRZ. Investigation was contributed by DD, HG, and DR. Resources were contributed by IHDB, JH, BK, and DR. Data curation was contributed by BR and LRZ. Writing the original draft was contributed by BR, IHDB, JAA, TRZ, and JLG. Writing the review and editing were contributed by BR, LRZ, IHDB, JAA, TRZ, JLG, JH, BK, SEK, KU, and DR. Visualization was contributed by BR and LZ. Supervision was contributed by IHDB, JH, BK, and DR. Project administration was contributed by IHDB and DR. Funding acquisition was contributed by IHDB, DR, JH, BK, KU, and SEK.
Supplementary Material
Acknowledgments
This project was funded in part by an unrestricted grant from the Northwest Kidney Centers and R01DK087726, R01DK087726-S1, 1K23DK099442 (BR, IHDB), R01DK087726, R01DK087726-S1, K01 DK102851 (JAA, IHDB), K23DK100533 (JLG), K24 DK096574 (TRZ), and P30 DK017047 (University of Washington Diabetes Research Center).
Version 1. 08/23/2018
Electronic publication
Footnotes
BR’s present address is: Department of Medicine/Division of Nephrology, UCD, Davis, California, USA.
Conflict of interest: The authors have declared that no conflict of interests exists.
Reference information: JCI Insight. 2018;3(16):e122219. https://doi.org/10.1172/jci.insight.122219.
Contributor Information
Baback Roshanravan, Email: bob.roshanravan.wrk@gmail.com.
Leila R. Zelnick, Email: LZelnick@Nephrology.washington.edu.
Haiwei Gu, Email: haiwei@uw.edu.
Jessica A. Alvarez, Email: jessica.alvarez@emory.edu.
Thomas R. Ziegler, Email: trztrz@me.com.
Bryan Kestenbaum, Email: brk@uw.edu.
Jonathan Himmelfarb, Email: himmej@u.washington.edu.
Steven E. Kahn, Email: skahn@uw.edu.
Daniel Raftery, Email: draftery@uw.edu.
Ian H. de Boer, Email: IDeBoer@Nephrology.washington.edu.
References
- 1.Shahim B, et al. The Prognostic Value of Fasting Plasma Glucose, Two-Hour Postload Glucose, and HbA1c in Patients With Coronary Artery Disease: A Report From EUROASPIRE IV: A Survey From the European Society of Cardiology. Diabetes Care. 2017;40(9):1233–1240. doi: 10.2337/dc17-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalyani RR, Metter EJ, Ramachandran R, Chia CW, Saudek CD, Ferrucci L. Glucose and insulin measurements from the oral glucose tolerance test and relationship to muscle mass. J Gerontol A Biol Sci Med Sci. 2012;67(1):74–81. doi: 10.1093/gerona/glr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barzilay JI, et al. Insulin resistance is associated with decreased quadriceps muscle strength in nondiabetic adults aged >or=70 years. Diabetes Care. 2009;32(4):736–738. doi: 10.2337/dc08-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Boer IH, et al. Impaired Glucose and Insulin Homeostasis in Moderate-Severe CKD. J Am Soc Nephrol. 2016;27(9):2861–2871. doi: 10.1681/ASN.2015070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeFronzo R, Smith D, Alvestrand A. Insulin action in uremia. Kidney Int Suppl. 1983;16:S102–S 14. [PubMed] [Google Scholar]
- 6.Thomas SS, Zhang L, Mitch WE. Molecular mechanisms of insulin resistance in chronic kidney disease. Kidney Int. 2015;88(6):1233–1239. doi: 10.1038/ki.2015.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deger SM, et al. Systemic inflammation is associated with exaggerated skeletal muscle protein catabolism in maintenance hemodialysis patients. JCI Insight. 2017;2(22):95185. doi: 10.1172/jci.insight.95185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey JL, Wang X, England BK, Price SR, Ding X, Mitch WE. The acidosis of chronic renal failure activates muscle proteolysis in rats by augmenting transcription of genes encoding proteins of the ATP-dependent ubiquitin-proteasome pathway. J Clin Invest. 1996;97(6):1447–1 53. doi: 10.1172/JCI118566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afshinnia F, et al. Impaired beta-Oxidation and Altered Complex Lipid Fatty Acid Partitioning with Advancing CKD. J Am Soc Nephrol. 2018;29(1):295–306. doi: 10.1681/ASN.2017030350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schefold JC, et al. Increased indoleamine 2,3-dioxygenase (IDO) activity and elevated serum levels of tryptophan catabolites in patients with chronic kidney disease: a possible link between chronic inflammation and uraemic symptoms. Nephrol Dial Transplant. 2009;24(6):1901–1908. doi: 10.1093/ndt/gfn739. [DOI] [PubMed] [Google Scholar]
- 11.Siew ED, Pupim LB, Majchrzak KM, Shintani A, Flakoll PJ, Ikizler TA. Insulin resistance is associated with skeletal muscle protein breakdown in non-diabetic chronic hemodialysis patients. Kidney Int. 2007;71(2):146–152. doi: 10.1038/sj.ki.5001984. [DOI] [PubMed] [Google Scholar]
