Abstract
Osteosarcoma (OS), a malignant tumor of bone, kills through aggressive metastatic spread almost exclusively to the lung. Mechanisms driving this tropism for lung tissue remain unknown, though likely invoke specific interactions between tumor cells and other cells within the lung metastatic niche. Aberrant overexpression of ΔNp63 in OS cells directly drives production of IL-6 and CXCL8. All these factors were expressed at higher levels in OS lung metastases than in matched primary tumors from the same patients. Expression in cell lines correlated strongly with lung colonization efficiency in murine xenograft models. Lentivirus-mediated expression endowed poorly metastatic OS cells with increased metastatic capacity. Disruption of IL-6 and CXCL8 signaling using genetic or pharmaceutical inhibitors had minimal effects on tumor cell proliferation in vitro or in vivo, but combination treatment inhibited metastasis across multiple models of metastatic OS. Strong interactions occurred between OS cells and both primary bronchial epithelial cells and bronchial smooth muscle cells that drove feed-forward amplification of IL-6 and CXCL8 production. These results identify IL-6 and CXCL8 as primary mediators of OS lung tropism and suggest pleiotropic, redundant mechanisms by which they might effect metastasis. Combination therapy studies demonstrate proof of concept for targeting these tumor-lung interactions to affect metastatic disease.
Keywords: Oncology
Keywords: Cancer
Tumor-derived IL6 and CXCL8 drive specific tumor-host interactions that are critical to lung colonization in osteosarcoma.
Introduction
Osteosarcoma (OS), the most common primary cancer of bone, kills approximately 40% of young people diagnosed with the malignancy (1). Despite 40 years of active drug development and extraordinary cooperation, including the formation of large international clinical trial consortia, OS remains as deadly today as it was in the 1970s (2–6). Lung metastasis remains the primary determinant of outcome. Whether patients initially present with metastasis or develop lung tumors years after therapy, 5-year survival rates plummet from a modest 70% (for patients with localized disease; see refs. 4, 5) to a sobering less than 20% once metastases occur (5, 7). Bones and lungs tend to be the only tissues involved; less than 2% of patients develop lesions in other organs (7, 8). There remains little known of the mechanisms that mediate this tissue tropism. As a disease where spread to a particular organ so clearly defines outcome, the study of OS’s lung tropism represents (a) a particularly useful system for the study of tumor-host interactions, (b) a disease model amenable to development of therapeutics that target specific tumor-host interactions, and (c) an opportunity to change the natural history of a particularly deadly disease in a way that significantly impacts patients (8, 9).
We have very specifically focused on the process of lung colonization, rather than the early steps in metastasis that result in dissemination (10–13). We have done this because dissemination has already occurred in most patients at diagnosis (14, 15), particularly in patients with OS (16). We have further focused on identifying the molecular interactions that license the growth of disseminated tumor cells within the otherwise hostile distant tissue (17). The tumor-host interactions that occur (or do not) within the early metastatic niche generate a metastatic bottleneck responsible for the attrition of almost all disseminated tumor cells (13). Identifying the mechanisms that facilitate survival through this bottleneck in OS may uncover targetable vulnerabilities that could be exploited to alter the natural course of disease.
In the studies outlined below, we have identified IL-6 and CXCL8 as 2 factors that mediate tumor-lung interactions and facilitate metastasis. Both play primary roles in cell-cell communication mediating innate immune response and have a number of pleiotropic effects. Our studies suggest that the broad range of activities affected by the activity of IL-6 and CXCL8 may, in fact, be one reason why these 2 factors play such a critical role in metastatic colonization. Importantly, this process seems to be targetable. The early preclinical work outlined below suggests a practical framework upon which feasible human (and/or canine) clinical trials could easily build.
Results
Metastasis enriches for expression of IL-6 and CXCL8 relative to matched primary tumors.
We hypothesized that, if primary tumors contain heterogeneous groups of cells with different characteristics (18, 19), then there must be enrichment in the metastatic process for cells that express factors that participate in lung tropism. Furthermore, samples taken from primary tumors that subsequently develop high rates of metastasis would also have enrichment for factors that drive tumor-lung interactions. We selected key manuscripts from the published literature that had already identified gene expression signatures associated with metastasis or outcome in OS (we took outcome to be a surrogate for metastasis in OS) (20–24). These publications each reported gene expression signatures in human samples, canine samples, or murine models and identified gene expression profiles that demonstrated strong correlation with metastasis or progression. We manually evaluated these published signatures gene by gene to develop a curated list of candidate genes. We selected genes likely to mediate cell-cell interactions based on known gene ontology/function, with special attention to cytokines, chemokines, genes known to regulate osteoblast/bone development (25, 26), and genes known to drive lung tropism in other tumor types (27). Our final candidate list (Figure 1 and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99791DS1) contained 56 genes whose expression within primary tumors correlated with metastasis/outcome in previous studies and whose known function suggested potential to mediate tumor-lung interactions important to lung tropism in OS (i.e., GO annotation “cell-cell signaling”).
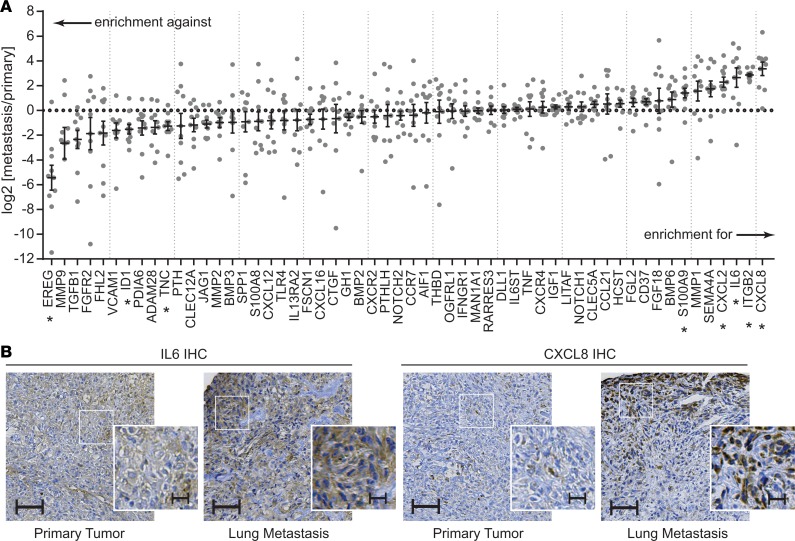
Figure 1. Candidate gene enrichment in primary tumor–lung metastasis pairs from patients with osteosarcoma.
(A) Gene expression for each of the 56 candidate genes was assessed in primary tumor–lung metastasis pairs using singleplex qPCR assays validated across multiple FFPE tissues. Expression within each lung metastasis sample was normalized to expression within the primary tissue, then log transformed. n = 10 sample pairs. Single-sample t tests against a theoretical mean of 0 (for normalization to primary tissue) were evaluated, controlling for a false discovery rate of 0.05 using the Benjamini-Hochberg method. *Denotes candidate genes whose expression is significantly different between primary tumors and metastases. (B) Representative IHC sections from 1 primary tumor–lung metastasis pair showing changes in the staining intensity and staining patterns from primary to metastasis. Scale bars: 100 μm and 25 μm (insets). Additional examples are shown in Supplemental Figure 2.
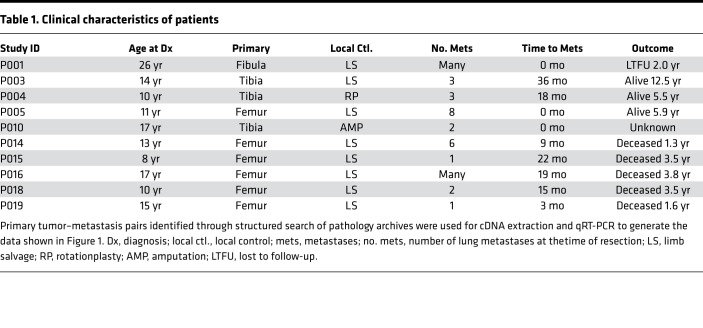
Using formalin-fixed paraffin-embedded (FFPE) primary tumor-lung metastasis pairs from tissues surgically excised from patients seen at our hospital, we determined the relative expression of each candidate gene using qRT-PCR assays specifically designed for and validated against archival FFPE tissues (Figure 1). Examples of hematoxylin and eosin–stained (H&E-stained) specimens and the specific tissues selected for RNA extraction are shown in Supplemental Figure 1. Of the candidate genes tested, IL-6 and CXCL8 were among the genes most reliably enriched for in the metastatic tumors. Expression of these genes was many fold higher in the metastatic lesions than in matched primaries. In IHC analysis, expression was heterogeneous and strongest for both IL-6 and CXCL8 along the leading edges (Figure 1B and Supplemental Figure 2). Select clinical characteristics of the sample population are shown in Table 1.
