Abstract
Defective DNA methylation in T cells leads to a series of T cell abnormalities in lupus; however, the full effect of T cell lineage–specific DNA methylation on disease expression has not been explored. Here, we show that 5-azacytidine, a DNA methyltransferase inhibitor, targeted to either CD4 or CD8 T cells in mice with established disease using a nanolipogel delivery system dramatically ameliorates lupus-related pathology through distinct mechanisms. In vivo targeted delivery of 5-azacytidine into CD4 T cells favors the expansion and function of Foxp3+ Tregs, whereas targeted delivery to CD8 T cells enhances the cytotoxicity and restrains the expansion of pathogenic TCR-αβ+CD4–CD8– double-negative T cells. Our results signify the importance of cell-specific inhibition of DNA methylation in the treatment of established lupus.
Keywords: Autoimmunity
Keywords: Cellular immune response, Lupus, T cells
5-azacytidine, targeted to either CD4 or CD8 T cells via a nanolipogel delivery system, ameliorates lupus-related pathology in lupus prone mice with established disease.
Introduction
Systemic lupus erythematosus (SLE) is a chronic debilitating autoimmune disease characterized by the production of autoantibodies targeted to a variety of nuclear antigens (1). Several cellular and biochemical pathways have been proposed to explain disease development (2, 3), but none has been identified so far as shared by the majority of patients who carry the clinical diagnosis of SLE. Because lupus patients exhibit global changes of DNA methylation patterns and two medications known to cause drug-induced lupus (procainamide and hydralazine) were found to inhibit methylation (4), it has been proposed that DNA hypomethylation contributes to the induction and progression of SLE (5–7).
T cell abnormalities are common in patients with SLE. Prominent among these are the activation and expansion of pathogenic Th1, Th17, and TCR-αβ+CD4–CD8– double-negative (DN) T cells and the failure of regulatory CD4/CD8 T lymphocytes (3, 8). The differentiation and function of each T cell subset is controlled by the cooperative action of different transcription factors (9). The commitment of different T cell lineages was originally viewed as a unidirectional process with nonreversible terminal differentiation, but it has become evident that T cells exhibit plasticity and are able to convert from one functional subtype to another (9, 10). DNA methylation represents a core epigenetic control mechanism that directs T cell activation and differentiation (11, 12). Generalized DNA hypomethylation has been linked to autoimmunity, presumably by favoring the expression of proinflammatory genes (13). However, both hypomethylated and hypermethylated cytosine-guanine sites have been found in T cells from patients with SLE compared with normal controls in several genome-wide DNA methylation studies (14, 15).
The global effect of DNA methylation in different cell types renders systemic treatment outcomes unpredictable, but the concept of precisely modulating DNA methylation in T cell subsets has transformative potential in the treatment of autoimmune disease and beyond (16). We loaded nanolipogels (nlg) (17, 18) with the most widely used epigenetic modulator, 5-azacytidine (5-Aza), a chemical analog of cytidine that inhibits DNA methylation (19), and, after tagging them with specific antibodies for delivery to T cell subsets, we found unexpectedly that 5-Aza promoted Treg expansion when delivered to CD4 T cells and inhibited the generation of DN cells when delivered to CD8 T cells. Both targeted deliveries, unlike the systemic delivery of 5-Aza (20, 21), resulted in disease suppression. Our studies reveal an unexpected therapeutic effect of DNA demethylation exercised precisely on either CD4 or CD8 T cells and its importance in reversing established disease.
Results
nlg-5-Aza targeted to CD4+ or CD8+ T cells but not free 5-Aza ameliorates disease manifestations in lupus-prone MRL/lpr mice.
Aberrant DNA methylation in T cells can induce dysregulation of the immune system and lead to autoimmunity. Although there is evidence that DNA methylation in T lymphocytes may contribute to the expression of SLE, it is unknown whether DNA methylation events in each T cell subset contribute independently to the expression of the disease. 5-Aza is a cytosine analog that is incorporated into DNA during the S phase of cell cycle, where it inhibits DNA methyltransferase (DNMT) (19). Unfortunately, the clinical application of this DNA-modulating drug is limited by its short half-life (22), the high doses required to accomplish the desirable effect (23), and possible unwanted effects on many types of T cells. To overcome these obstacles, we explored the possibility of delivering 5-Aza in a cell-targeted manner by loading the drug into nlg tagged with cell-specific antibodies (17, 18, 24).
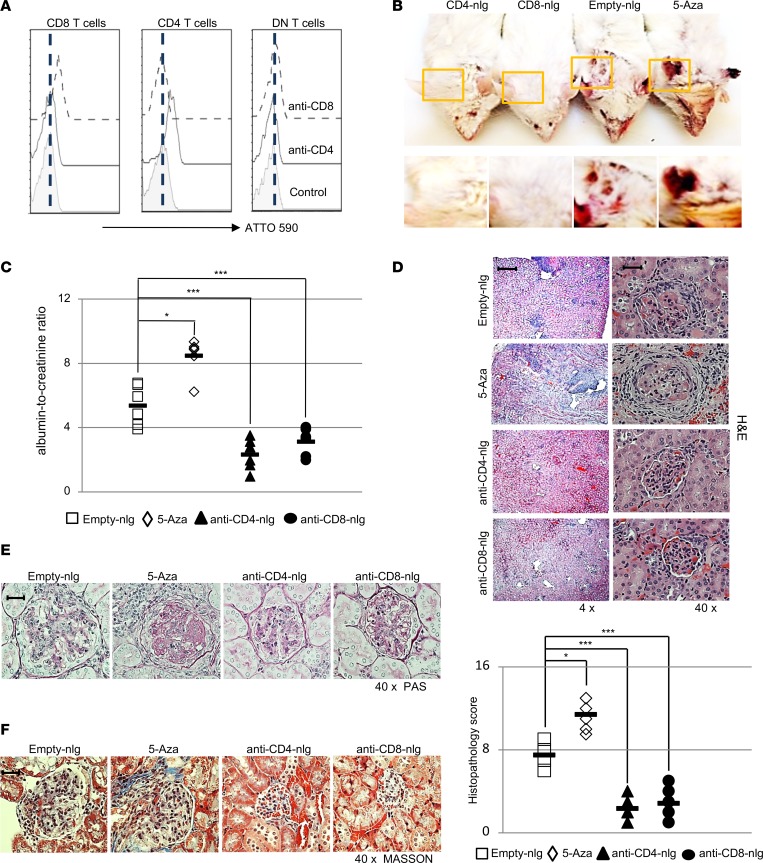
To test the delivery specificity of antibody-coated nlg in vivo, we injected ATTO590-labeled (a fluorescent dye derived from rhodamine) nondepleting mouse CD4 or mouse CD8 monoclonal antibody–coated nlg (Supplemental Figures 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.120880DS1) into MRL/lpr lupus-prone mice. Flow cytometry and confocal images revealed that only targeted T cells were positive for ATTO590 (Supplemental Figure 3A), whereas nontargeted T cells, including CD4−CD8− DN T cells (defined as CD3+TCR-β+TCR-γδ−CD49b−CD4−CD8− to exclude γδ T cells and NKT cells) remained negative 30 minutes after the administration of nlg (Figure 1A and Supplemental Figure 3B). In order to assess the independent contribution of DNA demethylation in CD4 or CD8 T cells to lupus pathogenesis in a well-established disease milieu, 5-Aza–loaded nlg coated with either CD4 or CD8 antibodies (15 μl nlg loaded with 5-Aza (nlg-5-Aza) per mouse, a dose comparable to 5 μg nlg-5-Aza) was administrated weekly into MRL/lpr mice, starting at 12 weeks of age (when both serum autoantibodies and proteinuria were observed). Age- and sex-matched mice treated with either unloaded nlg (empty-nlg) administered i.v. or free 5-Aza (5 μg/mouse) administered i.p. were used as controls. 60 days later, mice receiving 5-Aza systemically developed more severe facial rash and skin lesions than the control empty-nlg–treated mice (Figure 1B). Interestingly though, mice treated with anti-CD4– or anti-CD8–tagged nlg loaded with 5-Aza displayed less or no skin rash (Figure 1B). Similarly, administration of anti-CD4– or anti-CD8–tagged nlg-5-Aza reduced proteinuria (Figure 1C) and kidney pathology, as manifested by the reduced mesangial cell proliferation and crescent formation (Figure 1, D–F) and by limited presence of inflammatory cells (Supplemental Figure 4), whereas mice treated with free 5-Aza showed increased proteinuria and kidney pathology when compared with mice treated with empty-nlg.
