Abstract
Filoviruses, including ebolaviruses and marburgviruses, are the causative agents of highly lethal disease outbreaks. The 2013–2016 Ebola virus outbreak was responsible for >28,000 infections and >11,000 deaths. Although there are currently no licensed vaccines or therapeutics for any filovirus-induced disease, monoclonal antibodies (mAbs) are among the most promising options for therapeutic development. Hundreds of mAbs have been isolated from human survivors of filovirus infections that target the viral spike glycoprotein (GP). The binding, neutralization, and cross-reactivity of many of these mAbs has been determined. Several mAbs have been characterized structurally, and this information has been crucial for strategizing therapeutic and vaccine design. Here we present an overview of the structural features of the neutralizing/protective epitopes on filovirus glycoproteins.
Filoviruses cause severe disease in both humans and nonhuman primates. Outbreaks are unpredictable and occur with mortality rates between 25–90% [1,2]. Three genera comprise the family Filoviridae: Ebolavirus [which includes Ebola virus (EBOV), Sudan virus (SUDV), Bundibugyo virus (BDBV), Taï Forest virus, and Reston virus], Marburgvirus [which includes Marburg virus (MARV) and Ravn virus (RAVV)], and Cuevavirus [which includes Lloviu virus]. Ebolaviruses and marburgviruses cause the clinically similar Ebola Virus Disease (EVD) and Marburg Virus Disease (MVD), respectively.
Filoviruses form extended filamentous virions surrounded by a membrane envelope that is studded with copies of the surface glycoprotein (GP). GP is the only protein expressed on the viral surface, and serves to mediate entry into the target cell. Through GP, the virions first interact with target cells via lectins [3], membrane phosphatidylserine, or TIM-1 family members [4]. After internalization by macropinocytosis [5–7], the virions enter the endosome, where host cathepsins proteolytically process GP to remove the glycan cap and mucin-like domain, leaving behind GP cleaved (GPCL) [8–10]. In GPCL, the core of the protein is exposed and allows the receptor binding site (RBS) to recognize and engage domain C of the cholesterol transporter Niemann-Pick C1 (NPC1-C) [10–15]. Currently, GP is the primary target for antibodies and vaccines due to its prevalent exposure on the viral surface and its critical role in viral entry [16]. Given the complexity of antibody recognition and neutralization of filoviruses, analysis of structural differences in antibody-GP complexes and mechanisms of neutralization across the filovirus family is important for understanding antibody-mediated inhibition.
In the infected cell, GP is post-translationally processed by furin cleavage into GP1 and GP2 subunits [17]. The GP1 subunit facilitates host cell attachment and receptor recognition, whereas GP2 mediates fusion of the virus and host membranes [18–21]. Three GP1–GP2 heterodimers assemble into a trimeric peplomer, or “spike” on the viral surface [22–24]. The RBS is located beneath the glycan cap towards the top of the GP1 subunit and contains a hydrophobic pocket into which loop 2 of NPC1-C binds [11,12,15]. The C-terminus of GP1 has a heavily glycosylated mucin-like domain that is situated on the upper and outer portions of the peplomer [22]. The GP2 subunit contains an N-terminal peptide (released from GP1 by furin cleavage), an internal fusion loop (IFL), two heptad repeats (HR1 and HR2), a membrane proximal external region (MPER), and a C-terminal transmembrane domain [19,23]. HR1 wraps around the base of the GP1 receptor-binding core while HR2 forms a “stalk” that connects the GP core to the viral membrane [23]. Many portions of GP2 including the fusion loop, HR1, and the HR2 stalk are organized similarly between ebolaviruses and marburgviruses (Figure 1) [23,25].
Figure 1. Antibody epitopes on filovirus GPs.
