Abstract
Primary central nervous system lymphoma (PCNSL) is a rare and special type of non-Hodgkin lymphoma. The treatment of PCNSL is comprehensive, combining surgery, radiotherapy, and chemotherapy. However, the outcome is poor because of its high invasiveness and rate of recurrence. We analyzed 22 cases of PCNSL using next-generation sequencing (NGS) to detect 64 candidate genes. We used immunohistochemical methods to analyze gene expression in 57 PCNSL samples. NGS showed that recurrent mutations in KMT2D and CD79B, components of the NF-κB pathway, accounted for 65% of total mutations in PCNSL samples. The most frequent mutated gene was PIM1 (77.27%, 17/22), followed by MYD88 (63.64%, 14/22), CD79B (69.09%, 13/22), and KMT2D (50.00%, 11/22). Mutations of the CD79B gene were associated with an inferior progression-free survival (PFS), and GNA13 gene mutations were associated with a shorter PFS and overall survival (OS) in PCNSL patients (P < .05). PIM1 and MYD88 were highly expressed in PCNSL patients and were related to their OS time. MYD88 overexpression might be an independent and poor prognostic predictor of OS time. In summary, we identified highly recurrent genetic lesions in CD79B and KMT2D, components of the NF-κB pathway, in PCNSL and validated the expression of PIM1 and MYD88 related to poor survival, thereby providing novel insights into the pathogenesis and precision medicine of PCNSL.
Abbreviation: ABC, activated-B Cell; BTK, Bruton's tyrosine kinase; CNS, Central nervous system; CLL, chronic lymphocytic leukemia; DLBCL, Diffuse Large B-cell lymphoma; DAB, diaminobenzidin; DNMT, DNA methyltransferase; GO, gene ontology; GCB, germinal center cell like; HR, hazard ratio; HDAC, histone deacetylase; IHC, Immunohistochemistry; LDH, lactate dehydrogenase; MYD88, Myeloid Differentiation Factor 88; MCL, mantle cell lymphoma; NHL, Non-Hodgkin Lymphoma; NGS, next generation sequencing; OS, overall survival; ORR, overall response; PFS, progression-free survival; PIM1, proviral integration of moloney murine leukemia virus; PCNSL, Primary Central Nervous System Lymphoma; PKC, Protein kinase C; SNPs, single nucleotide polymorphisms
Introduction
Primary central nervous system lymphoma (PCNSL) is derived from the central nervous system (CNS) and can include the brain parenchyma, spinal cord, eyeball, cranial nerve, and meninges but does not include dural lymphomas (such as follicular cell lymphoma and mantle cell lymphoma), intravascular B-cell lymphoma, immunodeficiency, or secondary to CNS lymphoma. It is a special type of non-Hodgkin lymphoma (NHL), accounting for 4%-6% of extracellular lymphomas and 1% of adult NHLs. More than 95% of PCNSL cases are diffuse large B-cell lymphoma (DLBCL), and other rare types include T-cell lymphoma and Burkitt lymphoma [1], [2], [3] The incidence of PCNSL increases with age, and it is becoming more common in aging populations, with a median age of 55-65 years [4]. The etiology of PCNSL is unclear, but it was reported to be related to EB or HIV infection, organ transplantation, or other diseases leading to immunodeficiency [5]. PCNSL exists behind the blood-brain barrier and blood-cerebrospinal fluid barrier; therefore, traditional and single-treatment methods do not effectively control tumor development. Current treatment methods include high-dose methotrexate-based chemotherapy, radiotherapy, targeted therapy, and stem cell transplantation [3], [6], [7]. However, for most PCNSL patients, the overall prognosis is poor, with a median overall survival time of 1-4 years, and the prognosis is significantly worse than for other NHLs outside the brain [8], [9].