Figure 1. nlg-5-Aza targeted to CD4+ or CD8+ cells but not free 5-Aza ameliorates disease manifestations in lupus-prone MRL/lpr mice.
(A) 12-week-old MRL/lpr mice were treated i.v. with either anti-CD4 antibody– or anti-CD8 antibody–coated nanolipogel-ATTO590 (nlg-ATTO590) (a fluorescent dye derived from rhodamine), and isotype control antibody–coated nlg-ATTO590 was used as control. Mice were euthanized 30 minutes after nlg administration for analysis. n = 4 mice per group. (B–F) MRL/lpr mice were treated with either anti-CD4 antibody–coated nlg-5-Aza (15 μl nlg-5-Aza per mouse, a dose comparable to 5 μg 5-Aza per mouse) or anti-CD8 antibody–coated nlg-5-Aza (15 μl nlg-5-Aza per mouse) every 10 days for 60 days, starting at 12 weeks of age. Free-5-Aza (5 μg/mouse) or empty-nlg was applied to 2 control groups separately. n = 5–6 mice per group in 2 independent experiments. (A) Flow cytometry quantitation of ATTO590 intensity in different T cell subsets from spleens of mice subjected to the indicated treatment (CD3+TCRβ+TCR-γδ–CD49b– gated). (B) Representative images of facial skin from mice subjected to the indicated treatment. (C) The ratio of urine albumin to creatinine from mice subjected to the indicated treatment. (D) Representative images of H&E staining of kidneys from mice with the indicated treatment and histopathologic scoring of kidneys from mice with the indicated treatment. Original magnification, ×4 (left); ×40 (right). Scale bar: 160 μm (left); 20 μm (right). (E) Representative images of PAS staining of kidneys from mice with the indicated treatment. Original magnification, ×40. Scale bar: 20 μm. (F) Representative images of Masson staining of kidneys from mice with the indicated treatment. Original magnification, ×40. Scale bar: 20 μm. Data represent the mean ± SEM. *P < 0.05, ***P < 0.005 vs. control; 2-tailed Student’s t test.
nlg-5-Aza targeted to CD4+ or CD8+ cells suppresses systemic autoimmunity in lupus-prone MRL/lpr mice.
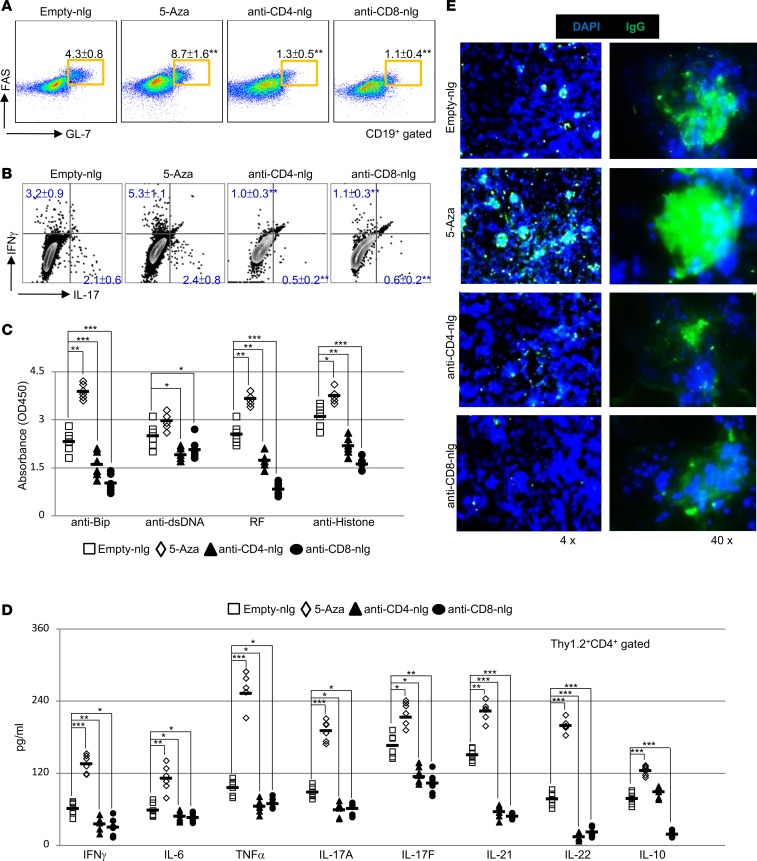
To determine the cellular mechanisms of targeted T cell delivery of 5-Aza, we first examined the formation of spontaneous germinal centers and autoantibody production. Flow cytometry analysis confirmed that systemic administration of 5-Aza promoted, whereas CD4- or CD8-targeted delivery of 5-Aza diminished, the frequency of germinal center B cells (Figure 2A) and inflammatory Th1/Th17 cells in peripheral lymphoid organs of treated MRL/lpr mice (Figure 2B and Supplemental Figure 5). We next assessed the titers of circulating autoantibodies and found that while free 5-Aza enhanced, nlg-5-Aza significantly reduced, the titers of various circulating autoantibodies, including those against dsDNA (Figure 2C). Similarly, bead-based ELISA assays confirmed that nlg-5-Aza delivered to either CD4 or CD8 T cells, in contrast to systemically administrated 5-Aza, reduced serum levels of multiple proinflammatory cytokines, including IFN-γ, IL-6, TNF-α, and IL-17 (Figure 2D). Interestingly, significant reduction of IL-10, an immunomodulatory cytokine, was only observed in mice treated with anti-CD8– but not anti-CD4–tagged nlg-5-Aza (Figure 2D). Consistently, immunofluorescent image analysis confirmed reduced IgG deposition in the kidneys of mice treated with nlg-5-Aza but not in the kidneys of animals treated with free 5-Aza, which instead showed increased IgG deposition (Figure 2E). Together, these findings suggest that enhanced DNA demethylation in either CD4 or CD8 T cells is clinically beneficial in mice with established disease, despite the fact that global demethylation orchestrates the development of lupus.
Figure 2. nlg-5-Aza targeted to CD4+ or CD8+ cells suppresses systemic autoimmunity in lupus-prone MRL/lpr mice.
(A–E) MRL/lpr mice were treated i.v. with either anti-CD4 antibody–coated nlg-5-Aza (15 μl nlg-5-Aza/mouse) or anti-CD8 antibody–coated nlg-5-Aza (15 μl nlg-5-Aza/mouse) every 10 days for 60 days, starting at 12 weeks of age. Free-5-Aza (5 μg/mouse) or empty-nlg were used in 2 control groups separately. (A) Flow cytometry quantitation of the percentage of peanut agglutinin+FAS+ germinal center B cells (CD19+ gated) in the spleens of mice subjected to the indicated treatment. (B) Flow cytometry quantitation of the percentage of IL-17+ and IFN-γ+ CD4 T cells (Thy1.2+CD4+ gated) in the spleens of mice subjected to the indicated treatment. (C) ELISA analysis of IgG autoantibodies in the sera from mice subjected to the indicated treatment. (D) Bead-based ELISA analysis of indicated cytokines in sera from mice subjected to the indicated treatment. (E) Representative immunofluorescent staining of IgG deposition in the kidney glomeruli from the mice with the indicated treatment. Original magnification, ×4 (left); ×40 (right). Scale bar: 100 μm (left); 20 μm (right). Data represent the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005 vs. control; 2-tailed Student’s t test. n = 5–6 mice per group for 2 independent experiments.