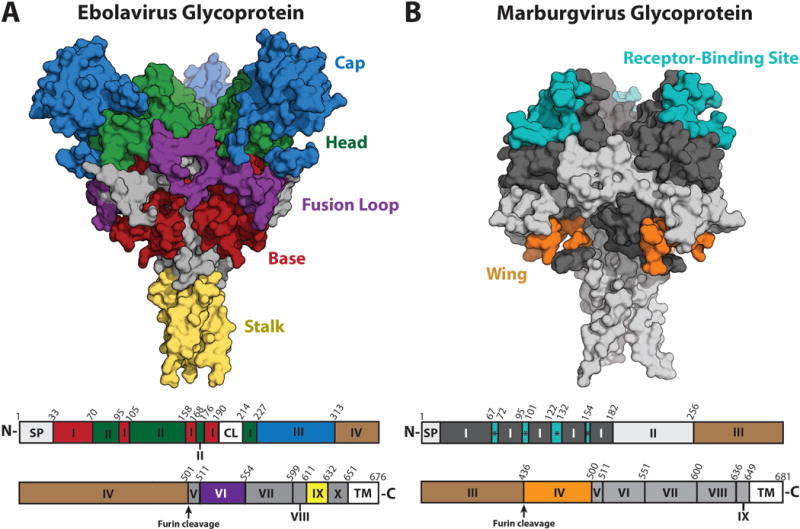
(A) Ebolavirus GP with antibody binding epitopes shown as patches of color on the GP surface (PDB: 5JQ7) [65] and a corresponding sequence map below. Labels for ebolaviruses: SP = Signal Peptide, I = Base, II = Head, CL = Cathepsin Cleavage Loop, III = Glycan Cap, IV = Mucin-like Domain (MLD), V = N-terminal Loop, VI = Fusion Loop, VII = Heptad Repeat 1 (HR1), VIII and IX are together Heptad Repeat 2 (HR2), of which IX = Stalk, X = Membrane Proximal External Region (MPER), and TM = Transmembrane domain. (B) Marburgvirus GP with antibody binding epitopes shown as patches of color on the GP surface (PDB: 6BP2) [25]. Labels for marburgviruses: SP = Signal Peptide, I = GP1, * = Receptor binding site, II = Glycan Cap, III = MLD, IV = Wing, V = N-terminal loop, VI = Fusion Loop, VII = HR1, VIII = HR2, IX = MPER, and TM = Transmembrane domain. The RBS is illustrated only on marburgvirus GP for clarity; on uncleaved ebolavirus GP, the glycan cap masks the RBS.
The GP2 of marburgviruses contains an additional domain, absent in ebolaviruses, termed the “wing” due to its outward projection and flexibility [26]. The wing results from an N-terminal shift in the relative position of the furin cleavage site between marburgviruses and ebolaviruses [27]. In marburgviruses, the mucin-like domain is attached to the C terminus of GP1, whereas the wing domain is attached to the N terminus of GP2 (Figure 1) [25,26]. Although the marburgvirus wing domain was thought to be analogous to the C-terminal portion of the ebolavirus mucin-like domain, recent structural information revealed otherwise. Part of the marburgvirus wing (residues 469–478 and 487–498) anchors itself to the GP core through a pair of beta strands that hug GP1 in an organization that is analogous to β1–β2 of ebolavirus GP1 [23,25]. The remaining portions of the marburgvirus wing, including residues 436–469 and those that connect the beta strand pair to the IFL, have yet to be defined structurally. Antibodies directed against the wing domain show neutralizing activity in cell culture and are protective in the mouse model [26]. However, whether the wing domain is involved in virus fusion and entry is unclear and is an important question for further investigation.
A broad array of neutralizing antibodies against ebolavirus GPs have been isolated from animal immunization studies as well as from human survivors of EVD [28–42]. Structural studies of such antibodies revealed neutralizing epitopes across the surface of GP [43,44]. Sets of antibodies can be grouped into epitope classes that are known to react to a GP1/GP2 containing region at the bottom of the GP core termed the “base”, the glycan cap of GP1, the head/apex of GP1, the IFL of GP2, the HR2 stalk of GP2, or to several linear epitopes within the mucin-like domain (Figure 1) [45]. At least one member of each epitope class offers in vivo protection in the mouse model of EBOV infection [29–32,35,42,44,45].
The mechanism of protection can be roughly divided according to the physical location of the epitope on the GP surface. Antibodies that recognize epitopes contained within GPCL (e.g., non-glycan cap or mucin-like domain) are typically neutralizing [11,29,30,33,35,37,39,41,43–46] and likely block infection by physically impeding virus entry through prevention of conformational changes required for fusion [8,47,48], GP cleavage [41], or receptor binding [25,49]. Meanwhile, antibodies targeting domains removed by cathepsin cleavage are less likely to be neutralizing [11,30,32,41,45,46], although exceptions exist [30,34].
Interestingly, some antibodies with limited neutralization capacity are nonetheless highly protective in an animal model [32,42,45,50], likely via immune-mediated clearance of the pathogen and infected cells [45,51,52]. The upper and outer location of these epitopes on the GP molecule could provide enhanced accessibility to FcR-bearing cells [22] and/or binding to these conformationally mobile parts of GP may facilitate IgG-IgG associations (Figure 2).
Figure 2. Visualization of the Mucin-Like Domain.
