Abstract
The extent to which evolution is reversible has long fascinated biologists.1–8 Most prior work on the reversibility of morphological and life-history evolution 9–13 has been indecisive, because of uncertainty and bias in the methods used to infer ancestral states for such characters.14,15 Further, despite theoretical work on the factors that could contribute to irreversibility,1,8,16 there is scant empirical evidence on its causes, because sufficient understanding of the mechanistic basis for the evolution of new or ancestral phenotypes is seldom available.3,8,17 By studying the reversibility of evolutionary changes in protein structure and function, these limitations can be overcome. Here we show, using the evolution of hormone specificity in vertebrate glucocorticoid receptors (GRs) as a case-study, that the evolutionary path by which GR acquired its new function soon became inaccessible to reverse exploration. Using ancestral gene reconstruction, protein engineering, and X-ray crystallography, we demonstrate that five subsequent “restrictive” mutations, which optimized GR’s new specificity, also destabilized elements of the protein’s structure that were required to support the ancestral conformation. Unless these ratchet-like epistatic substitutions are restored to their ancestral states, reversing the key function-switching mutations yields a non-functional protein. Reversing the restrictive substitutions first, however, does nothing to enhance the ancestral function. Our findings indicate that even if selection for the ancestral function were imposed, direct reversal would be extremely unlikely, suggesting an important role for historical contingency in protein evolution.
Evolutionary reversibility represents a strong test of the importance of contingency and determinism in evolution. If selection is limited in its ability to drive the reacquisition of ancestral forms, then the future outcomes available to evolution at any point in time must depend strongly on the present state and, in turn, on the past.2,4,8,18 Ready reversibility, in contrast, would indicate that natural selection can produce the same optimal form in any given environment, irrespective of history.19 The evolutionary reversibility of a protein can be evaluated at three levels: molecular sequence, protein function, and the structural/mechanistic underpinnings for that function. The latter is most relevant to understanding the roles of contingency and determinism in evolution. Exact molecular reversal to the ancestral amino acid sequence is possible but extremely unlikely and of trivial interest, because of the large number of sequences that code for the same structure and function. Selection will always produce adaptive functions or phenotypes in some form; however, if the underlying mechanism for a reversed function differs from that of the ancestor, then a novel, analogous state will have been achieved through onward evolution, not reversal -- a situation similar to false morphological reversal caused by convergent evolution.4 True reversal, involving restoration of the ancestral phenotype via the ancestral structure-function relations, would indicate that the forms of functional proteins can evolve deterministically, irrespective of contingent historical events.
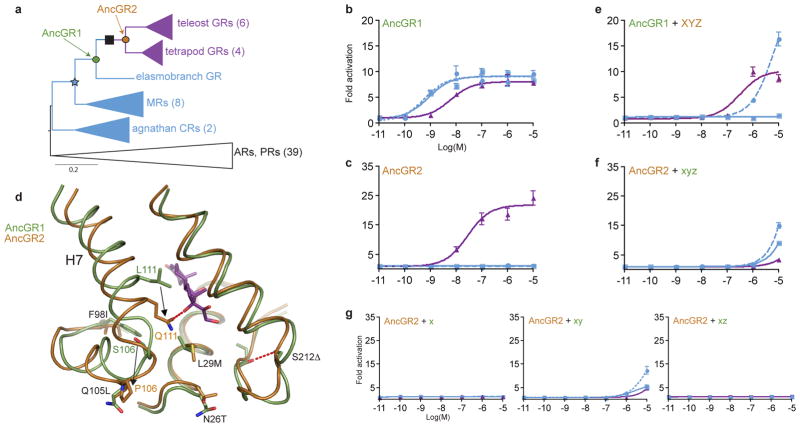
Recent developments in techniques for studying protein evolution allow the reversibility of protein structure and function to be studied directly. The intrinsic functions and atomic structures of ancestral genes can be determined by inferring their sequences using maximum likelihood phylogenetics, then biochemically synthesizing, expressing, and characterizing them using functional assays and X-ray crystallography.20 Moreover, the mechanisms by which new functions evolved can be identified by introducing historical substitutions into ancestral backgrounds and characterizing their effects on structure and function.21,22 Using these techniques, we recently established the mechanistic basis for the evolution of a novel function in the GR, a DNA-binding transcription factor that is specifically activated by the steroid hormone cortisol to regulate the long-term stress response and other processes in humans and other vertebrates.23,24 Specifically, we showed that the cortisol-specificity of the GR ligand-binding domain evolved from a more promiscuous ancient receptor that was activated by the