Specific targeting of nlg-5-Aza in CD4+ T cells suppresses autoimmunity by increasing Tregs.
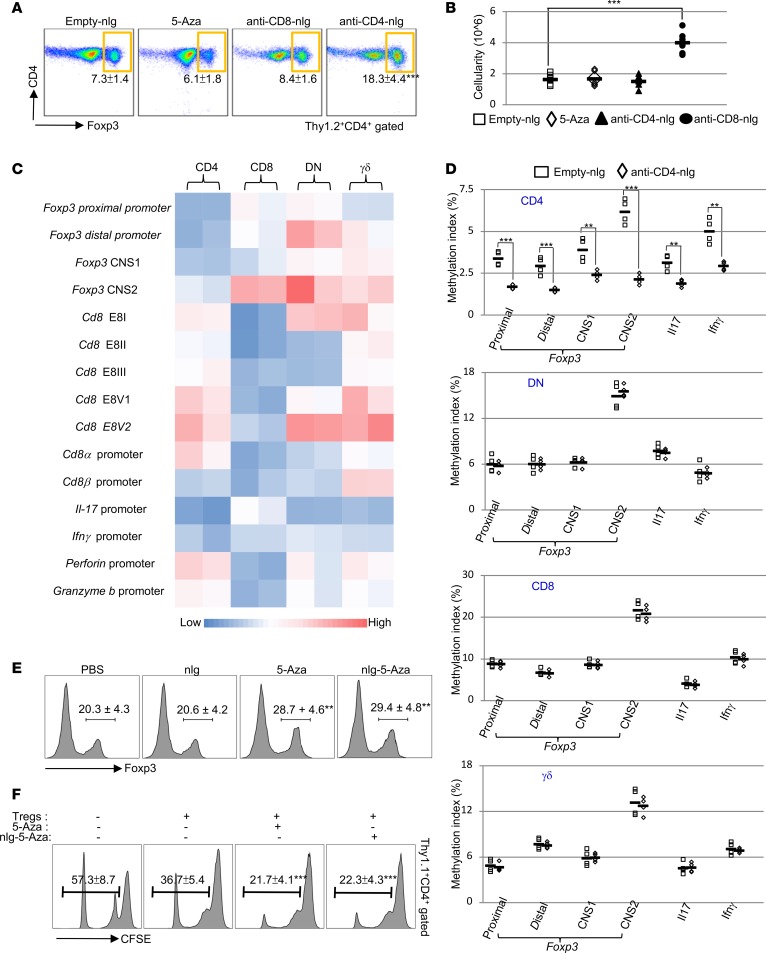
Because the production of IL-10 by Tregs is essential for the suppression of the immune response (25) and 5-Aza treatment has been reported to induce and stabilize Foxp3 expression in murine CD4 T cells (26), we asked whether there was an increase in the numbers of Tregs in mice treated with anti-CD4–tagged nlg–5-Aza (anti-CD4-nlg-5-Aza). Flow cytometry confirmed that, in anti-CD4-nlg-5-Aza– but not in anti-CD8-nlg-5-Aza–treated mice, Tregs were significantly expanded in the spleens (Figure 3, A and B) and cervical lymph nodes (Supplemental Figure 6). An important question arose as to whether the expansion of Tregs represents a direct cellular effect of anti-CD4-nlg-5-Aza. First, we performed quantitative real-time PCR (real-time qPCR) analysis on immunoprecipitated methylated DNA (MeDIP) to determine the DNA methylation index in different T lymphocytes from MRL/lpr mice. As expected, the 5 Cd8 enhancers, which individually or in combination direct CD8 expression, were highly methylated in CD4 and γδ T cells but not CD8 T cells (Figure 3C). In addition, lower levels of DNA methylation were observed in CD4 T cells on promoters and conserved noncoding DNA sequence elements at the Foxp3 locus (27) compared with other T lymphocytes (Figure 3C). As noted, increased DNA methylation was observed in DN T cells on Cd8 enhancer E8V (Figure 3C), deficiency of which could lead to CD8 variegation (28), but not on E8II and E8III (Figure 3C), which do not contribute to the expression of CD8 in peripheral T cells (29). To assess the targeting specificity of anti-CD4-nlg-5-Aza in vivo, different T lymphocytes were isolated from MRL/lpr mice 10 hours after administration of either anti-CD4-nlg-5-Aza or empty-nlg. MeDIP real-time qPCR assays revealed that a significant reduction of DNA methylation occurred in CD4 T cells, whereas no changes of DNA methylation were observed among non-CD4 T lymphocytes (Figure 3D), which demonstrated the delivery specificity.
Figure 3. Specific targeting of nlg-5-Aza in CD4+ T cells suppresses autoimmunity by increasing Tregs.
(A and B) MRL/lpr mice were treated i.v. with either anti-CD4 antibody–coated nlg-5-Aza (15 μl nlg-5-Aza/mouse) or anti-CD8 antibody–coated nlg-5-Aza (15 μl nlg-5-Aza/mouse). The treatment was provided every 10 days for 60 days, starting at 12 weeks of age. Free-5-Aza (5 μg/mouse) or empty-nlg was applied to 2 control groups separately. n= 5–6 mice per group in 2 independent experiments. (A) Flow cytometry analysis of the percentage of Foxp3+ cells in splenic CD4 T cells (Thy1.2+CD4+ gated) from mice subjected to the indicated treatment. (B) Quantitation of the absolute cell numbers of Foxp3+ Tregs (Thy1.2+CD4+gated) in the spleens of mice treated as indicated. (C) Heatmap analysis of DNA methylation index on indicated gene promoters or enhancers in splenic T lymphocytes from 12-week-old MRL/lpr mice. n= 2 mice per group in 2 independent experiments. (D) Quantitation of DNA methylation on indicated gene promoters or enhancers in indicated splenic T lymphocytes sorted from 12-week-old MRL/lpr mice 10 hours after anti-CD4 antibody–coated nlg-5-Aza (15 μl nlg-5-Aza/mouse) treatment. n= 4 mice per group. (E) Flow cytometry analysis shows the induction of Foxp3 in CD4 T cells polarized in vitro. Naive CD4 T cells from Foxp3-YFP-Tg mice were polarized under Treg-inducing conditions for 7 days with vehicle, 5-Aza (1 μM), or nlg-5-Aza (equivalent to 1 μM free 5-Aza) added 12 hours right before collection. n= 3–4 per group in 2 independent experiments. (F) Flow cytometry–based assay of in vitro Treg-mediated suppression (Thy1.1+CD4+ gated). Thy1.2+CD4+YFP+ T cells were sorted from Foxp3-YFP-Tg mice, treated with or without 5-Aza (1 μM) or nlg-5-Aza (equivalent to 1 μM free 5-Aza) for overnight, and then mixed with naive CFSE-labeled Thy1.1+CD4+ Tconv cells plus anti-CD3– and anti-CD28–stimulating antibodies for 72 hours at a 1:5 ratio. Proliferation of Tconv cells was determined by CFSE dilution. n= 3–4 for 2 independent experiments. Data represent the mean ± SEM. **P < 0.01, ***P < 0.005 vs. control; 2-tailed Student’s t test.