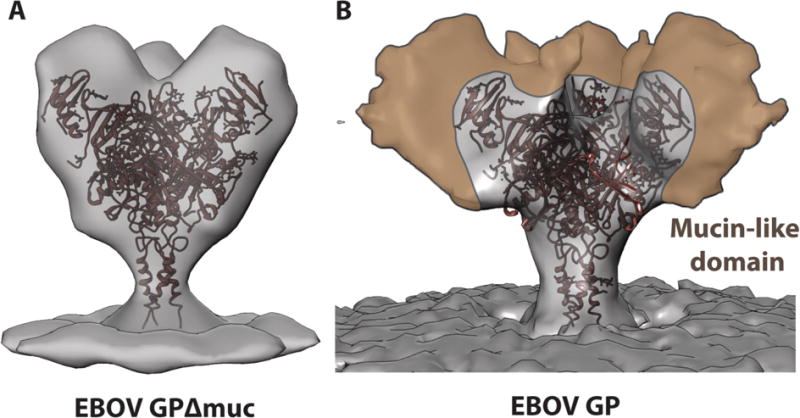
(A) The crystal structure of the mucin-deleted EBOV GP (PDB: 5JQ3) [65] is shown docked into a subtomogram averaged map of mucin-deleted EBOV GP [66] and a single-particle generated map of intact, mucin-containing EBOV GP (B) [22]. Although the observation of density for mobile regions is limited by technical factors in single particle reconstruction, the regions of the mucin-like domain that are visible appear to extend upwards and outwards from the glycan cap and base region, thereby shielding much of the GP core.
The primary product of the ebolavirus GP gene is not GP, but rather an abundantly produced soluble protein dimer termed secreted glycoprotein (sGP) [53,54]. The N-terminal 295 amino acids of GP and sGP are identical and include the RBS and the glycan cap domains (Figure 3). The C termini, however, differ. Only GP encodes the mucin-like domain and GP2. sGP instead ends in a short peptide with a cysteine that mediates a disulfide bond [54,55]. Antibodies that target the RBS and/or the glycan cap generally also bind sGP, sometimes with higher affinity than GP [28,29,52,55–61]. sGP has been hypothesized to serve several roles, including as an immune decoy, although its precise role in infection has not yet been fully elucidated [62]. Notably, the antibody termed mAb 114, which recognizes both GP and sGP, protected non-human primates against EBOV challenge when administered as a monotherapy [31,43], and 13C6, a key component of the ZMapp cocktail, also cross-reacts between sGP and GP [32,42]. However, in the mouse model, sGP cross-reactivity appears not have a significant impact on protection [45].
Figure 3. Structural similarities between sGP and GP.
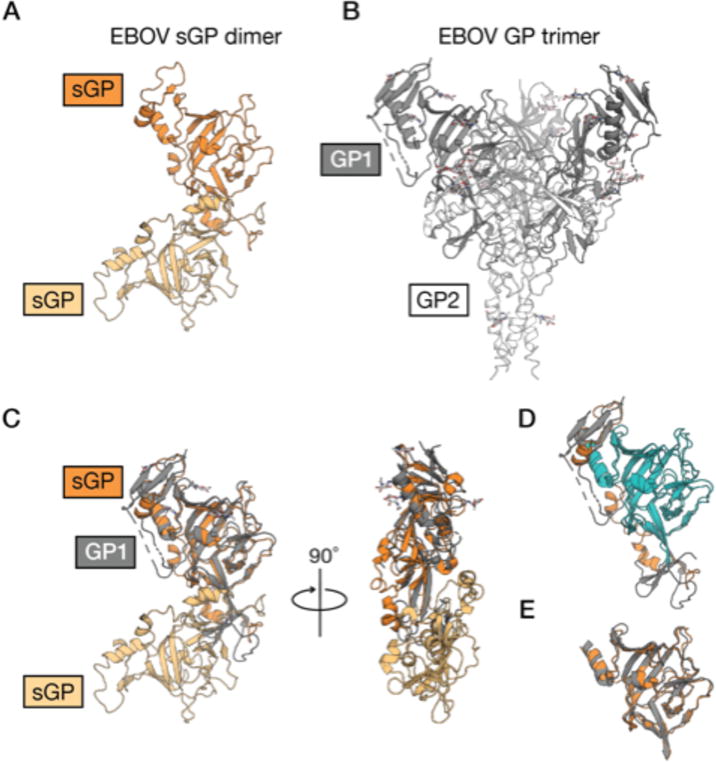
(A) The EBOV sGP dimer (PDB: 5KEM) [55] is shown as a cartoon with each monomer shaded orange. (B) The EBOV GP trimer (PDB: 5JQ3) [65] is shown as a cartoon with GP1 (dark gray) and GP2 (white). (C) GP1 and a single monomer of sGP align with a Cα r.m.s.d. of 3.87 Å over 217 aligned residues. sGP and GP share 100% sequence identity for their first 295 amino acids. (D) They are structurally similar between the core residues 66–184 and 216–259 (blue), although parts of these regions are likely obscured from immune surveillance by the dimerization interface. (E) The isolated cores (same as the blue region in (D)) align with a Cα r.m.s.d. of 1.71 Å over 163 residues.