Because of the rarity of PCNSL and the limited availability of biopsy tissues, the pathogenesis of PCNSL is still poorly understood, especially in the genomics research field in China. This has hindered the development and treatment of PCNSL. Advances in next-generation sequencing (NGS) technology have enabled the effective and comprehensive analysis of the molecular composition and function of various solid tumors and hematological malignancies, including PCNSL. Studies have shown that in PCNSL, MYD88 and CD79B are the most frequently mutated genes (30%-83%), and approximately 16% of the mutations are targeted to the CARD11 coiled helix domain leading to the inactivation of TFNAIP3 (3%) [10], [11], [12]. Transcriptome studies reported that disordered genes in PCNSL are involved in the IL-4/JAK/STAT6, cell adhesion-related, unfolding protein response, and apoptosis-related signaling pathways [13]. Replication of copy number variation studies found that PCNSL patients have frequent chromosome deletions, especially in chromosome regions 6q, 6p21.32, and 9p21 [14]. However, which mutant genes are present in PCNSL are still unclear.
The NF-κB signaling pathway belongs to the receptor protein hydrolase-dependent receptor signaling pathway. In recent years, many studies have shown that the NF-κB pathway has a significant role in tumor occurrence, development, proliferation, differentiation, apoptosis, invasion, and metastasis [15]. In DLBCL, NF-κB is a common downstream effector molecule of many signaling pathways, including the BCR, TLRs, NOTCH, and JAK-STAT signaling pathways, all of which can activate their downstream factor NF-κB. Furthermore, interactions between different pathways form a complex signaling network and jointly promote the proliferation and differentiation, apoptosis, angiogenesis, invasion, metastasis, resistance, and other pathophysiological processes of lymphoma cells [16], [17], [18]. Other studies reported high-frequency mutations and abnormal activation of the NF-κB pathway in lymphoma patients [19], [20], providing new insights to the pathogenesis and prognosis of lymphoma.
In this study, we investigated the genomic alterations and related gene expressions in PCNSL patients to provide new insights into the mechanisms of lymphomagenesis and potential prognostic factors or treatment opportunities for PCNSL.
Materials and Methods
Patients
A cohort of 57 specimens was obtained from Xiangya Hospital of Central South University from June 2012 to September 2016. Twenty-three of the latest 2-year diagnosis samples were used for DNA extraction and NGS. All cases of PCNSL were confined within the CNS (stage IE), were human immunodeficiency virus unrelated, and fulfilled the World Health Organization criteria for diagnosis. None of the patients had evidence of immunodeficiency. We also collected 20 cases diagnosed with lymphadenitis, which were used as the control group. All paraffin-embedded tumor specimens were collected in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national).
The clinical information included gender, age, PCNSL International Prognostic Index (IELSG), lactate dehydrogenase (LDH) level, type, treatment regimens, and survival time. All patients had complete clinical and follow-up data from the day of diagnosis to June 2017. Treatment response was evaluated using imaging techniques. The progression-free survival (PFS) and overall survival (OS) of these patients were calculated.
DNA Isolation
Twenty-three formalin-fixed paraffin-embedded (FFPE) tissue samples were obtained from archived material. FFPE material contained at least 70% tumor cells. DNA was extracted from a certain amount of wax roll samples using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). DNA concentrations were measured with Qubit Fluorometer 2.0 (Life Technologies, Darmstadt, Germany). Sufficient amounts of DNA for further analysis were isolated from all archived FFPE samples. One patient was excluded from further analysis because of poor DNA quality.
Next-Generation Sequencing
The sequencing platform was provided by Burning Rock Biomedical Company (Guangzhou, China) with a panel of 64 lymphoma-related genes (Supplementary Table 1), which were related to lymphoma pathogenesis and targeted therapy. We used the probe hybridization enrichment method to detect the exon regions of all the genes and the intron region of parts of the genes. According to the manufacturer's protocol, the library preparation was performed using approximately 200 ng of genomic DNA for sequencing on the Illumina MiSeq system (Illumina Inc., San Diego, CA). Library size and quality were demonstrated with the Agilent High sensitivity DNA Kit (Agilent, Santa Clara, CA).