Interestingly, although simultaneous DNA demethylation on both antiinflammatory and proinflammatory genes was observed, the expansion of Foxp3+ Tregs plays a dominant role and leads immune suppression. In parallel, CD4 T cells were isolated from MRL/lpr mice 10 hours after the administration of either anti-CD4- or anti-CD8-nlg-5-Aza (mice treated with either empty-nlg or free 5-Aza were used as controls). As expected, a significant reduction of DNA methylation occurred only in cells from mice treated with anti-CD4-nlg-5-Aza but not in cells from mice treated with anti-CD8-nlg-5-Aza, which further confirmed the specificity of targeted delivery. Of note, anti-CD4-nlg-5-Aza treatment led to more profound reductions in DNA methylation from CD4 T cells compared with free 5-Aza given systemically, which is consistent with our previous reports (17, 18, 24) that the nlg platform can efficiently reduce the administered dose of drug for equivalent therapeutic effects (Supplemental Figure 7).
It has been demonstrated that the Foxp3 enhancer, approximately 5 kb upstream of the transcriptional start site, is differentially methylated in different T cell lineages (27, 30). 5-Aza could demethylate and activate this element to induce Foxp3 expression in CD4 T cells (26), which subsequently allows the induction of Foxp3-dependent, Treg-restricted sets of genes (30). We confirmed that in vitro 5-Aza treatment could directly promote Foxp3 expression via epigenetic modulation. Flow cytometry revealed a significant upregulation of Foxp3 in CD4 T cells under Treg-polarizing conditions in the presence of 5-Aza (Figure 3E) (31). Similar enhanced induction of Foxp3 was also observed in 5-Aza–treated human CD4 T cells under Treg-polarizing condition (Supplemental Figure 8, A and B). We next investigated whether 5-Aza could enhance Treg-suppressive function. Indeed, 5-Aza–pretreated Tregs displayed an enhanced ability to suppress the proliferation of cocultured effector T cells (Figure 3F).
Specific targeting of nlg-5-Aza in CD8+ T cells significantly reduces the DN T cell pool.
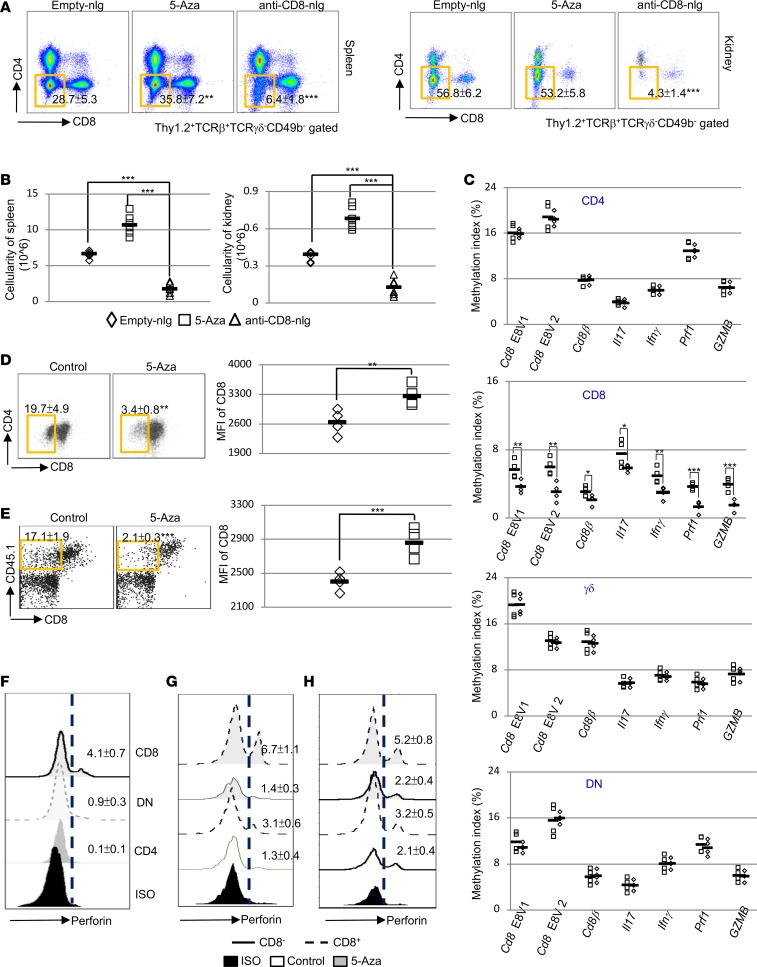
Although 5-Aza could efficiently induce and enhance Foxp3 expression through DNA demethylation, we did not observe induction of Foxp3 in CD8 T cells (Supplemental Figure 9) and it was still not clear how anti-CD8–tagged nlg-5-Aza conferred immune suppression in vivo. To address this question, we investigated the possible effects on the numbers of each T cell subset in lymphoid and nonlymphoid tissues of mice treated with nlg-5-Aza. Interestingly, flow cytometry revealed that there was a significant reduction of DN T cells (CD3+TCRβ+TCR-γδ–CD49b–CD4–CD8–) in the spleens, the kidneys, and the cervical lymph nodes of mice treated with anti-CD8-nlg-5-Aza (Figure 4, A and B, and Supplemental Figure 10), suggesting that the decrease of this subset of cells could be the direct outcome of DNA demethylation in CD8 T cells. Autoreactive DN T cells have been implicated in the pathogenesis of lupus in both patients and mice. We and others have previously presented evidence suggesting that DN T cells derive from CD8+ T cells upon antigenic stimulation (32–36). The intermediate levels of DNA methylation on 5 Cd8 enhancers (37, 38) in DN T cells reinforces our hypothesis that self-antigen–activated CD8 T cells can acquire inflammatory capacities (36) and become DN T cells through loss of CD8 expression mediated by DNA methylation (35, 36). To assess the targeting specificity of anti-CD8-nlg-5-Aza in vivo, anti-CD8-nlg-5-Aza or empty-nlg was administrated into MRL/lpr mice, and 10 hours after administration, cells were collected for MeDIP real-time qPCR analysis. Consistent with the results presented in the previous section, reduced DNA methylation was observed only in CD8 T cells but not in non-CD8 T lymphocytes, which further demonstrated the specificity of targeted delivery through the nlg carriers (Figure 4C). Parallel assessment on CD8 T cells isolated from MRL/lpr mice 10 hours after administration of either anti-CD4 or anti-CD8-nlg-5-Aza further confirmed the specificity of targeted delivery (Supplemental Figure 11). Furthermore, MeDIP real-time qPCR assays were performed to evaluate the DNA methylation levels on inflammation-associated genes; these assays revealed that a significant reduction of DNA methylation on many inflammation-associated genes occurred in macrophages and B cells from the mice treated with 5-Aza systemically, whereas no significant changes of DNA methylation were observed in these cells from mice treated with T cell–targeted delivery of 5-Aza (Supplemental Figure 12). Taken together, these data account for the discrepancies of final disease outcome between systemic vs targeted delivery of 5-Aza.
Figure 4. Specific targeting of nlg-5-Aza in CD8+ T cells significantly reduces DN T cells.