- 12.Conjard A, Ferrier B, Martin M, Caillette A, Carrier H, Baverel G. Effects of chronic renal failure on enzymes of energy metabolism in individual human muscle fibers. J Am Soc Nephrol. 1995;6(1):68–74. doi: 10.1681/ASN.V6168. [DOI] [PubMed] [Google Scholar]
- 13.May RC, Kelly RA, Mitch WE. Metabolic acidosis stimulates protein degradation in rat muscle by a glucocorticoid-dependent mechanism. J Clin Invest. 1986;77(2):614–621. doi: 10.1172/JCI112344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fazakerley DJ, et al. Mitochondrial CoQ deficiency is a common driver of mitochondrial oxidants and insulin resistance. Elife. 2018:7. doi: 10.7554/eLife.32111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoehn KL, et al. Insulin resistance is a cellular antioxidant defense mechanism. Proc Natl Acad Sci U S A. 2009;106(42):17787–17792. doi: 10.1073/pnas.0902380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440(7086):944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 17.Roshanravan B, et al. CKD and Muscle Mitochondrial Energetics. Am J Kidney Dis. 2016;68(4):658–659. doi: 10.1053/j.ajkd.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamboa JL, et al. Mitochondrial dysfunction and oxidative stress in patients with chronic kidney disease. Physiol Rep. 2016;4(9):e12780. doi: 10.14814/phy2.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung CK, et al. Coenzyme Q10 dose-escalation study in hemodialysis patients: safety, tolerability, and effect on oxidative stress. BMC Nephrol. 2015;16:183. doi: 10.1186/s12882-015-0178-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallan S, et al. Metabolomics and Gene Expression Analysis Reveal Down-regulation of the Citric Acid (TCA) Cycle in Non-diabetic CKD Patients. EBioMedicine. 2017;26:68–77. doi: 10.1016/j.ebiom.2017.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng L, et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat Commun. 2015;6:6001. doi: 10.1038/ncomms7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicholls DG, Ferguson SJ. Bioenergetics. Amsterdam: Academic Press; Elsevier: 2013. [Google Scholar]
- 23.Goncalves RL, Quinlan CL, Perevoshchikova, Hey-Mogensen M, Brand MD. Sites of superoxide and hydrogen peroxide production by muscle mitochondria assessed ex vivo under conditions mimicking rest and exercise. J Biol Chem. 2015;290(1):209–227. doi: 10.1074/jbc.M114.619072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan CL, Orr AL, Perevoshchikova, Treberg JR, Ackrell BA, Brand MD. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J Biol Chem. 2012;287(32):27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chouchani ET, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9(3):316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 28.Morita M, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18(5):698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Massudi H, Grant R, Guillemin GJ, Braidy N. NAD+ metabolism and oxidative stress: the golden nucleotide on a crown of thorns. Redox Rep. 2012;17(1):28–46. doi: 10.1179/1351000212Y.0000000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao WY, Zhang L, Sui MX, Zhu YH, Zeng L. Protective effects of sirtuin 3 in a murine model of sepsis-induced acute kidney injury. Sci Rep. 2016;6:33201. doi: 10.1038/srep33201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiner ID, Mitch WE, Sands JM. Urea and Ammonia Metabolism and the Control of Renal Nitrogen Excretion. Clin J Am Soc Nephrol. 2015;10(8):1444–1458. doi: 10.2215/CJN.10311013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiner ID, Verlander JW. Ammonia Transporters and Their Role in Acid-Base Balance. Physiol Rev. 2017;97(2):465–494. doi: 10.1152/physrev.00011.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chobanian MC, Hammerman MR. Insulin stimulates ammoniagenesis in canine renal proximal tubular segments. Am J Physiol. 1987;253(6 Pt 2):F1171–F1177. doi: 10.1152/ajprenal.1987.253.6.F1171. [DOI] [PubMed] [Google Scholar]
- 35.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16(10):619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang CH, et al. Chronic α-hydroxyisocaproic acid treatment improves muscle recovery after immobilization-induced atrophy. Am J Physiol Endocrinol Metab. 2013;305(3):E416–E428. doi: 10.1152/ajpendo.00618.2012. [DOI] [PubMed] [Google Scholar]
- 37.Hankard RG, Haymond MW, Darmaun D. Effect of glutamine on leucine metabolism in humans. Am J Physiol. 1996;271(4 Pt 1):E748–E754. doi: 10.1152/ajpendo.1996.271.4.E748. [DOI] [PubMed] [Google Scholar]