mineralocorticoids aldosterone and deoxycorticosterone (DOC) and more weakly by cortisol. The GR’s novel function (Fig. 1a–c) evolved because of a dramatic change in structure-function relations during the 40 million year period between GR in the last common ancestor of cartilaginous and bony fish (AncGR1, which had the ancestral phenotype) and GR in the last common ancestor of tetrapods and ray-finned fish (AncGR2, which was cortisol-specific). Of the 37 amino acid changes that occurred during this interval, two conserved substitutions (S106P and L111Q, called group X for convenience) were necessary and sufficient to switch the resurrected AncGR1’s preference from mineralocorticoids to cortisol. S106P radically repositioned helix 7 along the edge of the ligand pocket, reducing activation by all hormones but moving site 111 closer to the ligand. In this new position, the hydrophobic substitution L111Q generated a new hydrogen bond to the 17-hydroxyl group unique to cortisol, specifically restoring sensitivity to that hormone (Fig. 1d). Three more conserved substitutions (group Y) completed the loss of mineralocorticoid sensitivity to yield a cortisol-specific receptor; these changes further destabilized the receptor complex with mineralocorticoids but enhanced interaction with cortisol’s 17-hydroxyl. The protein could not tolerate group Y, however, without an additional pair of permissive substitutions (group Z), which added stability to the structural elements destabilized by group Y and conferred the full GR-like function upon AncGR1+XYZ (Fig. 1e and ref. 24).
Fig. 1.
Evolution and reversibility of GR function. a) Reduced phylogeny of corticosteroid receptors. Blue, receptors sensitive to both cortisol and mineralocorticoids; purple, sensitive to cortisol only; black, other steroid receptors (AR, androgen receptor; PR, progestagen receptor). Ancestral proteins AncGR1 and AncGR2 are labeled. 37 amino acid changes, including groups X, Y, and Z, occurred during the interval between these two proteins (black box; for complete list and alignment, see Fig. S1). Parentheses show number of sequences in each group. b, c) Ligand sensitivities of AncGR1 and AncGR2, shown as fold increase in expression of a luciferase reporter in the presence of increasing doses of cortisol (purple), aldosterone (solid blue), and deoxycorticosterone (DOC, dashed blue). Error bars, SEM. d) Conformational change causing cortisol-specificity in AncGR2 (see ref. 24). Partial structures of AncGR1 and AncGR2 are superimposed. Substitutions in group X (S106P and L111Q) are large effect mutations that reposition helix 7 (H7) and form a hydrogen bond to the 17-OH that is unique to cortisol (purple). Black arrows indicate change in position of these residues. Substitutions in groups Y (L29M, F98I, S212Δ), and Z (N26T, Q105L) optimize the derived function. e) When substitutions in sets X, Y, and Z are introduced into AncGR1, they recapitulate the evolution of a cortisol-specific activator. f) When these substitutions are reversed to the ancestral state (xyz) in the AncGR2 background, activation by all ligands is lost. g) All AncGR2 combinations in which group X is reversed also yield non-functional receptors.
The most direct pathway to reverse the evolution of AncGR2’s structure and function would be to reverse the key substitutions that generated the derived phenotype during the “forward” evolution of AncGR1. We used site-directed mutagenesis on AncGR2 to reverse the amino acids in groups X, Y, and Z to their ancestral states (x, y, and z) and employed a luciferase reporter gene assay to characterize their effect on receptor function (Fig. 1f). Surprisingly, AncGR2-xyz was unable to activate transcription in response to any ligand, even at very high concentrations. Reversing only the large-effect mutations in group X also produced a non-functional AncGR2-x receptor, as did all combinations that included restoration of group x (Fig. 1g). These results indicate that AncGR2’s structure and function are not reversible through this direct path, because the ancestral amino acids in group x -- and the change in conformation they cause -- are incompatible with the derived background. These states, however, were present in AncGR1 just 40 million years earlier and have been conserved in all mineralocorticoid receptors ever since, indicating that additional epistatic modifiers must have evolved between AncGR1 and AncGR2.