(A and B) MRL/lpr mice were treated i.v. with anti-CD8 antibody–coated nlg-5-Aza (15 μl nlg-5-Aza/mouse) every 10 days for 60 days, starting at 12 weeks of age. Free-5-Aza (5 μg/mouse) or empty-nlg was applied in 2 separated control groups. (E and H) OT-I TCR-Tg CD8 T cells from CD45.1 OT-I TCR-Tg Rag1–/– B6 mice were transferred i.v. (5 × 106 per mouse) into mOVA-Tg recipient mice. 12 hours after the transfer, recipient mice were administrated i.v. with either control nlg or anti-CD45.1–coated 5-Aza-nlg (15 μl nlg-5-Aza/mouse) and sacrificed with an addition 60 hours. (A) FACS analysis of T cell subsets (Thy1.2+TCR-γδ–TCRβ+CD49b– gated) in the spleens and kidneys from mice subjected to the indicated treatments. (B) Quantitation of the absolute cell numbers of CD4–CD8– DN T cells from the spleens or kidneys of mice subjected to the indicated treatment. (C) Quantitation of DNA methylation in indicated gene promoters or enhancers from splenic T lymphocytes sorted from 12-week-old MRL/lpr mice 10 hours after anti-CD8 antibody–coated nlg-5-Aza (15 μl nlg-5-Aza/mouse) treatment. n = 4 mice per group. (D) Flow cytometry analysis of cell surface CD8 downregulation on OT-I TCR-Tg T cells from CD45.1 OT-I TCR-Tg Rag1–/– B6 mice (CD45.1+TCRβ+ gated) cocultured with OVA257–264–loaded antigen-presenting cells for 12 hours with or without addition of 5-Aza (1 μM) and representative dot plots relative to the mean of fluorescence intensity (MFI) of CD8 expression. (E) Flow cytometry analysis of cell surface CD8 downregulation on transferred OT-I TCR-Tg T cells and representative dot plots relative to MFI of CD8 expression. (F) Flow cytometry analysis of intracellular perforin expression in splenic CD4, CD8, DN T cells from female MRL/lpr mice at 16 weeks of age. (G) Flow cytometry analysis of intracellular perforin expression in CD8+ or CD8– OT-I TCR-Tg T cells (CD45.1+TCRβ+ gated) cocultured with OVA257–264–loaded antigen-presenting cells for 12 hours with or without addition of 5-Aza (1 μM). (H) Flow cytometry analysis of intracellular perforin expression in CD8+ or CD8– OT-I TCR-Tg T cells. Data represent the mean ± SEM. n= 3–6 mice per group in 2 independent experiments for (A, B, and D–G). *P < 0.05, **P < 0.01, ***P < 0.005 vs. control; 2-tailed Student’s t test.
Of note, the DNA demethylation also occurred on promoters of perforin-1 (Prf1) and granzyme B (GZMB) in CD8 T cells, two critical molecules that have been shown to suppress disease in lupus-prone mice (39, 40). Previously, we provided evidence that DN T cells could originate from self-reactive CD8 T cells once they encounter antigens (36). Activated CD8 T cells have been found capable of limiting the expansion of autoreactive lymphocytes via targeted cytolysis, which relies on the interaction between CD8 and the MHC molecule connecting the CD8 T cell and the target cell together. Here, we hypothesized that activated CD8 T cells gave rise to DN T cells, which could not exert a cytolytic granule-dependent activity due to the reduction of CD8 expression. To test our hypothesis and further prove whether this conversion from CD8 to DN T cells is mediated by enhanced DNA methylation on the CD8 locus, we first isolated CD8+OT-I TCR (T cell receptor for chicken ovalbumin in the context of MHC class I) Tg T cells and cocultured them with OVA257–264–loaded antigen-presenting cells in vitro. The downregulation of CD8 expression on the vigorously proliferating OT-I T cells was observed 72 hours after stimulation, but the addition of 5-Aza preserved CD8 expression on the cell surface (Figure 4D). Similarly, 5-Aza could also significantly reduce activation-induced CD8 downregulation on CD8 T cells enriched from human peripheral blood (Supplemental Figure 13). We next transferred CD45.1+CD8+OT-I TCR-Tg T cells into CD45.2+ membrane-bound chicken ovalbumin–Tg (mOVA-Tg) mice treated with either anti-CD45.1-nlg-5-Aza or empty-nlg. In the recipients of empty-nlg, a larger fraction of the transferred CD45.1+OT-I T cells lost CD8 expression (Figure 4E). As expected, administration of anti-CD45.1-nlg-5-Aza efficiently prevented the generation and expansion of CD8–OT-I cells in vivo.
Work in mice supports the concept that therapeutic cytotoxic T cell lymphocyte enhancement is beneficial in lupus (39, 41, 42). During antigen-driven activation, CD8 T cells release cytolytic proteins, including perforin and granzymes, which eventually lead to the apoptosis of target cells. Here, we asked whether DN T cells also lose the ability to produce perforin and other cytolytic proteins. To test our hypothesis, we examined Prf1 expression in DN T cells from MRL/lpr mice. Flow cytometry analysis showed clearly that DN T cells expressed much lower Prf1 compared with their possible precursors (Figure 4F), the CD8T cells, which is consistent with slightly enhanced DNA methylation on the Prf1 promoter in DN T cells compared with CD8 T cells (Figure 3C). To further test our hypothesis, we examined the expression of perforin in OT-I TCR-Tg T cells after antigen stimulation in vitro. As expected, CD8– cells produce substantial lower levels of perforin compared with CD8+ cells, and the addition of 5-Aza into a cell coculture system efficiently enhanced perforin expression in CD8+ but not CD8– T cells (Figure 4G). Of note, 5-Aza could not induce perforin expression in naive cells but could enhance perforin expression in activated OT-I T cells (Supplemental Figure 14). To validate our in vitro observation, perforin expression was also assessed in CD8+ and CD8– T cells derived from OT-I TCR-Tg T cells transferred to mOVA-Tg recipients. As noted, the exact same cell behavior was revealed by flow cytometry ex vivo (Figure 4H), which is consistent with our observation in vitro. The simultaneous loss of both CD8 and perforin expression was observed. Of interest, the application of anti-CD8-nlg-5-Aza in vivo only promoted the perforin expression in CD8+ cells. Together, we provide compelling evidence that indicates the feasibility of modulating DNA methylation in CD8 T cells to prevent their conversion to DN cells and also maintain/increase their cytotoxic activity.
Discussion
DNA methylation regulates gene expression and takes place at the cytosine residues in cytosine-guanine dinucleotides located most often between 1,000 bp and 800 bp upstream of the transcription start site. Conversion of cytosine to 5-methylcytosines is vital in many cellular processes, such as chromatin structure, transcription, and embryonic development (5, 43). Aberrant DNA demethylation occurs actively in T cells from SLE patients (5, 14), and growing evidence indicates that these DNA methylation changes closely correlate with disease activity (13, 44). Despite this “guilt by association,” still little is known about the dynamic changes of DNA methylation in T cells in the context of SLE.
Here, we demonstrate that targeted delivery of 5-Aza to either CD4+ or CD8+ T cells in MRL/lpr mice with established disease was followed by significant improvement of autoimmunity and organ damage. At the molecular level, we showed that DNA demethylation in CD4 T cells augmented Foxp3 expression, whereas DNA demethylation in CD8 T cells reduced the activation-induced CD8 downregulation. Transient expression of Foxp3 in peripheral CD4+ T cells induces suppressive characteristics, but constitutive expression is required for stable suppressive function (45). High expression of DNMT1 in preactivated or differentiated T cells represses Foxp3 expression (46). Along these lines, we have for the first time to our knowledge demonstrated in lupus-prone MRL/lpr mice that pharmacological inhibition of DNA methylation strictly in CD4+ T cells promotes FoxP3+ Treg expansion and was clinically efficacious in restraining disease progression. This observation is in alignment with earlier reports that 5-Aza induced Foxp3 mRNA and protein expression in naive CD4+CD25− T cells in vitro (26) by epigenetically manipulating the Treg-specific locus.
Lupus-prone mice deficient in Cd8α (41) or Prf1 (39) display accelerated and severe disease, implying that CD8 T cells with normal cytotoxic capacity are important in the control of the expression of autoimmunity and lupus-like disease. In addition, CD8 T cells that encounter autoantigen become DN cells able to exacerbate autoimmunity by virtue of producing IL-17 (47). Our data are in support of these claims and provide additional insight on the importance of demethylation, which secures expression of CD8 and cytolysis-executing molecules.