Immunohistochemistry
The streptavidin-peroxidase–conjugated method was used for the detection of PIM1 and MYD88 expression. Resected tissue specimens of 4-μm thickness were deparaffinized in xylene and rehydrated through a gradient of alcohol and deionized water. Heat antigen retrieval was performed using ethylenediaminetetraacetic acid (pH 9.0) for 20 minutes followed by 3% hydrogen peroxide for 20 minutes and serum blocking for 30 minutes. Tissues were incubated overnight at 4°C with anti-PIM1 rabbit polyclonal antibody (Abcam, ab75776, USA, dilution 1:100) and anti-MYD88 rabbit monoclonal antibody (Abcam, ab33739, USA, dilution 1:500). Then, it was incubated with a biotin-conjugated secondary antibody for 30 minutes at 37°C. Slides were incubated in DAB for 1-2 minutes, and after 30 seconds of counterstaining with hematoxylin, the slides were dehydrated and mounted.
PIM1 and MYD88 immunohistochemistry expression was scored using a semiquantitative system based on the intensity and percentage of staining [21], [22]: negative, weak, moderate, or strong intensity. Negative and weak intensities were regarded as low expression; moderate and strong intensities were considered as high expression. We randomly selected five high-magnification (400×) areas of high-quality staining for each slide. Immunostaining results were independently evaluated by two pathologists who were blinded to the clinicopathological features. Appropriate positive and negative controls were included in the IHC assay.
Bioinformatics Analysis
Basic quality control was obtained using FastQC and raw data of the next-generation sequencing. We compared high-quality reads with the human genome (GRCH38, UCSC hg38) by using Burrows Wheeler Aligner software program. The indel realignment over the reads overlapping target regions was performed with the GATK Realigner Target Creator and Indel Realigner tools [23], [24]. In general, variants were accepted only if they occurred in at least 20% of reads; otherwise, variants with frequencies between 5% and 20% were analyzed manually using IGV [25]. InNels and single nucleotide polymorphisms were annotated by ANNOVARt37. Gene Ontology (GO) analysis was performed using the DAVID Bioinformatics Resources 6.8 database. GeneCards and the TCGA database were used for outcome comparisons and pathway analysis. We used R version 3.4.0 and GraphPad Prism 5 for figure rendering.
Statistical Analysis
All data were analyzed by SPSS 24.0 software. The relationship between gene mutation or expression and clinical data of PCNSL patients was analyzed by the Chi-square test. Wilcoxon's test was used to compare clinical pathological characteristics with PIM1 and MyD88 immunohistochemistry expression. Fisher's exact test was used to analyze GO analysis of different genes. Spearman's rank correlation analysis was used to detect correlations between the expressions of PIM1 and MyD88. The Kaplan-Meier and log-rank tests were used for survival analysis. The Cox proportional-hazard regression model was used for prognostic-related multivariate correlation analysis. All tests were performed using bilateral 95% confidence intervals (CI). A value of P < .05 was considered statistically significant.
Results
Clinical Information
This study included 57 cases of PCNSLs. The median age at diagnosis was 56 years, ranging from 17 to 78 years, and the male to female ratio was 1:0.9. Only 16 patients (28.1%) had an increased level of LDH. According to the PCNSL IELSG score standard [26], age >60 years, PS >1, LDH level higher than normal, higher protein concentration in cerebrospinal fluid, and tumor invasion of deep brain tissue (periventricular area, basal ganglia, brain stem, and cerebellum) are related to the prognosis index of risk. Eight patients (14.04%) were low risk (0-1 point), 35 (61.40%) were medium risk (2-3 point), and 14 (24.56%) were high risk (4-5 points). According to the Visco-Young algorithm [27], 49 patients (85.96%) had the activated B cell (ABC) type of lymphoma, and 8 had the germinal center B cell (GCB) subtype. Regarding initial surgical treatment, total tumor resection was performed in 30 patients (52.63%), and partial tumor resection was performed in 27 patients (47.37%). After initial treatment at the neurosurgery department and the histopathological confirmation of PCNSL, 22 patients were treated with chemotherapy and 9 received chemoradiotherapy.