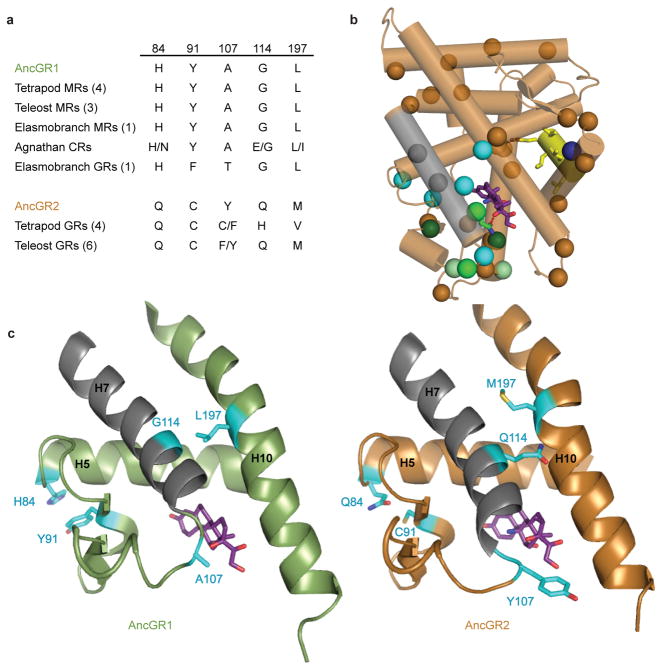
To identify candidate historical substitutions for this restrictive effect, we combined phylogenetic and structural analysis. Thirty substitutions in addition to X, Y, and Z occurred during the AncGR1-AncGR2 interval. We reasoned that amino acids required for the ancestral function should be conserved in the AncGR1-like state in extant receptors that retain the ancestral sensitivity to DOC and aldosterone; unlike X, Y, and Z, however, they would not be expected to be conserved in the GR-like receptors. Of these 30 sites, only six were invariant among all or all but one of the DOC/aldosterone-sensitive receptors. (Figs. 2a, S1). To predict which of these were most likely to enable the ancestral function, we expressed the AncGR2 ligand-binding domain and used X-ray crystallography (Table S1) to determine its empirical atomic structure at 2.5 Å resolution in complex with the synthetic glucocorticoid dexamethasone (Figs. 2b, S2, S3). The monomeric AncGR2 structure adopts the canonical active conformation for nuclear receptors 25 and is nearly identical to the GR2 structure previously predicted by homology modeling.24 Five of the six candidate substitutions identified by phylogenetic analysis (Group W: H84Q, Y91C, A107Y, G114Q, and L197M) are on or interact directly with the repositioned helix 7; the other (V234F) is far from the remodeled portion of the protein and does not appear to interact with it directly or indirectly (Fig. 2b). By comparing AncGR2’s structure with that of AncGR1,24 we predicted that the derived states at these five sites would be incompatible with the ancestral structure and function, because the AncGR1 states stabilize the active conformation with helix 7 in its ancestral position (Figs. 2b, 2c), but those in AncGR2 fail to support this conformation or actively clash with it. Specifically (Fig. 2c), two of these residues in AncGR1 -- Gly114 (on helix 7) and Leu197 (on helix 10) -- are close together and allow tight packing of H7 in the ancestral position against H10, with Leu197 packing into the core of the protein above the ligand pocket, stabilizing the ligand-bound conformation. AncGR2, in contrast, contains Gln and Met at these positions, the side chains of which are longer and less hydrophobic; the repositioning of helix 7 allows these two residues to be tolerated, but in the ancestral conformation their side chains would clash, pushing the two helices apart and away from the ligand. A second pair, the aromatic residues H84 and Y91, form a pi-stack in AncGR1, stabilizing the beta-strand that abuts the ligand and helix 7. Substitution of these residues to Gln and Cys (as in AncGR2) would destroy this interaction, increasing flexibility in the ligand pocket and destabilizing the active complex; AncGR2 can presumably tolerate this effect because of the additional stability contributed by the hydrogen bond between Gln111 and cortisol. The fifth candidate site, Ala107, lies near the base of helix 7, at the mouth of the ligand pocket where helices 3, 7, and 10 pack together. Replacement of Ala107 with AncGR2’s bulky tyrosine would clash with helices 3 and 10; however, the movement of helix 7 in AncGR2 repositions site 107, allowing a tyrosine to be tolerated.