Antibodies that cross-react among multiple pathogenic filoviruses would allow rapid treatment mobilization prior to the identification of the infecting strain. Moreover, which virus strain will emerge cannot be predicted, and production and stockpiling of distinct antibody therapies against each of the antigenically distinct filoviruses is cost-prohibitive. However, antibodies that cross-react with multiple filovirus GPs remain rare. The five known ebolavirus GPs differ by ~50% at the amino acid level, and EBOV and MARV GP sequences differ by ~70%. Much of the sequence variation, however, is concentrated in the mucin-like domain with less variation in the GP core, although conservation in the core does vary by epitope. For example, the IFL of GP2 is more conserved, and three broadly reactive antibodies have been identified with overlapping epitopes involving the tip or stem of the fusion loop [29,35,44]. In contrast, most antibodies against the base are virus-specific likely due to differences in amino acid composition and conformation in the GP2 N-terminal peptide [23,33,55]. Other broadly cross-reactive antibodies map to the glycan cap and the HR2 stalk [30], while broadly reactive non-neutralizing antibodies have been mapped to sites around the GP1 head and glycan cap [52,59]. High-resolution structures of the broadly reactive antibodies are needed to illuminate the determinants of their reactivity and to inform vaccine design.
In marburgviruses, the glycan cap appears to be less effective at shielding the RBS. Unlike survivors of ebolavirus infection, the majority of antibodies identified in a human survivor of MARV infection recognize the RBS, and can bind whether or not the GP is cleaved (i.e., whether or not the glycan cap is present) [63]. Two crystal structures of antibodies bound to the marburgvirus GP RBS are available [25,49] as are several low-resolution negative stain EM structures of other antibody complexes with this site [49,63] (Figure 4). Recent work describes a high resolution structure of a marburgvirus GP containing the glycan cap (PDB: 6BP2) [25]. In this structure, although the GP was intact, the cap was disordered and could not be observed structurally.
Figure 4. Neutralizing epitopes identified for marburgvirus.
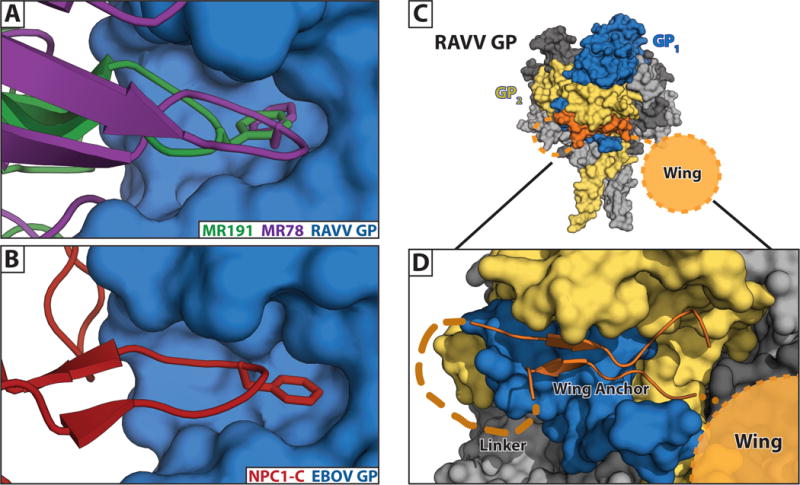
(A) Crystal structures of neutralizing antibodies MR191 [25] and MR78 [49] bound within the RBS of RAVV GP. (B) A phenylalanine at the apex of the CDR-H3s of both antibodies reaches into the hydrophobic pocket of the marburgvirus GP in a manner that structurally mimics interactions of ebolavirus GP both with its glycan cap and its host receptor, NPC1-C [12]. (C) A marburgvirus GP is shown with a single GP monomer colored in blue (GP1) and gold (GP2). The remaining two monomers are grey. The anchor of the wing domain (orange) is shown wedged underneath the base of GP. (D) Enlarged view of the wing illustrating the β-strand wing anchor region (connected by an 8 aa linker), and the relative position of the 33 aa wing targeted by antibodies.