Mutation Profile of PCNSL
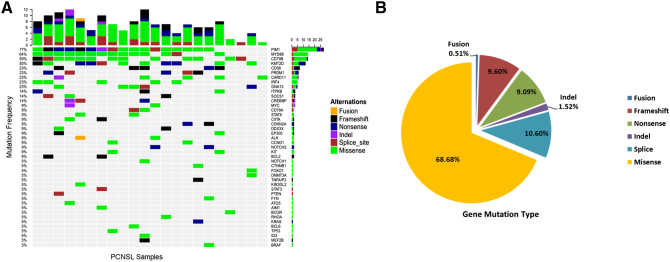
In this study, we detected and analyzed primary central nervous system lymphoma in 22 patients using a lymphoma-related gene panel of 64 targeted genes for targeted sequencing analysis (Supplementary Table 1). These genes were selected on the basis of their relationship to lymphoma pathogenesis and targeted therapy. The exon regions of all the genes and intron regions of parts of the genes were amplified. The mean sequence depth across all samples was over 1000, indicating the advantages of high-throughput sequencing. Overall, 198 mutations and 43 different gene mutations were detected. The mean number of mutated genes per case was 9. Of these, the most frequently mutated genes in our patient cohort were PIM1 (77.27%), MYD88 (63.64%), CD79B (59.09%), and KMT2D (50.0%) (Figure 1A, Supplementary Table 2). The gene mutation types of 22 samples were classified into six major categories: missense mutations, frameshift mutations, splice mutations, nonsense mutations, deletion mutations, and gene fusion. Among these, missense mutations were the most common (68.68%, 136/198), followed by splice mutations (10.60%), frameshift mutations (9.60%) and nonsense mutations (9.09%) (Figure 1B).
Figure 1.
Mutation frequencies in PCNSL cohort. (A) The distribution and frequency of genetic alterations in 22 primary central nervous system lymphoma patients. The types of mutation are labeled in different colors. (B) Pie chart showing the percentages of different types of somatic mutation in PCNSL.
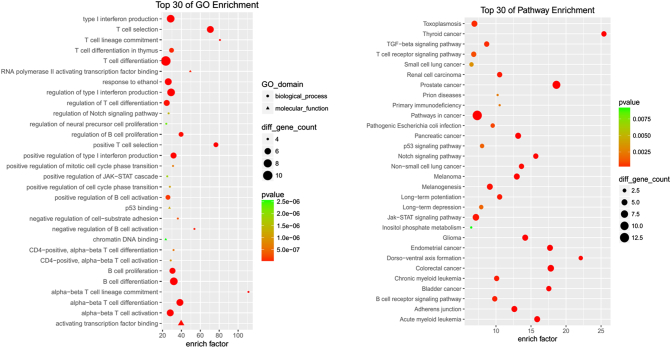
GO is widely used in the field of bioinformatics. It contains cellular components, molecular functions, and biological process. Bioinformatic analysis indicated that PCNSL-related mutation genes were mainly involved in the biological behavior of B cells and T cells and participated in the process of cancer formation (Figure 2).
Figure 2.
Bioinformatic analysis for gene ontology and KEGG in PCNSL cohort. Using Fisher accurate test, P < .05 as statistical significance. Enrich factor definition as the ratio of differential genes in a cohort group compared with the ratio in the database. The dots are sorted from the lower order of size according to the value of the enrich factor. (A) The top 30 GO enrichment in PCNSL. (B) The top 30 KEGG pathway enrichment in PCNSL.