Fig. 2.
Identification of “restrictive” substitutions that impede reversibility. a) Group W residues are conserved in the AncGR1-like state in virtually all extant receptors that retain the ancestral function. b) X-ray crystal structure of AncGR2 (bronze) with dexamethasone (purple). Repositioned Helix 7 is shown in grey. Residues substituted between AncGR1 and AncGR2 are marked with spheres at the α-carbon. Cyan, candidate restrictive substitutions (group W). Sites in groups X, Y, and Z are shown in medium, dark, and light green, respectively. Blue, V234F. c) Ligand pockets of AncGR1 (green, with cortisol) and AncGR2 (bronze, with dexamethasone). Group w residues (cyan) in their ancestral state in AncGR1 are predicted to support the ancestral conformation of helix 7 (grey), but to destabilize that conformation in the derived states of AncGR2.
To test the hypothesis that group W substitutions impede direct reversal of the key function-switching mutations, we used site-directed mutagenesis to reverse group W in the AncGR2-xyz background and determined their functional effect using a luciferase reporter assay (Fig. 3). As predicted, reversing all five group W mutations restored the ancestral phenotype, yielding a sensitive, promiscuous receptor with nanomolar response to both mineralocorticoids and cortisol and, like AncGR1, preference for aldosterone (Fig. 3a). All five group W substitutions contribute to AncGR2’s intolerance of the ancestral structure/function: Y107A alone partially rescued the transcriptional function of AncGR2-xyz and shifted it substantially towards the ancestral promiscuous phenotype, as did the pairs Q84H/C91Y and Q114G/M197L (Fig. 3b). Restoring the single mutations Q84H, C91Y, Q114G, and M197L had no or very weak effects (Fig. S4), presumably because of the structural interactions within each pair required to improve the receptor’s function.
Fig. 3.
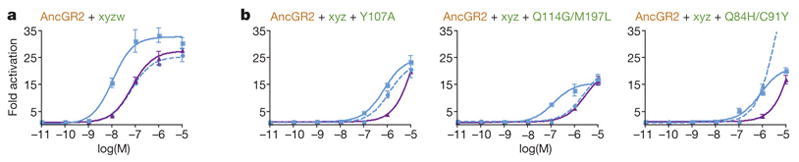
Restrictive substitutions impede evolutionary reversibility. a) When group W substitutions are restored to their ancestral state (w), the nonfunctional AncGR2-xyz is rescued, and the ancestral sensitivity to all three ligands is restored. Fold increase in luciferase reporter expression is shown with cortisol (purple), aldosterone (solid blue), and DOC (dashed blue). b) Group W substitutions all impede reversiblility: restoring the ancestral states singly (Y107A) or in structurally interacting pairs (Q84H/C91Y and Q114G/M197L) partially rescues AncGR2-xyz.