Table 1. Clinical characteristics of patients.
Production of IL-6 and CXCL8 correlates with metastatic potential in murine xenograft models of lung colonization.
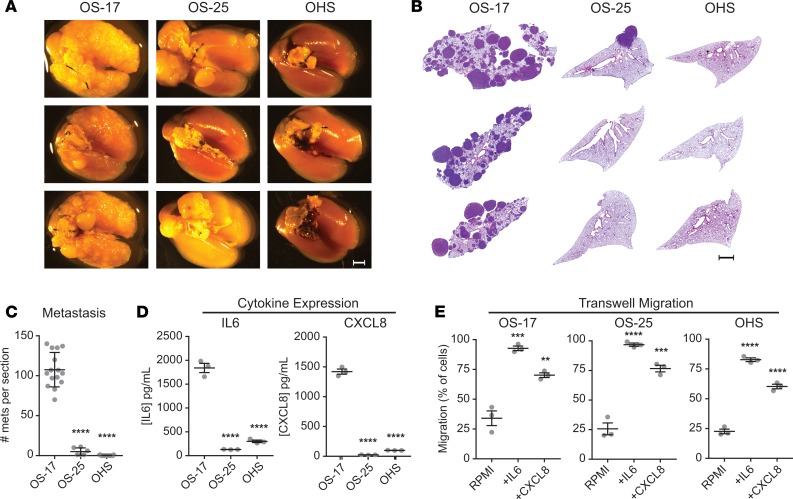
We next tested a panel of OS cell lines for their ability to colonize mouse lung. We found that OS-17 cells, when introduced into circulation via tail vein, develop metastatic foci with very high efficiency, while OHS and OS-25 cell lines demonstrate much lower metastatic efficiency (Figure 2). This effect remained consistent across multiple passages of cells and multiple assays. We tested these cell lines for production of IL-6 and CXCL8 by subjecting cell-free supernatants to ELISA (Figure 2D), which revealed a strong correlation between tumor cell production of both cytokines and the cell line’s capacity to colonize murine lung.
Figure 2. Expression of IL-6 and CXCL8 correlates with lung-colonization efficiency.
CB-17 SCID mice inoculated with 1 × 106 osteosarcoma cells were euthanized 49 days after inoculation. (A) Gross appearance of lung blocks taken from those mice suggests markedly greater efficiency of colonization by OS-17 relative to the other 2 cell lines. Scale bar: 2 mm. (B) H&E stains from sections of paraffin-embedded left lobes were counted to quantify the number of metastases per section. Scale bar: 2 mm. (C) Quantification reveals significantly higher numbers of metastases (mets) in the OS-17 sections relative to both OS-25 and OHS (n = 15 OS-17 and OHS, n = 6 OS-25). (D) Determination of IL-6 and CXCL8 concentrations in 72-hour supernatants from cultures of each cell line reveals significant expression of both cytokines in the metastatic OS-17 cells relative to either nonmetastatic cell line (n = 3 samples per cell line, run in triplicate). (E) Evaluation of capacity to respond to IL-6 and CXCL8 signals using transwell migration assay. Cells were plated in the top chamber and RPMI alone or RPMI containing 50 ng/mL IL-6 or 100 ng/ml IL-8 was placed in the bottom chamber. After 24 hours, plates were harvested and processed as described to quantify the number of cells migrating (n = 3 per condition). **P < 0.01; ***P < 0.001; ****P < 0.0001 relative to OS-17 (C and D) or RPMI (E); 1-way ANOVA with Tukey’s post hoc test.
IL-6 and CXCL8 stimulate chemokinesis and directional migration in OS cells, regardless of metastatic potential.
To begin to understand how IL-6 and CXCL8 might mediate metastasis, we first sought to determine whether these highly and poorly metastatic cell lines maintain features that facilitate response to these cytokines. We therefore performed both scratch assays (wound-healing assays) and transwell migration assays to assess response. Standardized wounds created in both OS-17 and OHS cell monolayers closed more effectively when cultured in media supplemented with IL-6 and/or CXCL8 (Supplemental Figure 3), demonstrating that either cytokine can stimulate chemokinesis (increased cell motility) in either cell line, irrespective of any basal production of that cytokine.
These cells exhibit similar results in assays testing directional migration. OS-17, OS-25, and OHS cells grown in the top chamber of a transwell system exhibit strong directional migration in response to chemotactic gradients of either IL-6 or CXCL8 (Figure 2E).
Combined blockade of IL-6 and CXCL8 pathways prevents formation of lung metastasis.
To evaluate the functional importance of these pathways to OS lung metastasis, we turned to our xenograft models of OS metastasis. CB17-SCID mice inoculated via tail vein with 1 × 106 luciferase-labeled OS-17 cells received treatment with sc144 (which stimulates degradation of the IL-6 signaling receptor IL-6ST through a novel mechanism; ref. 28), DF2156A (an allosteric inhibitor of the CXCL8 receptors CXCR1 and CXCR2; ref. 29), or both. Mice received treatment for 42 days, after which treatment stopped. Intravital imaging for in vivo assessment of tumor burden performed at 28 days after inoculation suggested markedly decreased tumor burden in the lungs of mice receiving combined therapy relative to those receiving no treatment or single-agent therapy (Figure 3A). Importantly, imaging did not show migration of tumor cells into other organs, but an overall loss of bioluminescence, suggesting decreased survival of circulating tumor cells. Two mice from each single-agent treatment group were euthanized 24 hours after the 14th dose of drug to perform pharmacodynamic assessment of target inhibition. Lungs from those mice stained with IHC for either p-FAK (CXCR1/2) or p-STAT3 (IL-6ST) showed sustained target inhibition at dosing trough (Supplemental Figure 4).
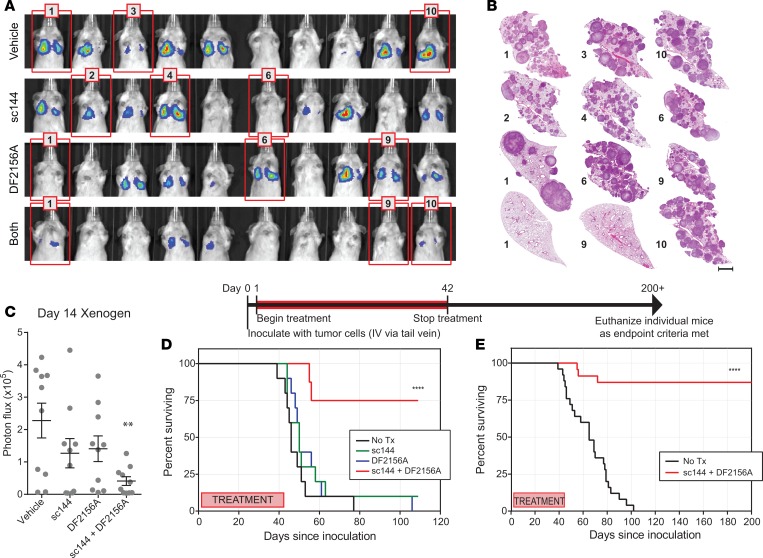
Figure 3. Combined disruption of IL-6 and CXCL8 signaling reduces or eliminates metastatic lung colonization.
Mice inoculated with 1 × 106 luciferase-expressing OS-17 cells were treated with pharmacologic inhibitors of IL-6 (sc144, inhibitor of IL-6ST), CXCL8 (DF2156A, inhibitor of CXCR1 and -2), or both. (A) Bioluminescent imaging completed at 28 days after inoculation. (B) Gross appearance of representative lung blocks taken from the mice identified with red frames in A at the time of euthanasia (endpoint criteria, or day 200 for mice 1 and 9 of combined treatment group) shows no microscopic evidence of tumor in long-term survivors. This included some mice, such as mouse 10 of the combined treatment group (endpoint at day 72) that showed response at early time points, but subsequently developed metastasis. Scale bar: 2 mm. (C) Quantification of the lung-field bioluminescence from day 14 (n = 10 mice per group). (D) Survival analysis of the mice shown in A. As noted, treatment continued until day 42, then was stopped. Mice that received combination therapy experienced significantly better outcomes than those who received no treatment or treatment with only 1 inhibitor (n = 10 mice per group, P value in comparison to vehicle control). (E) Validation studies confirm the results found in A. Mice surviving beyond 200 days were euthanized. Mice that died during the study, but had no evidence of metastatic tumor burden on necropsy (n = 3) were censored in this analysis (n = 25 mice per group). No Tx, no treatment. **P < 0.01; ****P < 0.0001 by 1-way ANOVA with Tukey’s post hoc test (C) or Mantel-Cox log-rank test (D and E).