The coexistence of hypomethylation of inflammatory elements and hypermethylation of regulatory elements in the context of ongoing disease explain our findings, which clearly show that precise DNA demethylation in T cell subsets suppresses established autoimmunity and related pathology in lupus-prone mice. Our data suggest that the role DNA methylation in the expression of lupus is rather complex and assessment of its global status is certainly not sufficient. Although the importance of hypomethylation is well recognized in SLE, the significance of DNA hypermethylation of genes or loci involved in the expression of the disease remains to be further explored. For example, CD3ζ, which is decreased in T cells from patients with SLE (48) and has been linked with severe SLE clinical manifestations, is hypermethylated (49).
Nanoparticle-based therapies are undoubtedly of great interest in the execution of precision medicine because they can accomplish cell/tissue-specific delivery of a drug and mitigate the risk of development of side effects (17, 18). Our data have substantial translational value, as they demonstrate the ability to reverse established disease by inhibiting DNA methylation in specific T cell subsets.
Methods
Mice.
OT1, CD45.1, Thy1.1, mOVAC57BL/6J, and MRL/lpr mice were obtained from The Jackson Laboratory. All mice were maintained under specific pathogen–free conditions.
Histology of frozen sections and immunohistochemistry imaging analysis.
Mouse kidneys were embedded in OCT tissue media (Tissue-Tek) and frozen on dry ice. Frozen sections (7-μm thickness) were fixed to slides in ice-cold acetone for 15 minutes and air dried for 30 seconds. The sections were blocked with 2% BSA for 30 minutes at room temperature and then stained for 30 minutes at room temperature in a humidified chamber with Hoechst 33258 (Life Technologies) and fluorescent-labeled goat anti-mouse IgG cross-adsorbed antibody (ThermoFisher Scientific). All tissue sections were mounted in Prolong Gold Antifade Mountant (Thermo Fisher Scientific).
Histopathology analysis.
Kidneys were cut in half longitudinally, embedded in paraffin, sectioned onto slides, stained with hematoxylin and eosin, examined microscopically, and scored as follows: (a) presence of crescents or fibrosis in glomeruli (1%–10%, grade 1; 10%–25%, grade 2; 25%–50%, grade 3; >50%, grade 4); (b) tubular cell changes (1%–10%, grade 1; 10%–25%, grade 2; 25%–50%, grade 3; >50%, grade 4); (c) lymphocyte infiltration (1%–10% of renal parenchyma, grade 1; 10%–25%, grade 2; 25%–50%, grade 3; >50%, grade 4); and (d) vasculitis (grade 1, small but definite perivascular infiltrates; grade 2, 1–3 foci of perivascular infiltrates without necrosis; grade 3, 3–5 foci of perivascular infiltrate, more extensive; grade 4, more than 5 foci of perivascular infiltrates). The sum of the 4 histopathology scores was calculated.
nlg preparations and characterization.
nlg were fabricated using a modified version of a previously reported preparation (17, 18). To prepare liposomes, a molar ratio mixture of 1.5:1.0:0.14:0.003 HSPC/cholesterol/DSPE-PEG (2000)-methoxy/DSPE-PEG (2000)-biotin (Avanti Polar Lipids) in chloroform was evaporated under a nitrogen gas stream and placed under vacuum overnight. The resulting dry lipid film was rehydrated with water and extruded through a series of filters with decreasing pore size (Whatman, GE Healthcare): 5 times with 1,000-nm pore size; 5 times with 400-nm pore size; and 10 times with 200-nm pore size. Sucrose (10 wt/v%) was added to the extruded liposome solution, and the solution was flash frozen in liquid nitrogen and lyophilized for several days. The loading procedure used was as follows: for each 60 mg of lipid in the lyophilized liposome powder, 6 mg 5-Aza-deoxycitidine precomplexed in a 1:1 molar ratio aminomethacrylate/succinylated β-cyclodextrin; 30 mg of a PEG oligomer consisting of 4,000-Da linear poly (ethylene glycol) flanked on each side by approximately 1–2 lactic acid monomers with a terminating acrylate group; and 1.25 mg Irgacure 2959 (Ciba) photoinitiator was added. For empty-nlg, the 6 mg 5-Aza was not added. All components were mixed in 2 ml PBS and added to the lyophilized liposomes. After 3 flash freeze-thaw cycles, the mixture was vigorously agitated by vortex for 30 minutes. 5 mg avidin was added, and the solution was further vortexed for 10 minutes at room temperature. Unencapsulated components/unbound avidin were then removed by repeated volume exchanges using centrifugal dialysis tubes with a 100-kDa MWCO. The complete volume was exchanged 3 times; after that, the volume was brought up to 4 ml and the solution was transferred evenly to 2 wells of a clear 6-well culture plate. The loaded liposomes were then cross-linked by placing the plate under a 430-W UV lamp with UVA light (315- to 400-nm transmission filter) for 8 minutes on ice to form the nlg. nlg were aliquoted and stored at –20°C until use. After dialysis, unbound avidin was quantified to ensure that the amount of avidin that remained on the particles was >90%. The final concentration of nlg was 13.7 mg/ml (24 μg 5-Aza/mg).
Conjugation of nlg with targeting antibodies.
A biotin-avidin-biotin bridge was applied to conjugate antibodies to the surface of liposomes. Biotin nondepletion anti-CD4 (clone RM4-4, Biolegend), anti-CD8 (clone YTS156.7.7, Biolegend), or anti-CD45.1 (clone A20, Biolegend) antibodies were added directly into prepared nlg (2 μg biotin antibody per 1 mg nlg) prior to use, mixed thoroughly for 5 minutes, and used immediately.
In vitro Treg differentiation.
Purified murine CD4+CD25– T cells were stimulated with antibodies to CD3 (1 μg/ml) and CD28 (1 μg/ml) under Treg differentiation conditions (20 ng/ml IL-2, 5 ng/ml TGF-β1, 10 μg/ml anti-IL-4, and 10 μg/ml anti-IFN-γ, 10 μg/ml anti-IL-6, and 10 μg/ml anti-IL-17).
ELISA-based assay of urinary albumin/creatinine ratio.
Twenty-four–hour urine samples were collected. Urine albumin was determined using a Mouse Albumin ELISA Quantitation Kit (Bethyl Laboratories) following the manufacture’s instructions. Urine creatinine was measured by the CreatinineParameter Assay Kit (R&D System) following the manufacture’s instructions.
Autoantibody detection.
Assays for serum autoantibodies were carried out as described previously (50). Immulon II plates (Dynatech) precoated with BSA were coated individually with indicated antigens (MilliporeSigma). The serum was diluted, and assayed for autoantigen reactivity against the plates described above by incubation for 2 hours at room temperature. Bound IgG was detected with a goat polyclonal HRP–anti-mouse IgG detection antibody (catalog 1030-50; Southern Biotech) and visualized at 450 nm using a tetramethylbenzidine substrate (MilliporeSigma).
Cytokine detection.
Assays for serum cytokines were carried out using the BioLegend LEGENDplex kit following the manufacture’s instruction. It is a bead-based immunoassay that utilizes the same basic principles of sandwich immunoassays to quantify multiple soluble analytes simultaneously in biological samples using a flow cytometer.
Flow cytometry.
Cell suspensions were prepared from spleens and stained at 4°C in PBS containing 2% FBS and 0.5% EDTA after FcγRII/III blocking. The following fluorescent antibodies were administrated according to the manufacturer’s instructions: anti-mouse Thy1.2 (clone 30H-12), anti-mouse CD4 (clone GK1.5), anti-mouse CD8 (clone 53-6.7), anti-mouse TCRβ (clone H57-597), anti-mouse TCR-γ/δ (clone GL3), anti-mouse CD49b (clone DX5), anti-mouse CD3 (clone 145-2C11), anti-mouse CD45.1 (clone A20), anti-mouse CD19 (clone 1D3/CD19), anti-mouse perforin (clone S16009A), anti-mouse Fas (clone SA367H8), and anti-mouse GL7 (clone GL-7). All antibodies were from BioLegend. Dead cells were excluded using Fixable Viability Dye staining (Life Technologies).