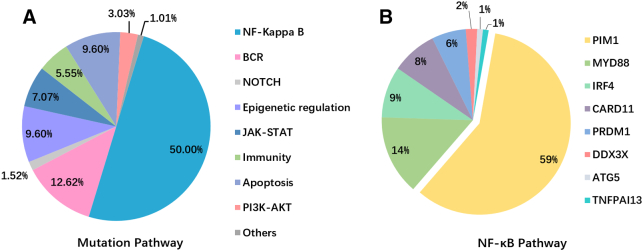
We further subdivided the 43 mutation genes targeted by the lymphoma panel into eight specific pathways (Figure 3A, Supplementary Table 3): NF-κB (PIM1, MYD88, IRF4, CARD11), B-cell receptor (CD79B, CD79A, ITPKB), epigenetic (KMT2D, CREBBP, MEF2B), JAK-STAT (CCND1, SOCS1, STAT6, STAT3), NOTCH1/2, immune-related (CD58, CIITA), and apoptotic signaling pathways (GNA13, MYC, RHOA, BCL2). As expected, the NF-κB signaling pathway had the most frequent mutations, accounting for 50.00% of cases, followed by the B-cell receptor signal pathway (12.62%), and apoptosis and epigenetic signal pathways (9.60%) (Figure 3A). Among the NF-κB signaling pathways, PIM1 (58.6%) and MYD88 (14.1%) were the most common mutations followed by IRF4 (9.1%), CARD11 (8.1%), and PRDM1 (6.1%) (Figure 3B).
Figure 3.
(A) Mutation pathway distribution among PCNSL specimens. Different color represents diverse mutation pathways and percentage. (B) The mutation genes frequency in NF-κB signaling pathway.
Potential Clinical Impact of Mutated Genes
We analyzed each mutated gene in the lymphoma panel to determine any correlation with clinical characteristics including age, gender, risk score, and LDH level. The CARD11 mutation was related to gender (P = .021), the IRF4 variant was significantly associated with age (P = .005), and the CD79B mutation correlated with risk score (P = .025).
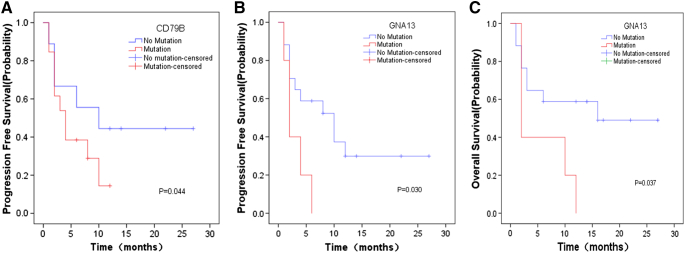
We investigated whether the presence or absence of the most prevalent variants was associated with clinical prognosis. We performed Kaplan-Meier analyses for the most frequently mutated genes using median PFS and median OS as readouts (Supplementary Table 4). Patients harboring a CD79B mutation in their lymphoma exhibited a significantly shorter PFS compared with patients with wild-type CD79B (Figure 4A, P = .044). Patients harboring a GNA13 mutation showed a favorable PFS and OS (Figure 4, B and C, P = .030 and P = .037, respectively).
Figure 4.
Mutation status and Kaplan-Meier survival curves of PCNSL patient cohort (n = 22 patients). (A) Progression-free survival for CD79B mutation status (P = .044). (B) Progression-free survival for GNA13 mutation status (P = .030). (C) Overall survival for GNA13 mutation status (P = .037).
Immunohistochemical Expression of PIM1 and MYD88 in PCNSL
We then performed PIM1 and MYD88 immunohistochemistry in samples from 57 PCNSL patients. PIM1 was mainly expressed in the nucleus, although cytoplasmic staining was found in a minority of patients (Figure 5A). MYD88 staining was predominantly cytoplasmic with little nuclear staining (Figure 5B). Positive expression was shown as yellow or brown staining. The positive expression rates of PIM1 and MYD88 in PCNSL were high (71.93%, 41/57 and 70.18%, 40/57, respectively) compared with the control group (30.00%, 6/20 and 15.00%, 3/20, respectively). The positive expression rate of PIM1 and MYD88 was statistically significant between PCNSL and lymphadenitis patients (P < .05, Table 1). Among PCNSL patients, the expression of PIM1 was positively correlated with the expression of MYD88 (r = 0.581, P = 2.0 × 10−6, Table 2).