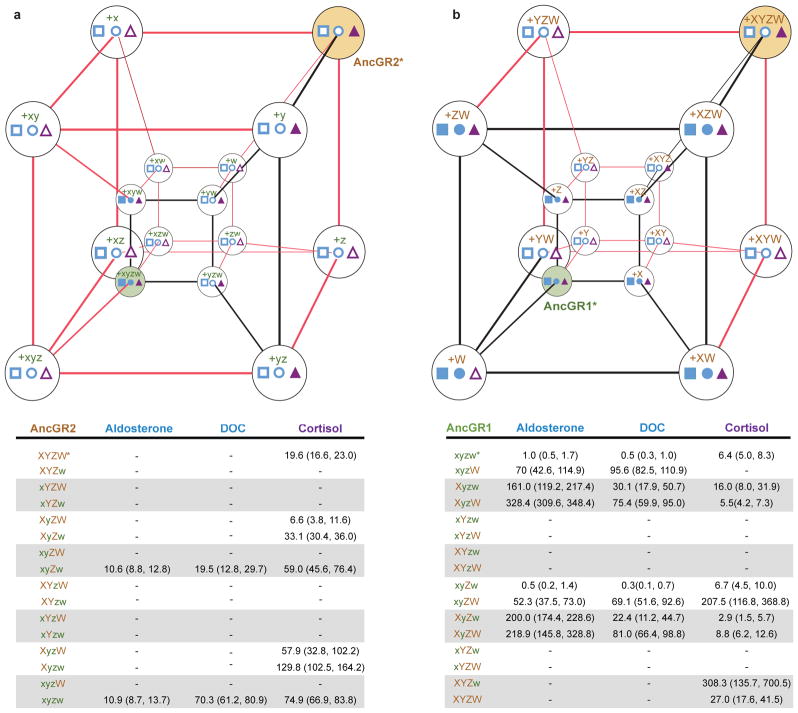
To test the hypothesis that group W substitutions specifically undermine the stability of the ancestral helix 7 conformation, we restored the ancestral state (w) in all possible combinations of x, y, and z, in the AncGR2 background (Fig. 4, S5). As predicted, reversal to x always impairs both the ancestral and derived function unless group w has been reversed first. Taken together, our experiments indicate that these five mutations prevent direct evolutionary reversal by weakening aspects of the receptor structure that were required to support the ancestral conformation. By reversing all of these restrictive substitutions, the ancestral structure and function can be largely restored. The reversed AncGR2-xyzw remains slightly less sensitive to hormone than AncGR1, indicating that some of the other 25 substitutions during the AncGR1-AncGR2 interval make additional, minor contributions to impeding direct evolutionary reversal (Figs., 4a, b). The restrictive effect of mutations in group W on reversal of group X does not depend on whether these other 25 substitutions are in their ancestral or derived states (Figs. 4a, b, S5). Although there are additional combinations of individual substitutions that we did not test, our results indicate that neither the restrictive effect of group W mutations nor the permissive effect of reversing them depends narrowly on a specific genetic background.
Fig. 4.
Epistasis limits trajectories of reverse and forward evolution. The corners of each hypercube represent states for residue sets X, Y, Z, and W. Edges show pathways between the derived (XYZW) and ancestral (xyzw) states. Red edges show unlikely evolutionary paths through nonfunctional intermediates; black paths pass through functional intermediates. Filled shapes at vertices indicate sensitivity to aldosterone (blue squares), DOC (blue circles), and cortisol (purple triangles); empty shapes, no activation by these hormones. Tables below each cube show sensitivity to each hormone as the EC50 (concentration required for half-maximal activation), with 95% confidence interval. Dashes, no activation. Asterisks, state combinations in AncGR2 and AncGR1. a) Limited evolutionary pathways to reverse AncGR2 (bronze) to the ancestral structure and function. Mutations were introduced in the AncGR2 background. b) Functional effect of substitutions during “forward” evolution when introduced into AncGR1 (green). The sets X, Y, Z, and W contain each more than one site, so the complete sequence space for each cube has 12 dimensions, 4096 vertices, and numerous additional trajectories.
The restrictive mutations that impede direct reversal may have been adaptive or neutral when they occurred. To characterize the “forward” effect of group W mutations on receptor function, we recapitulated them in the AncGR1 background with various combinations of groups X, Y, and Z (Fig. 4b). In AncGR1-XYZ and all other X-containing backgrounds, W mutations cause a moderate improvement or have no significant effect on receptor activation and cortisol-specificity, presumably by stabilizing the derived position of helix 7 and its interaction with that hormone. In the AncGR1 background and all other combinations that include x, however, W mutations dramatically reduce sensitivity to all hormones. Because selection makes evolutionary trajectories that pass through nonfunctional intermediates far less likely than those involving functional intermediates at every step,26 W mutations are therefore unlikely to have been complete before the remodeling and functional shift triggered by group X. Once X was in place, however, the W mutations that prevent direct evolutionary reversal are likely to have optimized the derived function or been neutral.