The fact that anti-MARV antibodies can bind the RBS of uncleaved marburgvirus GP suggests that the RBS of marburgviruses may be transiently exposed. We believe that the marburgvirus RBS is transiently rather than completely exposed because NPC1-C can not bind uncleaved GP, and instead only reacts with GPCL [25,26,49,63]. High-affinity antibodies may be able to displace the glycan cap, whereas lower affinity NPC1-C interactions may not; the affinity of NPC1-C for MARV GPCL at pH 7.4 is ~150 μM [12]. It is also possible that is that low levels of non-specific proteolysis of the glycan cap and mucin-like domain in vivo could facilitate exposure of the RBS to immune recognition.
For marburgviruses, protective antibodies have only been identified thus far against two epitopes on the GP: the RBS [25,49,63] and the marburgvirus-specific wing [26]. Murine antibodies against the wing are protective in the mouse model, but have not yet been evaluated in larger animal models [26]. The human antibody MR191 directed against the RBS has been evaluated in non-human primates and offered complete post-exposure protection five days after virus exposure [64]. Interestingly, escape mutants of MR191, uncovered using MARV GP-pseudotyped recombinant VSV, do not exist in the antibody’s RBS footprint, but instead, are in the glycan cap and the wing domains [25,63], suggesting that conformational adjustments in these flexible domains may affect the RBS through a mechanism that is not yet understood. Identification of additional protective antibodies against other sites will allow development of treatment cocktails to mitigate escape.
Antibodies that cross-react among multiple pathogenic filoviruses would allow rapid treatment mobilization prior to the identification of the infecting strain. Moreover, which virus strain will emerge cannot be predicted, and production and stockpiling of distinct antibody therapies against each of the antigenically distinct filoviruses is cost-prohibitive. However, antibodies that cross-react with multiple filovirus GPs remain rare. The five known ebolavirus GPs differ by ~50% at the amino acid level, and EBOV and MARV GP sequences differ by ~70%. Much of the sequence variation, however, is concentrated in the mucin-like domain with less variation in the GP core, although conservation in the core does vary by epitope. For example, the IFL of GP2 is more conserved, and three broadly reactive antibodies have been identified with overlapping epitopes involving the tip or stem of the fusion loop [29,35,44]. In contrast, most antibodies against the base are virus-specific. Although these antibodies may bridge the anchor point of the fusion loop, they also recognize other residues in GP that differ in sequence and conformation, such as the GP2 N-terminal peptide [23,33,55]. Other broadly cross-reactive antibodies map to the glycan cap and the HR2 stalk [30], while broadly reactive non-neutralizing antibodies have been mapped to sites around the GP1 head and glycan cap [52,59]. High-resolution structures of the broadly reactive antibodies are needed to illuminate the determinants of their reactivity and to inform vaccine design.
The first EBOV GP-antibody structure was determined ten years ago. Over the ensuing decade, additional structures of EBOV-, SUDV-, and MARV-reactive mAbs in complex with their cognate GPs have showcased the variety of antibody epitopes that lead to neutralization and protection, as well as differences among GP targets. Still needed are structures of BDBV GP, RESTV GP, and discovery, characterization or engineering of more mAbs that broadly react and broadly neutralize among the different filoviruses. Further, a better understanding of the Fc-mediated functions of these antibodies would support ongoing development of effective immunotherapeutics and vaccines to prevent filovirus disease.
Supplementary Material
Highlights.