Real-time qPCR analysis on MeDIP (MeDIP real-time qPCR assay).
DNA was extracted from the indicated lymphocytes and then subjected to sonication, which sheared it into random fragments, ranging from 300 to 1,000 base. The DNA fragments were denatured into single-stranded DNA and incubated with monoclonal 5-methylcytosine (5mC) antibodies. The MAGnify ChIP kit was used to purify anti-5-methylcytosine antibody–methylated DNA complex, which bound to conjugated magnetic beads, and unbound DNA was washed out in the supernatant. To further purify the DNA for real-time qPCR, proteinase K was added to digest the antibodies and released DNA from the complex. The methylation percentile was calculated by normalizing the amount of released DNA to total genomic DNA. The primers for different gene promoters and CD8 enhancers are as follows: Cd4 promoter forward, 5′-ACAGTTACATGCCCGTGCCC-3′; Cd4 promoter reverse, 5′-TGCCGTAAAAAGCTGAAACAGCC-3′; Cd8α promoter forward, 5′-GTTGACCCGCTTTCTGT-3′; Cd8α promote reverse, 5′-CCAAGTTCGGCGTCCATTTT-3′; Cd8β promoter forward, 5′-TGCCAGCTTTAGCCACCA-3′; Cd8β promoter reverse, 5′-GGCCGGAGTGTCATCTAGGC-3′; E8I forward, 5′-GCCTCTGCTACTTCTAGATG-3′; E8I reverse, 5′-CTCAGATGCCGATCTCATTT-3′; E8II forward, 5′-CGCATTCAGCTCCAACTAGG-3′; E8II reverse, 5′-CGCATTCAGCTCCAACTAGG-3′; E8IIIforward, 5′-GACAGTATCTCAAAGGCT-3′; E8III reverse, 5′-ATGCTCTAAGACAGAAGG-3′; E8V1 forward, 5′-GAAGCTAACACAGTGGAAGGT-3′; E8V1 reverse, 5′-GAAGATACTGTTTCAGCAACT-3′; E8V2 forward, 5′-CTGCACCCAAGATTCACCCG-3′; E8V2 reverse, 5′-ACTAGCCCTTGGATAGGTGC-3′; Il17 promoter forward, 5′-GCAAGGGATGCTCTCTAGCCA-3′; Il17 promoter reverse, 5′- TTGGGAGCTGAACAGAGATGCT-3′; Ifnγ promoter forward, 5′-GTTGATGTTGACTTGCTACGGTGAG-3′; and Ifnγ promoter reverse, 5′-CACCTCTCCAGCAAATAGCAA-3′.
In vitro antigen stimulation induced CD8 downregulation.
Splenocytes were collected from C57BL/6J mouse spleens, irradiated, and then loaded with OVA257–264 peptides (1 μg/ml) for 1 hour. OT1-Tg CD8 T cells were sorted from spleens of OT-1+Rag1–/– C57BL/6J mice and cocultured with OVA-pulsed irradiated C57BL/6Jsplenocytes. CD8 and perforin were examined by flow cytometry 3 days later.
Treg suppression assay.
CD4 FoxP3+ T cells were sorted from Foxp3-YFP-Tg mice treated with 5-Aza (5mM) for 2 hours (PBS as control). To assess suppressive capacity, naive CD4 T cells were sorted from Thy1.1 C57BL/6J mouse spleens, labeled with CFSE, stimulated with plate-bound anti-CD3 and anti-CD28 (2 μg/ml), and mixed with treated CD4 FoxP3+at 1:6 ratio for 72 hours. Cell proliferation was quantified 3 days later.
Adoptive transfers.
For adoptive transfers, CD45.1 Rag1–/–OT-I mice were used as donors and mOVA-Tg mice as recipients. T cells were isolated using negative isolation Dynabeads (Invitrogen) from spleen (purity ≥95%). 5 × 106 cells were transferred by tail vein injection. 12 hours after cell transfer, recipient mice were further administrated i.v. with empty-nlg or anti-CD45.1–coated 5-Aza-nlg (equivalent to 5-Aza/mouse) and sacrificed after an addition 60 hours.
Statistics.
All results are shown as the mean ± SEM. A 2-tailed, unpaired Student’s t test was used when only 2 groups were compared for statistical differences. For a comparison of more than 2 groups, the 1-way ANOVA was applied. P values of less than 0.05 were considered significant.
Study approval.
All animal procedures were approved by the Institutional Animal Care and Use Committee of Beth Israel Deaconess Medical Center, Harvard Medical School. Blood samples were obtained from 21 healthy platelet donors from the Kraft Family Blood Donor Center (Dana-Farber Cancer Institute, Boston, Massachusetts, USA). Studies and processes have been approved by the involved institutions. The blood samples were collected after informed consent was obtained.
Author contributions
HL and GCT designed experiments, analyzed data, and wrote the manuscript. HL performed the experiments. SB, PK, and TMF designed and prepared the engineered nlgs. MGT reviewed all mouse kidney section slides, provided the histopathology scores, and critically reviewed the manuscript. AS, YL, and VRM provided expertise and reviewed the manuscript.
Supplementary Material
Acknowledgments
This worked was supported by NIH grants R01 AI 42269 and AR 064350 (to GCT), R01 AR068974 (to VRM), and T32GM007205 (to SB).
Version 1. 08/23/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(16):e120880. https://doi.org/10.1172/jci.insight.120880.
Contributor Information
Hao Li, Email: hli13@bidmc.harvard.edu.
Maria G. Tsokos, Email: mtsokos@bidmc.harvard.edu.
Sean Bickerton, Email: sean.bickerton@yale.edu.
Amir Sharabi, Email: asharabi@bidmc.harvard.edu.
Yi Li, Email: yli16@bidmc.harvard.edu.
Philip Kong, Email: philip.kong@yale.edu.