- 38.Fujita S, et al. Nutrient signalling in the regulation of human muscle protein synthesis. J Physiol (Lond) 2007;582(Pt 2):813–823. doi: 10.1113/jphysiol.2007.134593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawahara K, Hohjoh H, Inazumi T, Tsuchiya S, Sugimoto Y. Prostaglandin E2-induced inflammation: Relevance of prostaglandin E receptors. Biochim Biophys Acta. 2015;1851(4):414–421. doi: 10.1016/j.bbalip.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 40.Roshanravan B, et al. Creatinine clearance, walking speed, and muscle atrophy: a cohort study. Am J Kidney Dis. 2015;65(5):737–747. doi: 10.1053/j.ajkd.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raphael KL, Carroll DJ, Murray J, Greene T, Beddhu S. Urine Ammonium Predicts Clinical Outcomes in Hypertensive Kidney Disease. J Am Soc Nephrol. 2017;28(8):2483–2490. doi: 10.1681/ASN.2016101151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamaki M, Miyashita K, Wakino S, Mitsuishi M, Hayashi K, Itoh H. Chronic kidney disease reduces muscle mitochondria and exercise endurance and its exacerbation by dietary protein through inactivation of pyruvate dehydrogenase. Kidney Int. 2014;85(6):1330–1339. doi: 10.1038/ki.2013.473. [DOI] [PubMed] [Google Scholar]
- 43.Cantó C, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15(6):838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang W, et al. Mitochondrial Sirtuin Network Reveals Dynamic SIRT3-Dependent Deacetylation in Response to Membrane Depolarization. Cell. 2016;167(4):985–1000.e21. doi: 10.1016/j.cell.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan Y, et al. Nicotinamide Mononucleotide, an NAD+ Precursor, Rescues Age-Associated Susceptibility to AKI in a Sirtuin 1-Dependent Manner. J Am Soc Nephrol. 2017;28(8):2337–2352. doi: 10.1681/ASN.2016040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran MT, et al. PGC1α drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature. 2016;531(7595):528–532. doi: 10.1038/nature17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long J, et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest. 2016;126(11):4205–4218. doi: 10.1172/JCI87927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suwa M, Egashira T, Nakano H, Sasaki H, Kumagai S. Metformin increases the PGC-1alpha protein and oxidative enzyme activities possibly via AMPK phosphorylation in skeletal muscle in vivo. J Appl Physiol. 2006;101(6):1685–1692. doi: 10.1152/japplphysiol.00255.2006. [DOI] [PubMed] [Google Scholar]
- 49.Coletta DK, et al. Pioglitazone stimulates AMP-activated protein kinase signalling and increases the expression of genes involved in adiponectin signalling, mitochondrial function and fat oxidation in human skeletal muscle in vivo: a randomised trial. Diabetologia. 2009;52(4):723–732. doi: 10.1007/s00125-008-1256-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura Y, et al. Effects of metformin on peripheral insulin sensitivity and intracellular lipid contents in muscle and liver of overweight Japanese subjects. Diabetes Obes Metab. 2008;10(9):733–738. doi: 10.1111/j.1463-1326.2007.00801.x. [DOI] [PubMed] [Google Scholar]
- 51.Irving BA, et al. Effect of insulin sensitizer therapy on amino acids and their metabolites. Metab Clin Exp. 2015;64(6):720–728. doi: 10.1016/j.metabol.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bao Y, et al. Metabonomic variations in the drug-treated type 2 diabetes mellitus patients and healthy volunteers. J Proteome Res. 2009;8(4):1623–1630. doi: 10.1021/pr800643w. [DOI] [PubMed] [Google Scholar]
- 53.Lee CG, et al. Insulin sensitizers may attenuate lean mass loss in older men with diabetes. Diabetes Care. 2011;34(11):2381–2386. doi: 10.2337/dc11-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pottel H, et al. Estimating glomerular filtration rate for the full age spectrum from serum creatinine and cystatin C. Nephrol Dial Transplant. 2017;32(3):497–507. doi: 10.1093/ndt/gfw425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22(9):1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 56.Wagner R, et al. A novel insulin sensitivity index particularly suitable to measure insulin sensitivity during gestation. Acta Diabetol. 2016;53(6):1037–1044. doi: 10.1007/s00592-016-0930-5. [DOI] [PubMed] [Google Scholar]
- 57.Ahmad I, et al. Chronic kidney disease and obesity bias surrogate estimates of insulin sensitivity compared with the hyperinsulinemic euglycemic clamp. Am J Physiol Endocrinol Metab. 2017;312(3):E175–E182. doi: 10.1152/ajpendo.00394.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu J, et al. Colorectal cancer detection using targeted serum metabolic profiling. J Proteome Res. 2014;13(9):4120–4130. doi: 10.1021/pr500494u. [DOI] [PubMed] [Google Scholar]
- 59.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ Accessed August 3, 2018.
- 61.Xia J, Wishart DS. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinformatics. 2016;55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.