Filovirus neutralization and protection is related to epitope on filovirus GPs
GP epitopes on marburgvirus and ebolavirus differ fundamentally at a structural level
Ebolavirus and marburgvirus GP have six and two, respectively, antibody epitope classes
Marburgvirus cleavage sites and glycosylation differ from those for ebolaviruses
The wing domain is unique to marburgvirus GP
Acknowledgments
We acknowledge NIH/NIAID U19-AI109762 to the Viral Hemorrhagic Fever Immunotherapeutic Consortium for support. This is manuscript number 29654 from The Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers have been annotated to indicate special interest (*) or outstanding interest (**)
- 1.Outbreaks Chronology: Ebola Virus Disease | Ebola Hemorrhagic Fever | CDC. 2017,
- 2.Outbreak Table | Marburg Hemorrhagic Fever | CDC. 2014,
- 3.Favier A-L, Gout E, Reynard O, Ferraris O, Kleman J-P, Volchkov V, Peyrefitte C, Thielens NM. Enhancement of Ebola Virus Infection via Ficolin-1 Interaction with the Mucin Domain of GP Glycoprotein. J Virol. 2016;90:5256–5269. doi: 10.1128/JVI.00232-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci U S A. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aleksandrowicz P, Marzi A, Biedenkopf N, Beimforde N, Becker S, Hoenen T, Feldmann H, Schnittler H-J. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J Infect Dis. 2011;204(Suppl 3):S957–67. doi: 10.1093/infdis/jir326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misasi J, Chandran K, Yang J-Y, Considine B, Filone CM, Côté M, Sullivan N, Fabozzi G, Hensley L, Cunningham J. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J Virol. 2012;86:3284–3292. doi: 10.1128/JVI.06346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Bornholdt ZA, Ndungo E, Fusco ML, Bale S, Flyak AI, Crowe JE, Chandran K, Saphire EO. Host-Primed Ebola Virus GP Exposes a Hydrophobic NPC1 Receptor-Binding Pocket, Revealing a Target for Broadly Neutralizing Antibodies. MBio. 2016;7 doi: 10.1128/mBio.02154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Wang H, Shi Y, Song J, Qi J, Lu G, Yan J, Gao GF. Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1. Cell. 2016;164:258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Côté M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Dye JM, Herbert AS, Kuehne AI, Barth JF, Muhammad MA, Zak SE, Ortiz RA, Prugar LI, Pratt WD. Postexposure antibody prophylaxis protects nonhuman primates from filovirus disease. Proc Natl Acad Sci U S A. 2012;109:5034–5039. doi: 10.1073/pnas.1200409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez A, Yang ZY, Xu L, Nabel GJ, Crews T, Peters CJ. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol. 1998;72:6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory SM, Harada E, Liang B, Delos SE, White JM, Tamm LK. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc Natl Acad Sci U S A. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, Nyenhuis DA, Nelson EA, Cafiso DS, White JM, Tamm LK. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc Natl Acad Sci U S A. 2017;114:E7987–E7996. doi: 10.1073/pnas.1708052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 21.Malashkevich VN, Schneider BJ, McNally ML, Milhollen MA, Pang JX, Kim PS. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci U S A. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beniac DR, Booth TF. Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 Å resolution. Sci Rep. 2017;7:46374. doi: 10.1038/srep46374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JE, Saphire EO. Neutralizing ebolavirus: structural insights into the envelope glycoprotein and antibodies targeted against it. Curr Opin Struct Biol. 2009;19:408–417. doi: 10.1016/j.sbi.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.King LB, Fusco ML, Flyak AI, Ilinykh PA, Huang K, Gunn B, Kirchdoerfer RN, Hastie KM, Sangha AK, Meiler J, et al. The Marburgvirus-Neutralizing Human Monoclonal Antibody MR191 Targets a Conserved Site to Block Virus Receptor Binding. Cell Host Microbe. 2018;23:101–109.e4. doi: 10.1016/j.chom.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Fusco ML, Hashiguchi T, Cassan R, Biggins JE, Murin CD, Warfield KL, Li S, Holtsberg FW, Shulenin S, Vu H, et al. Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPs. PLoS Pathog. 2015;11:e1005016. doi: 10.1371/journal.ppat.1005016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volchkov VE, Volchkova VA, Ströher U, Becker S, Dolnik O, Cieplik M, Garten W, Klenk HD, Feldmann H. Proteolytic processing of Marburg virus glycoprotein. Virology. 2000;268:1–6. doi: 10.1006/viro.1999.0110. [DOI] [PubMed] [Google Scholar]