Our findings indicate that epistatic modifiers, at least some of which evolved after GR’s new function emerged, acted as an evolutionary ratchet, making re-evolution of the ancestral structure-function relations far more difficult than it was initially. Reversal via a direct path that restores the key residues in group X became exceedingly unlikely, because features that once enabled the ancestral conformation of helix 7 had been modified. To restore the ancestral conformation by reversing group X, the restrictive effect of the substitutions in group W must first be reversed, as must group Y (Figs. 4a, b). Reversal to w and y in the absence of x, however, does nothing to enhance the ancestral function; in most contexts, reversing these mutations substantially impairs both the ancestral and derived functions (Figs. 4a, b). Further, the permissive effect of reversing four of the mutations in group W requires pairs of substitutions at interacting sites. Selection for the ancestral function would therefore not be sufficient to drive AncGR2 back to the ancestral states of w and x, because passage through deleterious and/or neutral intermediates would be required; the probability of each required substitution would be low, and the probability of all in combination would be virtually zero.
We have examined the sufficiency of selection to drive direct evolutionary reversal of the GR. There may be other potentially permissive mutations, of unknown number, that could compensate for the restrictive effect of group W and allow the ancestral conformation to be restored. Reversal by such indirect pathways could be driven by selection, however, only if these other mutations -- unlike those we studied -- could somehow relieve the steric clashes and restore the lost stabilizing interactions that make the ancestral position of helix 7 intolerable in AncGR2, and also independently restore the ancestral function when helix 7 is in its radically different derived conformation. Whether or not mutations that could achieve these dual ends exist, reversal to the ancestral conformation would require a considerably more complex pathway than was necessary before the ratchet effect of W evolved.
The extent to which our observations concerning GR’s evolutionary reversibility can be generalized to other proteins requires further research. We predict that future investigations, like ours, will support a molecular version of Dollo’s law4: as evolution proceeds, shifts in protein structure-function relations become increasingly difficult to reverse, whenever those shifts have complex architectures, such as requiring conformational changes and multiple epistatically interacting substitutions.2,8,16 Phenotypes at higher levels of genetic organization may also display ratchet-like modes of evolution if optimization of a derived phenotype causes changes in one gene, regulatory element, morphological structure, or physiological/developmental process that epistatically undermine the conditions that enabled the ancestral state at other such “loci.” In contrast, phenotypic shifts caused by single or additive genetic changes are likely to be readily reversible.1,27
Our observations suggest that history and contingency during GR evolution strongly limited the pathways that could be deterministically followed under selection. The “adaptive peak” represented by the promiscuous AncGR1 is a relatively close neighbor in sequence space to the more specific AncGR2. This peak was occupied in the ancestor of jawed vertebrates -- indicating that no intrinsic constraints prevent its realization -- but it became far more difficult to access just 40 million years later because of intervening epistatic mutations. Selection is an extraordinarily powerful evolutionary force;28 nevertheless, our observations suggest that, because of the complexity of GR architecture, low-probability permissive substitutions were required to open some mutational trajectories to exploration under selection,24,29 while restrictive substitutions closed other potential paths. Under selection, some kind of adaptation will always occur,30 but the specific adaptive forms that are realized depend on the historical trajectory that precedes them. The conditions that facilitated evolution of GR’s past forms were destroyed during realization of its present.2,4,7,16,18 The past is difficult to recover because it was built on the foundation of its own history, one irrevocably different from that of the present and its many possible futures.