Following treatment, mice were then observed until demonstrating signs of clinical deterioration, either weight loss greater than 10% or enhanced body condition score (eBCS) less than 8 (30), our defined endpoints. Mice were euthanized using approved methods and lungs were harvested, insufflated, fixed, embedded, sectioned, and stained. Survival analysis (Figure 3D) showed that nearly all mice receiving either no drug or single-agent therapy developed lethal lung metastasis by 60 days, whereas most mice receiving combined therapy (IL-6ST inhibitor + CXCR1/2 inhibitor) remained healthy at more than 100 days. Given the specific hypothesis tested, mice that did not demonstrate metastasis as a cause of endpoint symptomatology or death were censored from the survival analysis. This included 1 mouse censored from the vehicle group and 2 from the combined treatment group.
To ensure reproducibility, these experiments were repeated (omitting the single-agent treatment groups). Repeat experiments yielded essentially identical results. Figure 3E shows the combined data from the survival analyses. Lungs from mice surviving more than 200 days (more than 150 days after treatment) demonstrate no identifiable metastatic lesions (Figure 3B).
Combined IL-6 and CXCL8 inhibition prevents metastasis in multiple models of OS.
To ensure that the results obtained in these studies were broadly applicable and not unique to immunodeficient xenografts or to OS-17 cells, we repeated the treatment-related experiments using a number of different models. These included a syngeneic, immunocompetent model using a cell line derived from a spontaneously arising OS in a BALB/c mouse (K7M2; ref. 31), a syngeneic, immunocompetent model derived from a genetically engineered model of OS (F420; ref. 32), xenograft models of canine OS (OSCA-8 and OSCA-16; ref. 33), and additional xenograft models of human OS (143B). Mice inoculated with tumor cells were treated with either no drug or combined sc144 and DF2156A for 42 days. At the time that at least one mouse from any group (any cell line) reached endpoint with confirmed lung metastasis, all mice from that group were euthanized, lungs harvested, and metastatic lesions quantified. The ability of dual IL-6ST–CXCR1/2 inhibition to prevent the development of metastatic lung lesions remained consistent across models (Figure 4A).
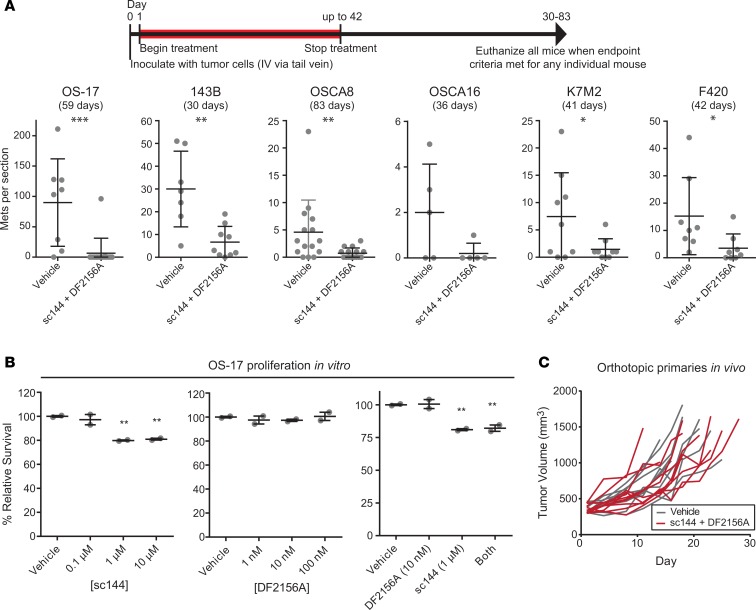
Figure 4. Combined therapy with anti–IL-6 and anti-CXCL8 treatment reduces metastatic lung colonization in many xenograft models of metastatic osteosarcoma (OS), but does not affect growth of primary orthotopic tumors.
(A) Validation of concept in multiple murine models of OS. To validate, studies were repeated in multiple OS models of metastasis including human OS xenografts (OS-17, 143B), canine OS xenografts (OSCA8 and OSCA16), and syngeneic/immunocompetent murine OS allografts (K7M2, F420). Subsequent to inoculation of OS cells, mice received either vehicle treatment or treatment with both sc144 and DF2156A for a period of 42 days. At the time that one mouse in either group met endpoint criteria, all mice within that study were euthanized and lungs harvested, metastases (Mets) counted. Representative lung sections shown (n = 5 to 15 mice per group as shown). (B) The effect of cytokine inhibitors on proliferation in vitro was evaluated using an Incucyte ZOOM live cell imaging system. Graphs represent number of cells per microscopic field in each culture condition, normalized to the average cell number in the vehicle control. (C) Mice bearing intratibial OS-17 tumors were treated with either vehicle or the combination of sc144 and DF2156A according to the same schedule used in the metastatic models. Tumor volume was calculated biweekly throughout treatment. All P values shown relative to control condition (vehicle). *P < 0.05; **P < 0.01; ***P < 0.001 by Mann-Whitney U test (A) or 1-way ANOVA with Tukey’s post hoc test (B and C).
Target validation using inducible shRNA-expressing cell lines.
In order to ensure that our phenotypic observations resulted from on-target effects of the inhibitors, we created OS-17–derived cell lines transduced with lentiviruses bearing tetracycline-inducible shRNA constructs targeting IL-6, CXCL8, CXCR1/2, and IL-6ST. We opted to use an inducible shRNA system rather than CRISPR, given the effect that the process of colony selection had on metastatic tendencies — an inducible system allowed for more equivalent control groups. After antibiotic selection and validation of target knockdown (Figure 5, A and B), these cells were inoculated into CB17-SCID mice via tail vein. One half of the mice inoculated with each cell line were maintained on regular water, the other half on water containing doxycycline. When any mouse in a group demonstrated signs of clinical deterioration, all mice from that group were euthanized. Harvested lungs stained with H&E were evaluated to quantify the number of metastatic tumors per section by an evaluator blinded to the treatment group. Results of those experiments showed significant decrease in the number and size of tumors with knockdown of IL-6 and IL-8, but not with CXCR1/2 and IL-6ST (Figure 5C). This both confirms the on-target effect of the drugs, consistent with the slight decrease in signal and delay seen in Figure 3, C and D, and suggests that the primary mechanism of action comes from the effect of tumor-derived cytokine on host lung cells, rather than host-derived cytokine on tumor cells. We caution, however, that this result does not necessarily rule out a role for autocrine stimulation.
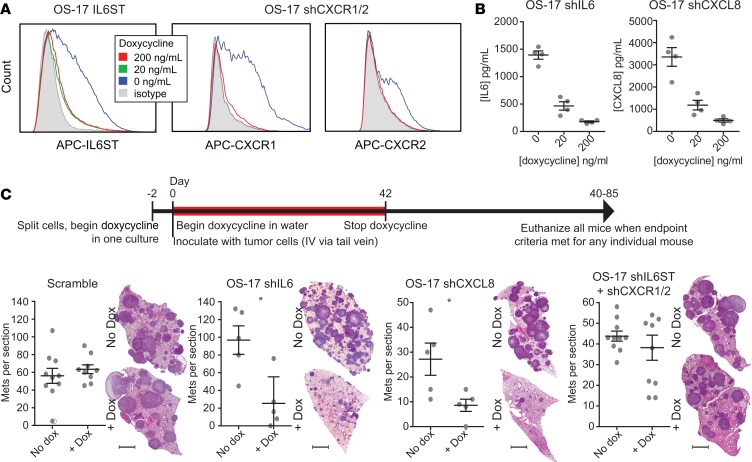
Figure 5. Tumor-derived IL-6 and CXCL8 drive osteosarcoma (OS) lung colonization.
(A) Flow cytometry for expression of surface receptors in cells modified with lentiviral constructs to inducibly express shRNAs targeting IL-6ST or both CXCR1 and -2 (single construct). (B) ELISA analysis of IL-6 and CXCL8 performed on 72-hour supernatants from OS-17 cell cultures bearing similar lentiviral constructs targeting cytokine production. (C) Quantification of metastatic colonies from lungs removed from mice inoculated with OS-17 cells bearing the inducible shRNA constructs. Mice were either maintained on water supplemented with doxycycline or with standard water. At the time that one mouse in either group met endpoint criteria, all mice within that study were euthanized and lungs harvested and metastases (Mets) counted. Scale bar: 2 mm. Representative lung sections shown (n = 5 or 10 mice per group). *P < 0.05 by Mann-Whitney U test.
ΔNp63 drives OS metastasis specifically through stimulation of IL-6 and CXCL8 production.