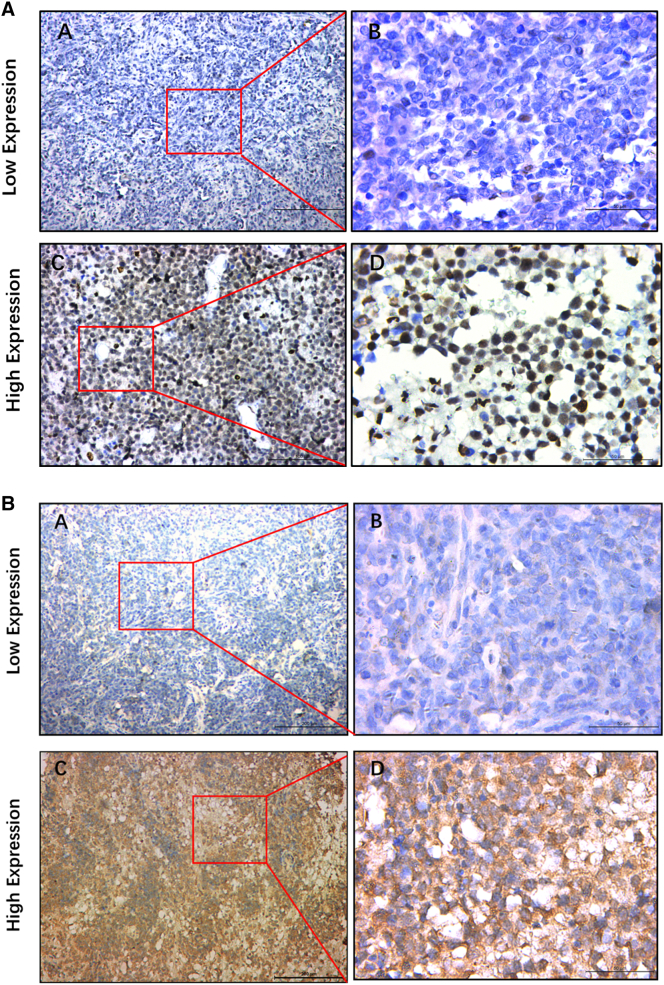
Figure 5.
Immunohistochemistry staining for PIM1(A) and MYD88 (B) in PCNSL specimens. (A) Low expression of PIM1/MYD88 in PCNSL (IHC, ×100). (B) Low expression of PIM1/MYD88 in PCNSL (IHC, ×400). (C) High expression of PIM1/MYD88 in PCNSL (IHC, ×100). (D) High expression of PIM1/MYD88 in PCNSL (IHC, ×400).
Table 1.
The Expression of PIM1 and MYD88 in PCNSL and Lymphadenitis Patients
| Histology | No. | PIM1 |
P Value | MYD88 |
P Value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | High (%) | Low | High | High (%) | ||||
| PCNSL | 57 | 16 | 41 | 71.9% | .001 | 17 | 40 | 70.2% | 1.2 × 10−5 |
| Lymphadenitis | 20 | 14 | 6 | 30.0% | 17 | 3 | 15.0% | ||
Table 2.
Correlation Between PIM1 and MYD88 Expression
| MYD88 |
r | P Value | |||
|---|---|---|---|---|---|
| Low | High | ||||
| PIM1 | Low | 12 | 5 | 0.581 | 2.0 × 10−6 |
| High | 5 | 35 | |||
PIM1 and MYD88 Expression Correlates with Clinical Characteristics and Prognosis
We investigated correlations of PIM1 and MYD88 expression with clinical characteristics including age, gender, risk score, type, and LDH level. The expression of PIM1 and MYD88 was related to the risk score of PCNSL patients (P = .023, P = 2.69 × 10−4, respectively). The high expression of PIM1 or MYD88 was correlated with a higher risk score, and the high expression of MYD88 was also correlated with the elevated level of LDH in our cohort (P = .015). However, there was no significant correlation with PCNSL patient age, gender, and/or type (P > .05) (Table 3).