- 28*.Bornholdt ZA, Turner HL, Murin CD, Li W, Sok D, Souders CA, Piper AE, Goff A, Shamblin JD, Wollen SE, et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science. 2016;351:1078–1083. doi: 10.1126/science.aad5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X, Howell KA, He S, Brannan JM, Wec AZ, Davidson E, Turner HL, Chiang C-I, Lei L, Fels JM, et al. Immunization-Elicited Broadly Protective Antibody Reveals Ebolavirus Fusion Loop as a Site of Vulnerability. Cell. 2017;169:891–904.e15. doi: 10.1016/j.cell.2017.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, Kuzmina N, et al. Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection. Cell. 2016;164:392–405. doi: 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corti D, Misasi J, Mulangu S, Stanley DA, Kanekiyo M, Wollen S, Ploquin A, Doria-Rose NA, Staupe RP, Bailey M, et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- 32**.Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 33*.Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, et al. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Gui M, Niu X, He S, Wang R, Feng Y, Kroeker A, Zuo Y, Wang H, Wang Y, et al. Potent neutralizing monoclonal antibodies against Ebola virus infection. Sci Rep. 2016;6:25856. doi: 10.1038/srep25856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furuyama W, Marzi A, Nanbo A, Haddock E, Maruyama J, Miyamoto H, Igarashi M, Yoshida R, Noyori O, Feldmann H, et al. Discovery of an antibody for pan-ebolavirus therapy. Sci Rep. 2016;6:20514. doi: 10.1038/srep20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holtsberg FW, Shulenin S, Vu H, Howell KA, Patel SJ, Gunn B, Karim M, Lai JR, Frei JC, Nyakatura EK, et al. Pan-ebolavirus and Pan-filovirus Mouse Monoclonal Antibodies: Protection against Ebola and Sudan Viruses. J Virol. 2015;90:266–278. doi: 10.1128/JVI.02171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynard O, Volchkov VE. Characterization of a Novel Neutralizing Monoclonal Antibody Against Ebola Virus GP. J Infect Dis. 2015;212(Suppl 2):S372–8. doi: 10.1093/infdis/jiv303. [DOI] [PubMed] [Google Scholar]
- 38.Sobarzo A, Groseth A, Dolnik O, Becker S, Lutwama JJ, Perelman E, Yavelsky V, Muhammad M, Kuehne AI, Marks RS, et al. Profile and persistence of the virus-specific neutralizing humoral immune response in human survivors of Sudan ebolavirus (Gulu) J Infect Dis. 2013;208:299–309. doi: 10.1093/infdis/jit162. [DOI] [PubMed] [Google Scholar]
- 39*.Takada A, Feldmann H, Stroeher U, Bray M, Watanabe S, Ito H, McGregor M, Kawaoka Y. Identification of protective epitopes on ebola virus glycoprotein at the single amino acid level by using recombinant vesicular stomatitis viruses. J Virol. 2003;77:1069–1074. doi: 10.1128/JVI.77.2.1069-1074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruyama T, Rodriguez LL, Jahrling PB, Sanchez A, Khan AS, Nichol ST, Peters CJ, Parren PW, Burton DR. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J Virol. 1999;73:6024–6030. doi: 10.1128/jvi.73.7.6024-6030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shedlock DJ, Bailey MA, Popernack PM, Cunningham JM, Burton DR, Sullivan NJ. Antibody-mediated neutralization of Ebola virus can occur by two distinct mechanisms. Virology. 2010;401:228–235. doi: 10.1016/j.virol.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Misasi J, Gilman MSA, Kanekiyo M, Gui M, Cagigi A, Mulangu S, Corti D, Ledgerwood JE, Lanzavecchia A, Cunningham J, et al. Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science. 2016;351:1343–1346. doi: 10.1126/science.aad6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Wec AZ, Herbert AS, Murin CD, Nyakatura EK, Abelson DM, Fels JM, He S, James RM, de La Vega M-A, Zhu W, et al. Antibodies from a Human Survivor Define Sites of Vulnerability for Broad Protection against Ebolaviruses. Cell. 2017;169:878–890.e15. doi: 10.1016/j.cell.2017.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45**.Saphire Erica Ollmann, Schendel Sharon L, Fusco Marnie L, Gangavarapu Karthik, Gunn Bronwyn M, et al. Erica Ollmann Saphire, Systematic analysis of monoclonal antibodies against Ebola virus GP defines features that contribute to protection. doi: 10.1016/j.cell.2018.07.033. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koellhoffer JF, Chen G, Sandesara RG, Bale S, Saphire EO, Chandran K, Sidhu SS, Lai JR. Two synthetic antibodies that recognize and neutralize distinct proteolytic forms of the ebola virus envelope glycoprotein. Chembiochem. 2012;13:2549–2557. doi: 10.1002/cbic.201200493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brecher M, Schornberg KL, Delos SE, Fusco ML, Saphire EO, White JM. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J Virol. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markosyan RM, Miao C, Zheng Y-M, Melikyan GB, Liu S-L, Cohen FS. Induction of Cell-Cell Fusion by Ebola Virus Glycoprotein: Low pH Is Not a Trigger. PLoS Pathog. 2016;12:e1005373. doi: 10.1371/journal.ppat.1005373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Hashiguchi T, Fusco ML, Bornholdt ZA, Lee JE, Flyak AI, Matsuoka R, Kohda D, Yanagi Y, Hammel M, Crowe JE, Jr, et al. Structural basis for Marburg virus neutralization by a cross-reactive human antibody. Cell. 2015;160:904–912. doi: 10.1016/j.cell.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettitt J, Zeitlin L, Kim DH, Working C, Johnson JC, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, et al. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci Transl Med. 2013;5:199ra113. doi: 10.1126/scitranslmed.3006608. [DOI] [PubMed] [Google Scholar]