Methods
Ancestral protein sequences
AncGR1 and AncGR2 sequences were inferred by maximum likelihood31 using PAML 3.15 software on the maximum likelihood phylogeny of 60 amino acid sequences of extant steroid and related receptors (see refs. 23 and 24 for details). In brief, the likelihood of each possible amino acid state was calculated given the extant sequence data, the maximum likelihood phylogeny, the Jones-Taylor-Thornton amino acid replacement model (which had 100% posterior probability in a Bayesian evaluation of numerous protein models), and a gamma distribution of among-site rate variation. The maximum likelihood amino acid sequences of the ligand-binding domains of AncGR1 and AncGR2, including the carboxy-terminal extension (CTE), were back-translated assuming human codon bias. Coding DNAs were then synthesized (GenScript, Piscataway, NJ), verified by sequencing, and cloned into pSG5-Gal4DBD with the human GR hinge domain for expression and characterization. The functions of AncGR1 and AncGR2 LBD fusion proteins, assayed as described below, were robust to statistical ambiguity in the ancestral sequence inference.24
Molecular biology
The hormone-dependent transcriptional activity of resurrected ancestral receptors and their variants was assayed using a luciferase reporter system. Chinese hamster ovary (CHO-K1) cells were grown in 96-well plates and transfected with1 ng of receptor plasmid, 100 ng of a UAS-driven firefly luciferase reporter (pFRluc), and 0.1 ng of the constitutive phRLtk Renilla luciferase reporter plasmid, using Lipofectamine and Plus Reagent in OPTIMEM (Invitrogen, Carlsbad, CA). After 4 hours, transfection medium was replaced with phenol-red-free αMEM supplemented with 10% dextran-charcoal-stripped fetal bovine serum (Hyclone, Logan, UT). Following overnight recovery, cells were incubated in triplicate with aldosterone, cortisol or 11-deoxycorticosterone from 10−11 to 10−5 M for 24 hours, then assayed using Dual-Glo luciferase (Promega, Madison, WI). Firefly luciferase activity was normalized by Renilla luciferase activity. Dose-response relationships were estimated using non-linear regression in Prism4 software (GraphPad Software, Inc., San Diego, CA); fold increase in activation was calculated relative to vehicle-only control. Mutagenesis to recapitulate historical substitutions was performed using QuikChange (Stratagene, La Jolla, CA) and verified by sequencing.
Structural biology
The atomic structure of AncGR2-LBD was determined using X-ray crystallography. AncGR2-LBD cDNA (residues 1–248) was cloned into pMALCH10T (a gift of J. Tesmer) and expressed as a maltose-binding protein/TEVfusion protein in BL21(DE3) pLys cells in the presence of 50 μM dexamethasone using standard methods. Expressed protein was purified using affinity chromatography. Following TEV cleavage, the tagged fusion protein was removed using a nickel affinity column, polished via gel filtration, dialyzed (200 mM sodium chloride, 50 μM HEPES (pH 7.8) and 50 μM CHAPS), and concentrated to 3–5 mg/mL. Crystals of AncGR2-LBD with dexamethasone were grown by hanging drop vapor diffusion at 22 °C from solutions containing 0.75 μL of protein at 3–5 mg/mL protein and 0.75 μL of crystallant (0.5–0.75 M Ammonium, pH = 7.4), and a 21 amino acid NR box-3 peptide of GR coactivator human TIF2 (+H3N-PVSPKKKENALLRYLLDKDDT-CO2−, Synbiosci, Livermore, CA). Crystals were cryoprotected in crystallant with 25% glycerol and flash-cooled in liquid N2. Data to 2.5 Å resolution were collected at 100 K at the South East Regional Collaborative Access Team at the Advanced Photon Source, and were processed and scaled with HKL200032 (Table S1). Initial phases for the AncGR2-cortisol complex were determined using PHASER33 in the CCP4 software suite. The previously described homology model of AncGR224 was used as a molecular replacement search model. All structures were refined using COOT34 and CNS.35 The X-ray crystal structure of AncGR2 (PDB identifier 3GN8) was compared to the model of AncGR1,24 which was previously generated by homology modeling based on the X-ray crystal structure of its evolutionary precursor AncCR (PDB ID 2Q1V), with which it is identical at 90% of sites.
Supplementary Material
Acknowledgments
Supported by National Science Foundation IOB-0546906, National Institutes of Health R01-GM081592 and F32-GM074398, and a Sloan Foundation Fellowship to J.W.T. We thank members of the Thornton, Cresko, and Phillips laboratories for comments.
Footnotes
Atomic coordinates and structure factors for AncGR2 have been deposited and assigned PDB identifier 3GN8.
J.T.B. and J.W.T. conceived the experiments. J.T.B. performed the functional experiments, E.A.O. the structural analysis, and J.W.T. the phylogenetic analysis. J.T.B, E.A.O, and J.W.T. interpreted the results. J.T.B. and J.T. wrote the paper.
The authors have no competing financial interests to declare.