As our previous work suggested a link between the aberrant expression of ΔNp63 in OS and production of IL-6/CXCL8 (34), we sought to understand whether activation of this axis might drive the metastatic phenotypes observed. To test this hypothesis, we used a lentivirus-mediated inducible shRNA system designed to target specifically the ΔN isoform of p63. OS-17 cells express high levels of ΔNp63 at baseline. OS-17 cells transduced with the inducible shRNA exhibit marked knockdown of ΔNp63 upon administration of isopropyl β-D-1-thiogalactopyranoside (IPTG) (Figure 6, A and C) (See full uncut gels in Supplemental Materials). Mice inoculated with these cells exhibited significantly less metastatic lung colonization when maintained on IPTG-containing water than those receiving regular water.
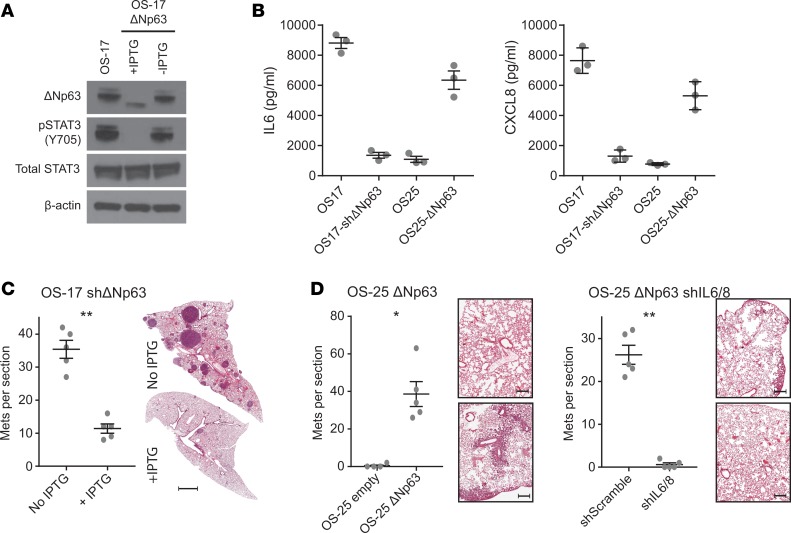
Figure 6. Tumor-derived IL-6 and CXCL8 driven by aberrant ΔNp63 expression is critical for osteosarcoma (OS) lung colonization.
(A) Knockdown of ΔNp63 using an inducible shRNA construct demonstrates strong reduction in p63 expression. (B) This reduction correlates with a loss of both IL-6 and CXCL8, while lentivirus-transduced cells overexpressing ΔNp63 increase production of IL-6 and CXCL8. (C) Mice inoculated with inducible-knockdown ΔNp63 constructs demonstrate decreased lung colonization when maintained on IPTG-containing water. Scale bar: 2 mm. (D) Mice inoculated with OS-25 cells transduced with a vector expressing ΔNp63 show a gain of lung colonizing capacity relative to empty vector–transduced cells. When those same cells are also transduced with shRNAs targeting both IL-6 and CXCL8, the increased lung colonization capacity is lost. Scale bar: 500 μm. Representative lung sections shown (n = 5 or 10 mice per group). *P < 0.05; **P < 0.01 by Mann-Whitney U test. Mets, metastases.
To determine whether ΔNp63 expression alone was sufficient to endow otherwise poorly metastatic cells with a highly metastatic phenotype, we introduced a construct driving expression of ΔNp63 into OS-25 cells. When tested using our in vivo lung colonization methods, cells expressing ΔNp63 acquired significant capacity for metastatic lung colonization (Figure 6, B and D). To determine the degree to which this phenotype depends on IL-6 and CXCL8 expression, we further modified those OS-25 cells overexpressing ΔNp63 with our IL-6– and CXCL8-targeting shRNA constructs. Suppression of these 2 cytokines in cells overexpressing ΔNp63 abolished their capacity to colonize mouse lungs (Figure 6, B and D). These results suggest not only a mechanism driving the production of IL-6 and CXCL8 in OS, but also argue strongly for a bona fide mechanistic role in lung colonization.
IL-6 and CXCL8 pathway activation does not significantly impact proliferation of OS cells.
As many of these results, including migration, invasion, and metastatic colonization assays, can sometimes be confounded by the effects of experimental conditions on cellular proliferation, we evaluated the effects of IL-6 and CXCL8 pathway inhibition on in vitro proliferation. Inhibition of these pathways did not have a meaningful effect on proliferation, neither when used alone nor in combination (Figure 7A), suggesting that the above results represent true migration and invasion.
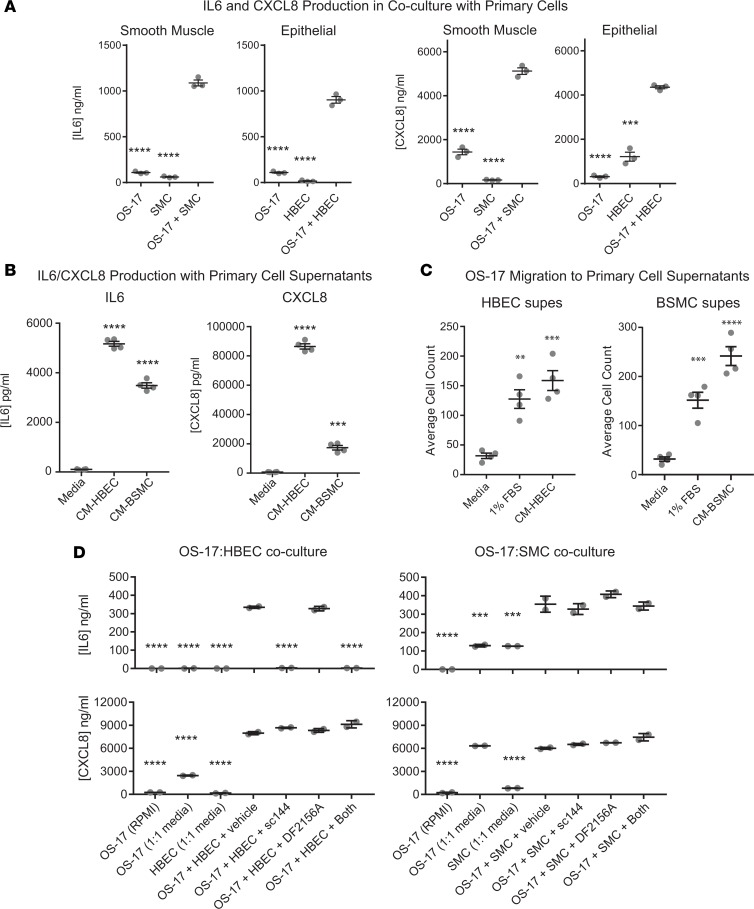
Figure 7. Interactions of OS-17 cells with bronchial epithelial and smooth muscle cells stimulate IL-6 and CXCL8 production and migration/invasion.
(A) Supernatants from cell cultures of serum-starved OS-17 grown alone or together with human bronchial smooth muscle cells (SMCs) or human bronchial epithelial cells (HBECs) for 48 hours were analyzed by ELISA for IL-6 or CXCL8. Coculture stimulated production of both IL-6 and CXCL8. (B) Supernatants from serum-starved OS-17 cultured for 48 hours in fresh mixed media or in media containing cell-free supernatants from HBECs or SMCs were likewise evaluated using ELISA. Both sets of conditioned supernatants stimulated IL-6 and CXCL8 production far above baseline. (C) Transwell migration using FBS or conditioned supernatants (supes) from HBECs or BSMCs as the chemoattractant. (D) Further refinement of tumor–primary cell interactions that drive production of IL-6 and CXCL8. After serum starvation, osteosarcoma cells were exposed to the respective media alone or to this plus the primary cells in the presence or absence of inhibitors of IL-6 and CXCL8 signaling. P values shown relative to coculture condition (A), to fresh media (B and C), or to coculture plus vehicle (D). **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA with Tukey’s post hoc test.
To ensure that no cell-autonomous mechanism intrinsic to growing tumors was affected by anti–IL-6ST or anti-CXCR1/2 therapies, we evaluated the effects of the same sc144 and DF2156A treatments utilized above on mice bearing orthotopic OS-17 tumors. After implantation of tumor cells into the proximal tibia, mice were monitored until tumors reached 400 mm2. At that time, mice were randomized to receive treatment with either vehicle or a combination of sc144 and DF2156A at the same dose and schedule as previously used. In contrast to the results seen in our models of metastatic colonization, orthotopic tumors treated with the combination of inhibitors showed no difference in growth relative to the vehicle-treated controls (Figure 4, B and C).
Coculture of OS cells with primary lung cells identifies strong interactions with bronchial epithelial cells and bronchial smooth muscle cells.