Table 3.
Correlation of PIM1 and MYD88 Expression with Clinicopathological Features
| Characteristics | No. | PIM1 |
P Value | MYD88 |
P Value | ||
|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||
| Age | .206 | .206 | |||||
| <60 | 33 | 12 | 21 | 12 | 21 | ||
| ≥60 | 24 | 5 | 19 | 5 | 19 | ||
| Gender | .976 | .542 | |||||
| Male | 30 | 9 | 21 | 10 | 20 | ||
| Female | 27 | 8 | 19 | 7 | 21 | ||
| Risk group | .023 | 2.69 × 10−4 | |||||
| Low-risk group | 8 | 5 | 3 | 7 | 1 | ||
| Median-risk group | 35 | 11 | 24 | 9 | 26 | ||
| High-risk group | 14 | 1 | 13 | 1 | 13 | ||
| Type | .748 | .609 | |||||
| ABC | 49 | 15 | 34 | 14 | 35 | ||
| GCB | 8 | 2 | 6 | 3 | 5 | ||
| LDH level | .619 | .015 | |||||
| Normal | 41 | 13 | 28 | 16 | 25 | ||
| High | 16 | 4 | 12 | 1 | 15 | ||
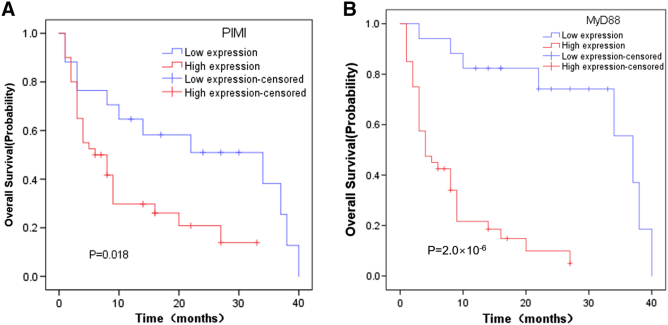
Among 57 PCNSL patients, the median OS for all patients was 16 months. The correlations of clinicopathological features and the expressions of PIM1 and MYD88 with overall survival time were determined (Table 4). We found that risk score, LDH level, and treatment method were statistically significant predictors for OS (P < .05), whereas age, gender, and type were not statistically significant (P > .05). Kaplan-Meier analysis also showed that the high expression of PIM1 and MYD88 was a statistically significant unfavorable prognostic factor for PCNSL patients, with a median OS of 11 months vs 23 months (P = .018) and 8 months vs 31 months (P = 2.0 × 10−6), respectively (Figure 6). Multivariate Cox regression model analysis including risk score, LDH level, treatment method, and PIM1 and MYD88 expression status indicated that the expression status of MYD88 was an independent predictor of OS with a hazard ratio (HR) of 0.004 (Table 5).
Table 4.
Association of OS and Clinicopathological Features
| Characteristics | Median OS (Months) | P Value |
|---|---|---|
| Age | .63 | |
| <60 | 17 | |
| ≥60 | 15 | |
| Gender | .347 | |
| Male | 14 | |
| Female | 17 | |
| Risk group | .001 | |
| Low-risk group | 34 | |
| Median-risk group | 15 | |
| High-risk group | 5 | |
| Type | .083 | |
| ABC | 14 | |
| GCB | 22 | |
| LDH level | .015 | |
| Normal | 18 | |
| High | 6 | |
| Treatment | 1.2 × 10−6 | |
| Surgery | 4 | |
| Surgery+chemotherapy | 25 | |
| Surgery+radiohemotherapy | 23 |
Figure 6.