Supplementary Information accompanies the paper on www.nature.com/nature.
References
- 1.Muller HJ. Reversibility in evolution considered from the standpoint of genetics. Biological Reviews. 1939;14:261–280. [Google Scholar]
- 2.Simpson GG. The major features of evolution. Columbia University Press; New York: 1953. [Google Scholar]
- 3.Crick FH. The origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 4.Gould SJ. Dollo on Dollo’s law: irreversibility and the status of evolutionary laws. J History Biol. 1970;3:189–212. doi: 10.1007/BF00137351. [DOI] [PubMed] [Google Scholar]
- 5.Dobzhansky TG. Genetics of the evolutionary process. Columbia University Press; New York: 1971. [Google Scholar]
- 6.Macbeth N. Reflections on Irreversibility. Systematic Zoology. 1980:402–404. [Google Scholar]
- 7.Bull JJ, Charnov EL. On irreversible evolution. Evolution. 1985:1149–1155. doi: 10.1111/j.1558-5646.1985.tb00455.x. [DOI] [PubMed] [Google Scholar]
- 8.Teotonio H, Rose MR. Perspective: reverse evolution. Evolution. 2001;55:653–660. doi: 10.1554/0014-3820(2001)055[0653:pre]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Collin R, Cipriani R. Dollo’s law and the re-evolution of shell coiling. Proc Biol Sci. 2003;270:2551–2555. doi: 10.1098/rspb.2003.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domes K, Norton RA, Maraun M, Scheu S. Reevolution of sexuality breaks Dollo’s law. Proc Natl Acad Sci U S A. 2007;104:7139–7144. doi: 10.1073/pnas.0700034104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohlsdorf T, Wagner GP. Evidence for the reversibility of digit loss: a phylogenetic study of limb evolution in Bachia (Gymnophthalmidae: Squamata) Evolution. 2006;60:1896–1912. [PubMed] [Google Scholar]
- 12.Whiting MF, Bradler S, Maxwell T. Loss and recovery of wings in stick insects. Nature. 2003;421:264–267. doi: 10.1038/nature01313. [DOI] [PubMed] [Google Scholar]
- 13.Chippindale PT, Bonett RM, Baldwin AS, Wiens JJ. Phylogenetic evidence for a major reversal of life-history evolution in plethodontid salamanders. Evolution. 2004;58:2809–2822. doi: 10.1111/j.0014-3820.2004.tb01632.x. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg EE, Igic B. On phylogenetic tests of irreversible evolution. Evolution. 2008 doi: 10.1111/j.1558-5646.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 15.Collin R, Miglietta MP. Reversing opinions on Dollo’s Law. Trends Ecol Evol. 2008 doi: 10.1016/j.tree.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Wagner GP. The logical structure of irreversible systems transformations: a theorem concerning Dollo’s law and chaotic movement. J Theor Biol. 1982;96:337–346. doi: 10.1016/0022-5193(82)90114-x. [DOI] [PubMed] [Google Scholar]
- 17.Zufall RA, Rausher MD. Genetic changes associated with floral adaptation restrict future evolutionary potential. Nature. 2004;428:847–850. doi: 10.1038/nature02489. [DOI] [PubMed] [Google Scholar]
- 18.Lewontin RC. Is nature probable or capricious? Bioscience. 1966;16:25. [Google Scholar]
- 19.Pagel M. Limpets break Dollo’s Law. Trends Ecol Evol. 2004;19:278–280. doi: 10.1016/j.tree.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 20.Thornton JW. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 21.Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci U S A. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1681/01.asn.0000926836.46869.e5. [DOI] [PubMed] [Google Scholar]
- 24.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wurtz JM, et al. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:206. doi: 10.1038/nsb0296-206. [DOI] [PubMed] [Google Scholar]
- 26.Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 27.Majerus MEN. Melanism: evolution in action. Oxford University Press; Oxford; New York: 1998. [Google Scholar]
- 28.Dawkins R. Blind watchmaker. W.W. Norton; New York: 1994. [Google Scholar]
- 29.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 30.Dennett DC. Darwin’s dangerous idea: evolution and the meanings of life. Simon & Schuster; New York: 1995. [Google Scholar]
- 31.Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.