Given the exquisite tropism of OS metastases for lung tissue, we postulated that if IL-6 and/or CXCL8 play critical roles in the biology underlying lung metastasis, there might exist feed-forward interactions between OS cells and primary lung cells that drive production of these cytokines. To look for such interactions, we performed culture of OS cells both alone and in combination with primary lung cells, including human lung fibroblasts (hLFs), vascular endothelial cells (HUVECs), bronchial epithelial cells (HBECs), bronchial smooth muscle cells (BSMCs), and macrophages, then subjected cell-free supernatants to ELISA to measure expression of IL-6 and CXCL8 (Supplemental Figure 5, most relevant results shown in Figure 7A). We then looked for combinations that produced higher levels of these cytokines in coculture than in either monoculture and that exhibited these patterns in a highly metastatic cell line (OS-17), but not in a poorly metastatic line (OHS). Results suggested strong interactions with BSMCs and HBECs for both IL-6 and CXCL8. To determine whether this interaction was driven by a soluble factor or some contact-mediated signaling interaction, we cultured OS-17 cells in conditioned cell-free supernatants harvested from either HBECs or BSMCs. Both sets of supernatants drove very high levels of expression of IL-6 and CXCL8 in the OS cells (Figure 7B), and stimulated robust chemotaxis of OS cells in transwell assays (Figure 7C).
A more detailed and controlled evaluation of these interactions yielded some interesting results. Figure 7D illustrates the results of similar coculture experiments with additional control conditions and with the addition of the IL-6 and CXCL8 pathway inhibitors. In the production of IL-6, neither serum-starved OS cells nor HBECs produce large quantities of cytokine alone, but coculture drives significant IL-6 production. IL-6 production is inhibited by IL-6 blockade, suggesting the existence of a paracrine feedback loop. In contrast, while the same cocultures also drive production of CXCL8, levels of this chemokine do not decrease with inhibition of IL-6 nor of CXCL8, suggesting that a different factor drives its production and that it does not form part of a paracrine loop. We observed similar results for IL-6 production on interaction with smooth muscle cells. Interestingly, the addition of 50% smooth muscle cell growth medium in and of itself drove production of CXCL8, suggesting that this previously noted response did not occur because of cell-cell interactions, but because of factors contained in the growth medium.
To evaluate the capacity of these primary cells to respond to IL-6 and CXCL8, we performed flow cytometry for each of the relevant cell surface receptors (IL-6R, IL-6ST, CXCR1, and CXCR2) on both HBECs and BSMCs. Results (Supplemental Figure 6) showed that both cell types express receptors for IL-6. A minor subpopulation of BSMCs express CXCR2 and, to a lesser degree, CXCR1. HBECs do not express receptors for CXCL8.
Discussion
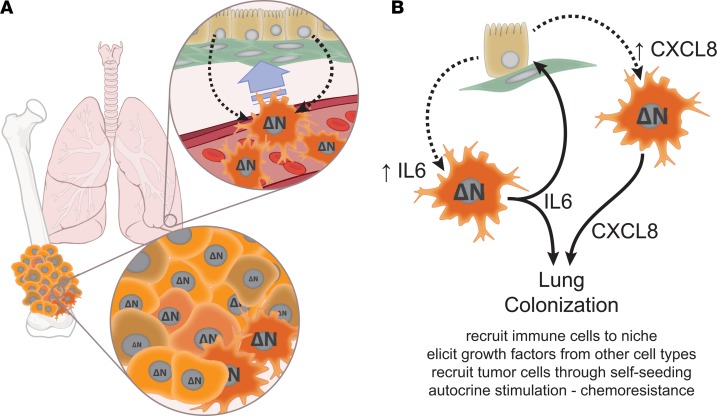
These studies illustrate what we believe are novel mechanisms that mediate tissue-specific colonization of lung by OS cells. These data can be integrated into a coherent model describing some of the OS–lung cell interactions that facilitate metastasis (Figure 8). They suggest that aberrant expression of ΔNp63 endows OS cells with lung-colonizing capacity by priming them for production of IL-6 and CXCL8. Upon arrest in the vascular beds of the lung, interactions occur that stimulate migration of the OS cells toward the micrometastatic niche. Tumor-derived IL-6 and CXCL8 increase dramatically within that niche, both in response to factors produced by the host lung cells and by the establishment of specific paracrine loops (in the case of IL-6). The downstream mechanisms by which these soluble factors mediate survival and proliferation of OS cells at this point remain poorly understood. Given the known biology of IL-6 and CXCL8, one can reasonably postulate several potential mechanisms. These factors could affect metastasis by recruiting other critical cell types to the niche, recruiting additional OS cells to the niche through a self-seeding mechanism (35), or triggering paracrine interactions that cause other host cells to produce prosurvival and growth-promoting factors. Indeed, the unidentified second factor in the OS-HBEC paracrine loop could itself also support survival and proliferation. Identification of this second signal remains an active area of investigation. Each of these potential effects would support the establishment of the micrometastatic niche.
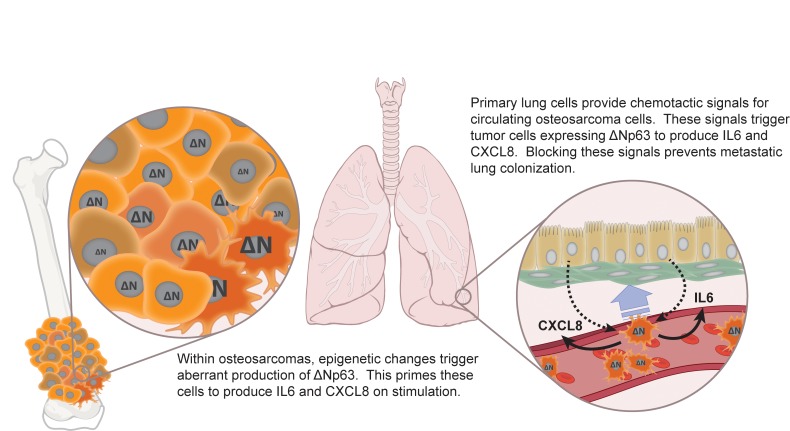
Figure 8. IL-6 and CXCL8 mediate tumor-host interactions that facilitate lung colonization by osteosarcoma (OS) cells.
(A) Our data suggest that within a primary tumor, epigenetic mechanisms activated within some tumor cells trigger aberrant expression of ΔNp63. As these cells intravasate and travel through the bloodstream, they will arrest within the capillary beds of the lung, where they can interact with multiple host cell types. Interactions between circulating OS cells and lung epithelial/smooth muscle cells drives chemotaxis into lung tissues, likely facilitating extravasation. (B) ΔNp63 expression primes select tumor cells to respond to signals provided by lung cells with robust IL-6 and CXCL8 production. At least one of these interactions (OS cells interacting with bronchial epithelium) generates a feed-forward paracrine loop, which amplifies IL-6 production. This lung cell–provoked production of IL-6 and CXCL8 facilitates colonization of lung tissue by those cells, likely through multiple mechanisms. These mechanisms might include recruitment of other cell types to the micrometastatic niche, stimulation of other cells to provide necessary growth and survival signals in a reciprocal interaction, tumor-tumor signaling to recruit additional circulating cells to the micrometastatic niche, or autocrine signaling that could provide early survival signals and could contribute to chemoresistance. See Discussion section for detailed narrative. ΔN, ΔNp63.
The metastatic trigger is likely tripped by an epigenetic event that drives aberrant expression of ΔNp63, endowing a subset of cells with increased metastatic efficiency. This primes these cells to respond to exogenous signals with exaggerated IL-6 and CXCL8 production (34). As OS cells arrest within the capillary beds of the lung, they begin interacting with the surrounding lung parenchyma. Soluble factors produced by bronchial epithelial and smooth muscle cells provide chemotactic signals and amplify IL-6 and CXCL8 production. This sets feed-forward intercellular signaling loops in motion that drive production of factors and/or recruitment of additional cell types that facilitate survival and proliferation of OS cells within the now-established metastatic niche. Ongoing experiments within our lab aim to determine whether these trigger phenomena are clonal or dynamic, to identify the proximal epigenetic events that set this metastatic module in motion, and to identify the lung-derived second signals that support the tumor cells within the niche.
Interestingly, the evidence for cell-cell interaction proved much stronger for bronchial epithelial cells and smooth muscle cells (cell types rarely implicated in malignant progression) than for more commonly implicated cell types like lung fibroblasts and macrophages. While we note that this does not prove that these are the only cells involved in the OS metastatic niche, this finding should prompt further consideration of the role these cells might play within the metastatic niche.
We note that data contained within this study show that OS tumor cells both produce large amounts of soluble IL-6 and CXCL8 and exhibit strong chemotaxis to those factors. This may suggest that at least one element of this mechanism works through self-seeding — a concept that would fit well with much of the emerging literature suggesting that metastases arise from polyclonal sources, with multiple subclones present in the primary tumor represented in distant metastases (11, 36).