George C. Tsokos, Email: gtsokos@bidmc.harvard.edu.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Wigren M, Nilsson J, Kaplan MJ. Pathogenic immunity in systemic lupus erythematosus and atherosclerosis: common mechanisms and possible targets for intervention. J Intern Med. 2015;278(5):494–506. doi: 10.1111/joim.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moulton VR, Suarez-Fueyo A, Meidan E, Li H, Mizui M, Tsokos GC. Pathogenesis of human systemic lupus erythematosus: a cellular perspective. Trends Mol Med. 2017;23(7):615–635. doi: 10.1016/j.molmed.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woosley RL, Drayer DE, Reidenberg MM, Nies AS, Carr K, Oates JA. Effect of acetylator phenotype on the rate at which procainamide induces antinuclear antibodies and the lupus syndrome. N Engl J Med. 1978;298(21):1157–1159. doi: 10.1056/NEJM197805252982101. [DOI] [PubMed] [Google Scholar]
- 5.Altorok N, Sawalha AH. Epigenetics in the pathogenesis of systemic lupus erythematosus. Curr Opin Rheumatol. 2013;25(5):569–576. doi: 10.1097/BOR.0b013e328364206f. [DOI] [PubMed] [Google Scholar]
- 6.Teruel M, Sawalha AH. Epigenetic variability in systemic lupus erythematosus: what we learned from genome-wide DNA methylation studies. Curr Rheumatol Rep. 2017;19(6):32. doi: 10.1007/s11926-017-0657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol Med. 2011;17(12):714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moulton VR, Tsokos GC. T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity. J Clin Invest. 2015;125(6):2220–2227. doi: 10.1172/JCI78087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YK, Mukasa R, Hatton RD, Weaver CT. Developmental plasticity of Th17 and Treg cells. Curr Opin Immunol. 2009;21(3):274–280. doi: 10.1016/j.coi.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30(5):646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Sawalha AH. Epigenetics and T-cell immunity. Autoimmunity. 2008;41(4):245–252. doi: 10.1080/08916930802024145. [DOI] [PubMed] [Google Scholar]
- 12.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282(9):5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. J Autoimmun. 2013;41:92–99. doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6(5):593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung SA, et al. Genome-wide assessment of differential DNA methylation associated with autoantibody production in systemic lupus erythematosus. PLoS One. 2015;10(7):e0129813. doi: 10.1371/journal.pone.0129813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Sawalha AH, Lu Q. Epigenetics in the treatment of systemic lupus erythematosus: potential clinical application. Clin Immunol. 2014;155(1):79–90. doi: 10.1016/j.clim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Otomo K, et al. Cutting Edge: Nanogel-based delivery of an inhibitor of CaMK4 to CD4+ T cells suppresses experimental autoimmune encephalomyelitis and lupus-like disease in mice. J Immunol. 2015;195(12):5533–5537. doi: 10.4049/jimmunol.1501603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Look M, et al. Nanogel-based delivery of mycophenolic acid ameliorates systemic lupus erythematosus in mice. J Clin Invest. 2013;123(4):1741–1749. doi: 10.1172/JCI65907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christman JK. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida H, Yoshida M, Merino R, Shibata T, Izui S. 5-Azacytidine inhibits the lpr gene-induced lymphadenopathy and acceleration of lupus-like syndrome in MRL/MpJ-lpr/lpr mice. Eur J Immunol. 1990;20(9):1989–1993. doi: 10.1002/eji.1830200917. [DOI] [PubMed] [Google Scholar]
- 21.Quddus J, et al. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest. 1993;92(1):38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notari RE, DeYoung JL. Kinetics and mechanisms of degradation of the antileukemic agent 5-azacytidine in aqueous solutions. J Pharm Sci. 1975;64(7):1148–1157. doi: 10.1002/jps.2600640704. [DOI] [PubMed] [Google Scholar]
- 23.Derissen EJ, Beijnen JH, Schellens JH. Concise drug review: azacitidine and decitabine. Oncologist. 2013;18(5):619–624. doi: 10.1634/theoncologist.2012-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fahmy TM, Fong PM, Park J, Constable T, Saltzman WM. Nanosystems for simultaneous imaging and drug delivery to T cells. AAPS J. 2007;9(2):E171–E180. doi: 10.1208/aapsj0902019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28(4):546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Lal G, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182(1):259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463(7282):808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garefalaki A, et al. Variegated expression of CD8 alpha resulting from in situ deletion of regulatory sequences. Immunity. 2002;16(5):635–647. doi: 10.1016/S1074-7613(02)00308-4. [DOI] [PubMed] [Google Scholar]
- 29.Feik N, Bilic I, Tinhofer J, Unger B, Littman DR, Ellmeier W. Functional and molecular analysis of the double-positive stage-specific CD8 enhancer E8III during thymocyte development. J Immunol. 2005;174(3):1513–1524. doi: 10.4049/jimmunol.174.3.1513. [DOI] [PubMed] [Google Scholar]
- 30.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114(18):3727–3735. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198(12):1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bristeau-Leprince A, et al. Human TCR alpha/beta+ CD4-CD8- double-negative T cells in patients with autoimmune lymphoproliferative syndrome express restricted Vbeta TCR diversity and are clonally related to CD8+ T cells. J Immunol. 2008;181(1):440–448. doi: 10.4049/jimmunol.181.1.440. [DOI] [PubMed] [Google Scholar]
- 33.Brooks EG, Balk SP, Aupeix K, Colonna M, Strominger JL, Groh-Spies V. Human T-cell receptor (TCR) alpha/beta + CD4-CD8- T cells express oligoclonal TCRs, share junctional motifs across TCR V beta-gene families, and phenotypically resemble memory T cells. Proc Natl Acad Sci USA. 1993;90(24):11787–11791. doi: 10.1073/pnas.90.24.11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han M, Harrison L, Kehn P, Stevenson K, Currier J, Robinson MA. Invariant or highly conserved TCR alpha are expressed on double-negative (CD3+CD4-CD8-) and CD8+ T cells. J Immunol. 1999;163(1):301–311. [PubMed] [Google Scholar]
- 35.Crispín JC, Tsokos GC. Human TCR-alpha beta+ CD4- CD8- T cells can derive from CD8+ T cells and display an inflammatory effector phenotype. J Immunol. 2009;183(7):4675–4681. doi: 10.4049/jimmunol.0901533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-Rodríguez N, et al. Programmed cell death 1 and Helios distinguish TCR-αβ+ double-negative (CD4-CD8-) T cells that derive from self-reactive CD8 T cells. J Immunol. 2015;194(9):4207–4214. doi: 10.4049/jimmunol.1402775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellmeier W, Sunshine MJ, Losos K, Littman DR. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9(4):485–496. doi: 10.1016/S1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- 38.Ellmeier W, Sunshine MJ, Maschek R, Littman DR. Combined deletion of CD8 locus cis-regulatory elements affects initiation but not maintenance of CD8 expression. Immunity. 2002;16(5):623–634. doi: 10.1016/S1074-7613(02)00309-6. [DOI] [PubMed] [Google Scholar]
- 39.Peng SL, Moslehi J, Robert ME, Craft J. Perforin protects against autoimmunity in lupus-prone mice. J Immunol. 1998;160(2):652–660. [PubMed] [Google Scholar]
- 40.Graham KL, Thibault DL, Steinman JB, Okeke L, Kao PN, Utz PJ. Granzyme B is dispensable for immunologic tolerance to self in a murine model of systemic lupus erythematosus. Arthritis Rheum. 2005;52(6):1684–1693. doi: 10.1002/art.21092. [DOI] [PubMed] [Google Scholar]
- 41.McPhee CG, et al. MHC class I family proteins retard systemic lupus erythematosus autoimmunity and B cell lymphomagenesis. J Immunol. 2011;187(9):4695–4704. doi: 10.4049/jimmunol.1101776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki M, Konya C, Goronzy JJ, Weyand CM. Inhibitory CD8+ T cells in autoimmune disease. Hum Immunol. 2008;69(11):781–789. doi: 10.1016/j.humimm.2008.08.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hedrich CM, Mabert K, Rauen T, Tsokos GC. DNA methylation in systemic lupus erythematosus. Epigenomics. 2017;9(4):505–525. doi: 10.2217/epi-2016-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes T, Sawalha AH. The role of epigenetic variation in the pathogenesis of systemic lupus erythematosus. Arthritis Res Ther. 2011;13(5):245. doi: 10.1186/ar3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 46.Josefowicz SZ, Wilson CB, Rudensky AY. Cutting edge: TCR stimulation is sufficient for induction of Foxp3 expression in the absence of DNA methyltransferase 1. J Immunol. 2009;182(11):6648–6652. doi: 10.4049/jimmunol.0803320. [DOI] [PubMed] [Google Scholar]
- 47.Crispín JC, et al. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J Immunol. 2008;181(12):8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liossis SN, Ding XZ, Dennis GJ, Tsokos GC. Altered pattern of TCR/CD3-mediated protein-tyrosyl phosphorylation in T cells from patients with systemic lupus erythematosus. Deficient expression of the T cell receptor zeta chain. J Clin Invest. 1998;101(7):1448–1457. doi: 10.1172/JCI1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong KM, et al. CD3Z hypermethylation is associated with severe clinical manifestations in systemic lupus erythematosus and reduces CD3ζ-chain expression in T cells. Rheumatology (Oxford) 2017;56(3):467–476. doi: 10.1093/rheumatology/kew405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez J, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17(7):893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.