- 51.Liu Q, Fan C, Li Q, Zhou S, Huang W, Wang L, Sun C, Wang M, Wu X, Ma J, et al. Antibody-dependent-cellular-cytotoxicity-inducing antibodies significantly affect the post-exposure treatment of Ebola virus infection. Sci Rep. 2017;7:45552. doi: 10.1038/srep45552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duehr J, Wohlbold TJ, Oestereich L, Chromikova V, Amanat F, Rajendran M, Gomez-Medina S, Mena I, tenOever BR, García-Sastre A, et al. Novel Cross-Reactive Monoclonal Antibodies against Ebolavirus Glycoproteins Show Protection in a Murine Challenge Model. J Virol. 2017;91 doi: 10.1128/JVI.00652-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci U S A. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volchkov VE, Becker S, Volchkova VA, Ternovoj VA, Kotov AN, Netesov SV, Klenk HD. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 55*.Pallesen J, Murin CD, de Val N, Cottrell CA, Hastie KM, Turner HL, Fusco ML, Flyak AI, Zeitlin L, Crowe JE, Jr, et al. Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nat Microbiol. 2016;1:16128. doi: 10.1038/nmicrobiol.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hood CL, Abraham J, Boyington JC, Leung K, Kwong PD, Nabel GJ. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J Virol. 2010;84:2972–2982. doi: 10.1128/JVI.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bale S, Dias JM, Fusco ML, Hashiguchi T, Wong AC, Liu T, Keuhne AI, Li S, Woods VL, Jr, Chandran K, et al. Structural basis for differential neutralization of ebolaviruses. Viruses. 2012;4:447–470. doi: 10.3390/v4040447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Murin CD, Fusco ML, Bornholdt ZA, Qiu X, Olinger GG, Zeitlin L, Kobinger GP, Ward AB, Saphire EO. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc Natl Acad Sci U S A. 2014;111:17182–17187. doi: 10.1073/pnas.1414164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keck Z-Y, Enterlein SG, Howell KA, Vu H, Shulenin S, Warfield KL, Froude JW, Araghi N, Douglas R, Biggins J, et al. Macaque Monoclonal Antibodies Targeting Novel Conserved Epitopes within Filovirus Glycoprotein. J Virol. 2015;90:279–291. doi: 10.1128/JVI.02172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang B, Kluwe CA, Lungu OI, DeKosky BJ, Kerr SA, Johnson EL, Jung J, Rezigh AB, Carroll SM, Reyes AN, et al. Facile Discovery of a Diverse Panel of Anti-Ebola Virus Antibodies by Immune Repertoire Mining. Sci Rep. 2015;5:13926. doi: 10.1038/srep13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howell KA, Qiu X, Brannan JM, Bryan C, Davidson E, Holtsberg FW, Wec AZ, Shulenin S, Biggins JE, Douglas R, et al. Antibody Treatment of Ebola and Sudan Virus Infection via a Uniquely Exposed Epitope within the Glycoprotein Receptor-Binding Site. Cell Rep. 2016;15:1514–1526. doi: 10.1016/j.celrep.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de La Vega M-A, Wong G, Kobinger GP, Qiu X. The multiple roles of sGP in Ebola pathogenesis. Viral Immunol. 2015;28:3–9. doi: 10.1089/vim.2014.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63**.Flyak AI, Ilinykh PA, Murin CD, Garron T, Shen X, Fusco ML, Hashiguchi T, Bornholdt ZA, Slaughter JC, Sapparapu G, et al. Mechanism of Human Antibody-Mediated Neutralization of Marburg Virus. Cell. 2015;160:893–903. doi: 10.1016/j.cell.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Mire CE, Geisbert JB, Borisevich V, Fenton KA, Agans KN, Flyak AI, Deer DJ, Steinkellner H, Bohorov O, Bohorova N, et al. Therapeutic treatment of Marburg and Ravn virus infection in nonhuman primates with a human monoclonal antibody. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aai8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65**.Zhao Y, Ren J, Harlos K, Jones DM, Zeltina A, Bowden TA, Padilla-Parra S, Fry EE, Stuart DI. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tran EE, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S. Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. J Virol. 2014;88:10958–10962. doi: 10.1128/JVI.00870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