At a mechanistic level, we find the pleiotropic nature of IL-6/CXCL8 and the apparent redundancy that exists within the system somewhat intriguing. While many of the detailed mechanisms underlying the phenomena outlined here remain an active area of investigation, numerous clues may be found within the existing literature. Others have previously identified ΔNp63, CXCL8, and IL-6 as potential players in lung metastasis of this and other tumors (20, 26, 35, 37, 38). Especially intriguing is the possibility that mechanisms described here could serve as a proximal event, leading to the recruitment of mesenchymal stem cells, which would further amplify these same systems (39, 40). Data presented here suggest a framework for both the mechanistic function of these pathways in lung metastasis and demonstrate a potentially novel therapeutic implication, whereby redundancy between the 2 systems requires cotargeting for ultimate efficacy.
Importantly, the effects of IL-6 and CXCL8 inhibition were not limited to any particular cell line or model. Findings recapitulated well across human, murine, and canine models of disease, suggesting broad applicability to OS, despite a well-recognized interpatient heterogeneity. We do note the importance of understanding differences in cytokine/chemokine biology between the mouse and human for accurate interpretation of xenograft experiments such as these. The known xenobiology of both IL-6 and CXCL8 supports the above hypothesis — human IL-6 readily activates the murine receptor complex and human CXCL8 activates murine CXCR1 and -2 with efficiency similar to that seen in the human receptors. A more detailed discussion of IL-6 and CXCL8 xenobiology, including supporting data, is provided in the supplemental materials.
While this work is, on one level, immediately translatable to the clinical realm, some work remains to develop clinically viable treatment regimens. sc144, specifically, is not a viable clinical candidate and we are not aware of any highly attractive clinically viable alternative for targeting IL-6ST. While viable IL-6–specific targeting agents exist, we have very intentionally chosen a therapeutic approach that targets the receptors, rather than the cytokines. This decision is based on data suggesting that ΔNp63 also drives expression of other ligands that activate both IL-6ST (IL-11, oncostatin M) and CXCR1 and -2 (CXCL1), which provides inherent resistance to ligand-targeted approaches (data not shown). Regimens targeting signaling machinery downstream of the receptors may identify very translatable approaches (PI3K, MAPK, JAK/STAT, etc.), and our group is actively exploring these alternatives. STAT3 does not appear to be the relevant intracellular signal (41). The effect that such inhibitors might have on established metastases and primary tumor likewise remains to be seen.
Methods
Human primary tumor–metastasis pairs.
Matched surgical specimens were identified through a structured search of the pathology database at Nationwide Children’s Hospital. H&E slides from each pair were reviewed by a pathologist to verify diagnosis and ensure a sufficient quantity of viable tumor. Samples showing evidence of decalcification were excluded. Paraffin blocks from identified pairs were recut at 5 μm to make 10 fresh unstained slides with an eleventh slide from the center of the stack stained with H&E. The stained slides were used to identify gross areas of viable tumor. Nontumor and necrotic tissues were scraped off of the unstained slides. These slides were then processed to extract RNA using the Qiagen RNeasy FFPE kit according to the manufacturer’s recommendations. Purified RNA was used to generate cDNA (Invitrogen VILO cDNA synthesis kit, ThermoFisher Scientific, 11754250) with pooled sequence-specific primers (Supplemental Table 1).
qPCR evaluation of candidate genes.
qPCR assays were developed for each of the 56 candidate genes identified through curation of published metastatic expression signatures as well as for a set of 4 internal control genes. Primers targeting each gene were designed using NCBI Primer-BLAST(42) to meet the following criteria: amplicon smaller than 125 bp, pair must be separated by at least 1 intron (where possible), preference for 5′ mRNA targets, no nonsynonymous target recognition for potential amplicons smaller than 1,500 bp. Primer pairs were validated using cDNA extracted from a panel of archival FFPE tissues (lung, liver, tonsil, gastric wall). Amplification was performed using RT2 SYBR Green qPCR Mastermix (Qiagen, 11754250) on an Applied Biosystems HT 7900 thermocycler. Accepted primer pairs demonstrated efficiency greater than 92% (as calculated using “window of linearity” methods; ref. 43), single, narrow curves on melting curve analysis, and consistency across all 4 validation specimens. Sequences for each of the primer pairs are listed in Supplemental Table 1. cDNAs from our primary tumor-lung metastasis pairs were then analyzed by qPCR using each primer set separately, run in triplicate. Dissociation curves were run for every assay well. Less than 5% of wells were censored from further analysis due to signs of nonspecific amplification in the dissociation curves. Cycle threshold (Ct) values were determined visually for each primer set using Applied Biosystems SDS software. Relative quantification was determined and then normalized to expression within the primary tumor for each primer set.
Immunohistochemistry for IL-6 and CXCL8.
Deparaffinized slides underwent microwave antigen retrieval with citrate AR. Slides were blocked using a background sniper and a power block for 30 minutes each. Slides were incubated with primary IL-6 antibody (Abcam, ab6672) at 1:500 or primary CXCL8 antibody (Abcam, ab7747) at 1:500 in Dako diluent overnight at 4°C, then washed and finally incubated in MACH2 (Biocare Medical, MALP521) for 20 minutes at room temperature. Sections were developed with a DAB Kit (Vector, SK-4100), counterstained with Mayer’s hematoxylin, rinsed, dehydrated, dried, and coverslipped.
Cell lines and primary cell cultures.
OS-17 was derived from an early passage of the OS-17 patient–derived xenograft, grown from a primary femur biopsy performed at St. Jude’s Children’s Research Hospital in Memphis and was a gift from Peter Houghton (44). OS-25 and OHS (45) were a gift from Øystein Fodstad’s lab at the Radium Hospital in Oslo. All were maintained in RPMI (Corning, 10-040-CV) supplemented with 10% FBS (Atlanta Biologicals, S11150H). 143B and K7M2 cells were obtained from the American Type Culture Collection (ATCC, CRL8303 and CRL2836) and grown in DMEM (Corning, 10-013-CV) supplemented with 10% FBS. OSCA-8 and OSCA-16 (33) were provided by Jamie Modiano the the University of Minnesota in Minneapolis and grown in RPMI with 10% FBS. Lung smooth muscle cells (ATCC, PCS-130-10) were grown in vascular cell basal medium (ATCC, PCS-100-030) supplemented with the vascular smooth muscle cell growth kit (ATCC, PCS-100-042). HUVECs (Lonza, CC-2517) were grown in endothelial basal medium (Lonza, CC-5036) supplemented with the EGM-plus singlequot (Lonza, CC-4542). Human lung fibroblasts (ATCC, PCS-201-013) were grown in Eagle’s Minimum Essential Medium (EMEM) (ATCC, 30-2003) supplemented with 10% FBS. HBEC3-KT cells (ATCC, CRL-4051) were grown in airway epithelial cell basal medium (ATCC, PCS-300-030) supplemented with the bronchial epithelial cell growth kit (ATCC, PCS-300-040). Macrophages were derived from monocytes isolated from whole blood using a CD14 magnetic bead selection system (Miltenyi Biotec, 130-050-201) followed by 72 hours of culture in XVIVO serum-free medium (Lonza, 04-380Q) supplemented daily with 20 ng/ml recombinant human M-CSF (BioLegend, 574802). For coculture experiments, cultures within each group (coculture and related monocultures) were performed using a 1:1 mixture of the 2 corresponding growth media to control for differences in media components.
IL-6 and CXCL8 ELISA.
Cell-free supernatants from 72-hour cultures of each cell line performed in 24-well plates were evaluated for IL-6 and CXCL8 concentrations using DuoSet ELISA Development Kits (R&D Systems, DY206 and DY208), used according to the manufacturer’s recommendations.
Scratch (wound-healing) assays.
Monolayer cultures of OS-17 or OHS cell lines were disrupted using an Essen Incucyte WoundMaker (Essen Cell Migration Kit, 4493). Individual wells were then serially imaged using an Essen Incucyte Zoom. Analysis was performed and wound width quantified using Essen’s Integrated Cell Migration Analysis Module (Essen, 9600-0012).
Transwell migration and invasion assays.
OS cells (1 × 104) were plated into transwell inserts (either Falcon 353097 for migration or Corning 354483 for Matrigel invasion assays) containing appropriate chemotactic factors. After 24 hours of incubation, transwells were drained and upper chambers/membrane upper surfaces scraped using a polyester swab. Membranes were stained using a Dif-Quik Stain Set (Siemens, B4132-1A) and dried, then imaged on an inverted microscope. Cells were quantified using Adobe Photoshop counting tools. For experiments involving IL-6 and CXCL8 chemotaxis, media contained 1% FBS in both chambers, with recombinant protein added to the bottom chamber to make 50 ng/ml IL-6 (BioLegend, 570804) or 100 ng/ml CXCL8 (BioLegend, 574204). For experiments using serum as a chemoattractant, top chambers contained RPMI only, while bottom chambers contained 1% or 2.5% serum. Where noted, 20 μg/ml of neutralizing antibodies against either IL-6 (Abcam, AB6672), CXCL8 (Abcam, AB18672), or both were added to both upper and lower chambers. In experiments testing the ability of small molecules to block serum-induced migration/invasion, 10 μM sc144 (a small molecule inhibitor of IL-6ST, the signaling receptor for IL-6; ref. 28; Sigma-Aldrich, SML0763) and/or 100 nM DF2156A (ladarixin, a small-molecule inhibitor of CXCR1 and -2, the receptors for CXCL8; ref. 29; kindly provided by Dompé Farmaceutici, L’Aquila, Italy) was added to the media.
OS cell proliferation.
Cells plated at 20% confluence were cultured in growth medium as above containing inhibitors as noted in each figure. Proliferation was serially quantified using an Essen Biosciences Incucyte Zoom over the time period noted in each figure.
Colony formation.
OS cells (1 × 104) were plated in 1.5 ml of 0.5% soft agar (Lonza SeaPlaque GTG Agarose, 50111 in Gibco powdered RPMI 430-1800) over a 1.5-ml bed of 1% soft agar in 6-well plates, then covered with 500 μl RPMI. Where noted, drug was added to the RPMI layer sufficient to generate the stated concentration when diffused throughout both media and agar.
Inducible shRNA-expressing cell lines.
OS-17 cells were transduced using lentiviral particles designed to express shRNA constructs targeting specific mRNAs. The tet-pLKO-neo/puro system (46) (a gift from Dmitri Wiederschain, Addgene plasmid 21915) combined with targeting sequences as suggested by the RNAi consortium (47) (and relying heavily on the use of their published protocols) was used to generate the lentiviruses containing sequences outlined in Supplemental Table 2. For each target, 3–6 unique hairpins were tested in OS-17 cells, then evaluated for the efficiency with which they knocked down their intended targets and the degree to which they did not interfere with expression of any of the other analytes (for instance, shRNAs targeting IL-6ST were also stained for CXCR1 and -2 and had ELISAs performed for IL-6 and CXCL8). Those shown and used represent the best performers in each group. For validation experiments, cells were stained with APC-conjugated antibodies (BioLegend, 320612 [CXCR1], 320710 [CXCR2], 362005 [IL-6ST], or 400221 [isotype]) and evaluated by flow cytometry and cell-free supernatants were subjected to IL-6 and CXCL8 ELISA as outlined above.
Xenograft survival studies.
Six- to 8-week-old CB17-SCID (Envigo C.B-17/IcrHsd-Prkdcscid) mice inoculated via tail vein with 1 × 106 OS-17 cells (day 0) received daily injections of sc144 (10 mg/kg s.c. once daily, Sigma-Aldrich, SML0763), DF2156A (30 mg/kg i.p. once daily), or both beginning 24 hours after inoculation. sc144 was prepared by dissolving and warming in DMSO to make a 40 mg/ml solution, which was immediately diluted to 2 mg/ml using 40% propylene glycol/1% Tween-20 in water. An average 20-g mouse received 100 μl per dose. Doses of sc144 were prepared fresh each day. DF2156A was prepared by dissolving in PBS to create a 6 mg/ml solution for similar 100-μl doses in a 20-g mouse (20 mg/kg). Treatments continued for 42 days, then stopped. Mice were monitored with twice weekly weights and eBCS (30). Mice demonstrating greater than 10% weight loss or eBCS less than 8 were euthanized and tissues harvested, lungs insufflated, fixed in 10% neutral buffered formalin, then embedded and processed as above. Mice not demonstrating metastatic disease burden (presumably dying from other causes) were censored in the survival analysis. This includes 2 mice receiving combined therapy, 1 receiving sc144, and 1 control mouse.
Time point treatment studies.
Six- to 8-week-old CB17-SCID mice were inoculated with 1 × 106 143B, OSCA-8, OSCA-16, or K7M2 cells (for K7M2 cells, immunocompetent BALB/c mice were used). Twenty-four hours after inoculation, mice began treatment with daily sc144 and/or DF2156A, which continued for 42 days as above. Mice were then observed as above until one mouse from any given cell line group reached endpoint. If lungs taken from this sentinel mouse showed signs of metastatic disease, all mice from that group were euthanized, lungs harvested, insufflated, fixed, embedded, and stained. A central section of the left lobe stained with H&E was reviewed using microscopy to count metastatic lesions by an experienced, blinded reviewer.
shRNA-knockdown metastasis studies.
Ten 6- to 8-week-old CB17-SCID mice were inoculated with 1 × 106 OS-17 cells transduced with lentivirus as above. For each cell line, one half of the mice were maintained on water containing 1 mg/ml doxycycline, and the other half received plain water. Mice were then observed as above until any mouse from that group reached endpoint, at which time all mice from that group were euthanized. Lungs were sectioned and analyzed as above.
Coculture of OS and primary lung cells.
OS cells (2 × 105), primary lung cells (2 × 105 HBEC3-KT or SMC cells), or a combination of the two were plated in a 24-well plate (Corning, 3524) containing a 1:1 mix of RPMI with 1% FBS and the appropriate primary cell medium along with inhibitors as shown in each figure. After 48 hours, cell-free supernatants were collected and stored at –20°C until detection of IL-6 and CXCL8 by ELISA.
Flow cytometry.
Immunostaining of HBEC3-KT and SMC cells was performed with the following antibodies: PE anti–human CD130 (IL-6ST) (Biolegend, 362003); PE mouse IgG2a, κ isotype control (Biolegend, 400213); FITC anti–human CD181 (CXCR1) (Biolegend, 320605); FITC mouse IgG2b, κ isotype control (Biolegend, 400309); APC anti–human CD126 (IL-6Rα) (Biolegend, 352805); PAC mouse IgG1, κ isotype control (Biolegend, 400121); PerCP anti–human CD182 (CXCR2) (Invitrogen, 46-1829-42); and mouse IgG1 κ isotype control, PerCP-eFluor 710 (Invitrogen, 46-4714-82). Flow cytometry analysis was performed on a BD Biosciences LSR II.
Statistics.
Data were graphed and analyzed using Prism 7 (GraphPad Software, Inc.). The qPCR gene profiling data were analyzed using a 1-sample t test; adjustment for multiple comparisons was performed using the Benjamini-Hochberg method to control for a false discovery rate (FDR) of 0.05. For experiments involving comparisons between more than 2 groups, data were subjected to analysis of variance (ANOVA) followed by Tukey’s post hoc testing. For metastasis data (which do not follow a normal distribution), groups were compared using Mann-Whitney U test. Time to lethal metastasis and overall survival data were analyzed using Cox’s log-rank methodologies. For all plots shown with error bars, the data represent the mean ± the standard error of the mean (SEM). A P value of less than 0.05 was considered significant.
Study approval.
Approval for all studies involving the use of human tissues was obtained from the Nationwide Children’s Hospital Institutional Review Board. Where required, written informed consent was received from participants prior to inclusion in the study (tumor samples and blood samples for macrophage isolation). Ethical approval for all animal studies presented was obtained from the Institutional Animal Care and Use Committee of the Research Institute at Nationwide Children’s Hospital prior to conducting experiments.
Author contributions
ACG conducted experiments, acquired data, analyzed data, and assisted in writing the manuscript. HC designed research studies, conducted experiments, acquired data, provided reagents, and assisted in writing the manuscript. DAP, AJS, HKB, MC, SAW, and JMH conducted experiments, acquired data, and analyzed data. CAL, LB, and PJH designed research studies and provided reagents. MAA identified specimens and provided pathology review. XM assisted with statistical analysis and writing the manuscript. RDR oversaw the conduct of this work, designed research studies, conducted experiments, acquired data, analyzed data, prepared graphics, and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the contributions of several high school and undergraduate students for their contributions supporting the production of this manuscript, including Collin Campbell, Brian Lucero, Alexander Salmons, and Sophia Vatelle.
This work was supported by grants from the NCI (K08CA201638 to R.D. Roberts, P01CA165995 to P.J. Houghton, C.A. London, H. Cam, and R.D. Roberts), St. Baldrick’s Foundation Scholar Award (to R.D. Roberts), a 2014 QuadW Foundation-AACR Fellowship for Clinical/Translational Sarcoma Research (14-40-37-ROBE to R.D. Roberts), the Research Institute at Nationwide Children’s Hospital (to R.D. Roberts), and a CTSA award from NIH (UL1TR001070).
Version 1. 08/23/2018
Electronic publication
Footnotes
Conflict of interest: Laura Brandolini is an employee of Dompé Farmaceutici SpA, Italy. The company has interests in the development of CXCR1/2 inhibitors for the treatment of CXCL8-associated pathologies.
Reference information: JCI Insight. 2018;3(16):e99791. https://doi.org/10.1172/jci.insight.99791.
Contributor Information
Hakan Cam, Email: hakan.cam@nationwidechildrens.org.
Maren Cam, Email: maren.cam@nationwidechildrens.org.
Laura Brandolini, Email: laura.brandolini@dompe.com.
Xiaokui Mo, Email: xiaokui.mo@osumc.edu.
References
- 1.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis IJ, et al. Improvement in histologic response but not survival in osteosarcoma patients treated with intensified chemotherapy: a randomized phase III trial of the European Osteosarcoma Intergroup. J Natl Cancer Inst. 2007;99(2):112–128. doi: 10.1093/jnci/djk015. [DOI] [PubMed] [Google Scholar]
- 3.Meyers PA, et al. Osteosarcoma: the addition of muramyl tripeptide to chemotherapy improves overall survival--a report from the Children’s Oncology Group. J Clin Oncol. 2008;26(4):633–638. doi: 10.1200/JCO.2008.14.0095. [DOI] [PubMed] [Google Scholar]
- 4.Bielack SS, et al. Methotrexate, doxorubicin, and cisplatin (MAP) plus maintenance pegylated interferon alfa-2b versus MAP alone in patients with resectable high-grade osteosarcoma and good histologic response to preoperative MAP: First results of the EURAMOS-1 Good Response Randomized Controlled Trial. J Clin Oncol. 2015;33(20):2279–2287. doi: 10.1200/JCO.2014.60.0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marina NM, et al. Comparison of MAPIE versus MAP in patients with a poor response to preoperative chemotherapy for newly diagnosed high-grade osteosarcoma (EURAMOS-1): an open-label, international, randomised controlled trial. Lancet Oncol. 2016;17(10):1396–1408. doi: 10.1016/S1470-2045(16)30214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allison DC, et al. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma. 2012;2012:704872. doi: 10.1155/2012/704872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aljubran AH, Griffin A, Pintilie M, Blackstein M. Osteosarcoma in adolescents and adults: survival analysis with and without lung metastases. Ann Oncol. 2009;20(6):1136–1141. doi: 10.1093/annonc/mdn731. [DOI] [PubMed] [Google Scholar]
- 8.Khanna C, et al. Toward a drug development path that targets metastatic progression in osteosarcoma. Clin Cancer Res. 2014;20(16):4200–4209. doi: 10.1158/1078-0432.CCR-13-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treat Rev. 2014;40(4):523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turajlic S, Swanton C. Metastasis as an evolutionary process. Science. 2016;352(6282):169–175. doi: 10.1126/science.aaf2784. [DOI] [PubMed] [Google Scholar]
- 12.Sethi N, Kang Y. Unravelling the complexity of metastasis - molecular understanding and targeted therapies. Nat Rev Cancer. 2011;11(10):735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanharanta S, Massagué J. Origins of metastatic traits. Cancer Cell. 2013;24(4):410–421. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 15.Stott SL, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2(25):25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruland OS, Høifødt H, Saeter G, Smeland S, Fodstad O. Hematogenous micrometastases in osteosarcoma patients. Clin Cancer Res. 2005;11(13):4666–4673. doi: 10.1158/1078-0432.CCR-05-0165. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Cao X. Characteristics and significance of the pre-metastatic niche. Cancer Cell. 2016;30(5):668–681. doi: 10.1016/j.ccell.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 19.Powell AA, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7(5):e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paoloni M, et al. Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genomics. 2009;10:625. doi: 10.1186/1471-2164-10-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanna C, et al. Metastasis-associated differences in gene expression in a murine model of osteosarcoma. Cancer Res. 2001;61(9):3750–3759. [PubMed] [Google Scholar]
- 22.Leonard P, et al. Gene expression array profile of human osteosarcoma. Br J Cancer. 2003;89(12):2284–2288. doi: 10.1038/sj.bjc.6601389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buddingh EP, et al. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res. 2011;17(8):2110–2119. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 24.Endo-Munoz L, et al. Loss of osteoclasts contributes to development of osteosarcoma pulmonary metastases. Cancer Res. 2010;70(18):7063–7072. doi: 10.1158/0008-5472.CAN-09-4291. [DOI] [PubMed] [Google Scholar]
- 25.Javed A, Chen H, Ghori FY. Genetic and transcriptional control of bone formation. Oral Maxillofac Surg Clin North Am. 2010;22(3):283–293. doi: 10.1016/j.coms.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slootweg MC, Most WW, van Beek E, Schot LP, Papapoulos SE, Löwik CW. Osteoclast formation together with interleukin-6 production in mouse long bones is increased by insulin-like growth factor-I. J Endocrinol. 1992;132(3):433–438. doi: 10.1677/joe.0.1320433. [DOI] [PubMed] [Google Scholar]
- 27.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17(7):867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu S, Grande F, Garofalo A, Neamati N. Discovery of a novel orally active small-molecule gp130 inhibitor for the treatment of ovarian cancer. Mol Cancer Ther. 2013;12(6):937–949. doi: 10.1158/1535-7163.MCT-12-1082. [DOI] [PubMed] [Google Scholar]
- 29.Bertini R, et al. Receptor binding mode and pharmacological characterization of a potent and selective dual CXCR1/CXCR2 non-competitive allosteric inhibitor. Br J Pharmacol. 2012;165(2):436–454. doi: 10.1111/j.1476-5381.2011.01566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paster EV, Villines KA, Hickman DL. Endpoints for mouse abdominal tumor models: refinement of current criteria. Comp Med. 2009;59(3):234–241. [PMC free article] [PubMed] [Google Scholar]
- 31.Khanna C, Prehn J, Yeung C, Caylor J, Tsokos M, Helman L. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clin Exp Metastasis. 2000;18(3):261–271. doi: 10.1023/A:1006767007547. [DOI] [PubMed] [Google Scholar]
- 32.Zhao S, et al. NKD2, a negative regulator of Wnt signaling, suppresses tumor growth and metastasis in osteosarcoma. Oncogene. 2015;34(39):5069–5079. doi: 10.1038/onc.2014.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott MC, et al. Molecular subtypes of osteosarcoma identified by reducing tumor heterogeneity through an interspecies comparative approach. Bone. 2011;49(3):356–367. doi: 10.1016/j.bone.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bid HK, et al. ΔNp63 promotes pediatric neuroblastoma and osteosarcoma by regulating tumor angiogenesis. Cancer Res. 2014;74(1):320–329. doi: 10.1158/0008-5472.CAN-13-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim MY, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139(7):1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gundem G, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520(7547):353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez Calleja L, et al. ΔNp63α silences a miRNA program to aberrantly initiate a wound-healing program that promotes TGFβ-induced metastasis. Cancer Res. 2016;76(11):3236–3251. doi: 10.1158/0008-5472.CAN-15-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thanapprapasr K, Nartthanarung A, Thanapprapasr D, Jinawath A. pFAK-Y397 overexpression as both a prognostic and a predictive biomarker for patients with metastatic osteosarcoma. PLoS One. 2017;12(8):e0182989. doi: 10.1371/journal.pone.0182989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baglio SR, et al. Blocking tumor-educated MSC paracrine activity halts osteosarcoma progression. Clin Cancer Res. 2017;23(14):3721–3733. doi: 10.1158/1078-0432.CCR-16-2726. [DOI] [PubMed] [Google Scholar]
- 40.Tu B, Du L, Fan QM, Tang Z, Tang TT. STAT3 activation by IL-6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett. 2012;325(1):80–88. doi: 10.1016/j.canlet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Yu PY, et al. Target specificity, in vivo pharmacokinetics, and efficacy of the putative STAT3 inhibitor LY5 in osteosarcoma, Ewing’s sarcoma, and rhabdomyosarcoma. PLoS One. 2017;12(7):e0181885. doi: 10.1371/journal.pone.0181885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339(1):62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 44.Houghton PJ, et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr Blood Cancer. 2007;49(7):928–940. doi: 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 45.Fodstad O, Brøgger A, Bruland O, Solheim OP, Nesland JM, Pihl A. Characteristics of a cell line established from a patient with multiple osteosarcoma, appearing 13 years after treatment for bilateral retinoblastoma. Int J Cancer. 1986;38(1):33–40. doi: 10.1002/ijc.2910380107. [DOI] [PubMed] [Google Scholar]
- 46.Wiederschain D, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8(3):498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 47.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3(9):715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.