Kaplan-Meier analysis of PCNSL patients with differential PIM1 and MYD88 expression (n = 57 patients). (A) Overall survival for PIM1 high and low expression in PCNSL patients (P = .018). (B) Overall survival for MYD88 high and low expression in PCNSL patients (P = 2.0 × 10−6).
Table 5.
Multivariate Analysis of Prognostic Factors Affecting OS of PCNSL Patients
| Parameters | HR | 95% CI | P Value |
|---|---|---|---|
| Risk group | 0.386 | 0.086-1.736 | .215 |
| Treatment methods | 3.922 | 1.198-12.837 | .054 |
| LDH Level | 1.098 | 0.492-2.451 | .16 |
| MYD88 expression | 0.143 | 0.037-0.546 | .004 |
| PIM1 expression | 2.102 | 0.777-5.691 | .144 |
Discussion
Recently, NGS has redefined the genetic landscape of lymphomas, especially DLBCL, by identifying recurrent somatic mutations. In some cases, these mutations affect potential therapeutic targets and provide new opportunities for personalized therapies. In the era of precision medicine, it is vital to effectively combine new technology with clinical practice and develop new targeted therapies to achieve individualized treatment. Schmitz et al. [28] reported the genetic subtype of DLBCL with diverse genotypic, epigenetic, and clinical characteristics, providing a potential nosology for precision-medicine strategies in DLBCL. The current study used NGS technology to detect the gene mutation status of PCNSL patients. We found a multilocus gene mutation phenomenon, which will help future pathogenesis studies and provide a new concept for the treatment of PCNSL.
In our study, a lymphoma-related gene panel covering 64 genes closely related to PCNSL pathogenesis and targeted therapy of lymphoma was used for NGS. In all 22 PCNSL samples, 198 mutations and 43 different genes mutations were detected. Overall, 65% mutations were found in NF-κB pathway components, including CD79B and KMT2D. The mutation frequency of PIM1 was the highest with 58 mutations, followed by MyD88, CD79B, and KMT2D. The diverse mutation profiles indicated that PCNSL has heterogeneous tumor characteristics, and its heterogeneity may be related to tumor invasion, metastasis, disease resistance, and relapse. There were some differences in the mutation map compared with other studies [11], [29], [30], [31], [32], [33], which may be related to the analysis methods, sample types, heterogeneity of the tumor tissues, and/or differences in the races of patients. Of these, regional differences might mostly explain the variance because our patients and samples were from Chinese patients compared with other studies that involved patients from North America or Europe. Another explanation might be that most of our cases were of the ABC subtype of PCNSL, which is characterized by activation of the BCR/NF-κB signaling pathway. This explains why we observed a high mutation frequency for MYD88, PIM1, CD79A/B, and CARD11.
Lymphoma is a group of diseases with significant heterogeneity and different clinical manifestations, morphological features, immunophenotypes, molecular subpopulations, and clinical prognosis. Previous studies showed that the mutational characterization of many somatic mutation targeted genes was highly variable and that they had a key role in B cell–related signaling pathways (BCR, NF-κB, NOTCH, and Toll-like receptor signaling pathways), immunity, and cell cycle– or apoptosis-related signaling pathways. Most of the genetic alterations could be grouped into 20 related signaling pathways [34], [35], [36]. In our study, we found that mutations of the NF-κB signaling pathway predominated (50% of total variance), followed by B-cell receptor (12.62%), epigenetically related (9.60%), and apoptotic signaling pathways (9.60%). Bruno [29] and Fukumura [37] found a similar mutation distribution in these pathways. However, the mutation frequencies of different genes and related pathways are distinctive. These may be related to the different methods of research and sequencing, as well as ethnic, geographical, and regional differences.
The relationship between mutated genes and a patient's clinicopathological features indicated that some genes were significantly correlated with patient gender, age, and risk score. Furthermore, the CD79B and GNA13 mutations were associated with an unfavorable prognosis, and GNA13 was reported to have mutations in follicular and Burkitt lymphoma and GCB DLBCLs (<25%). Some of the mutation sites negatively influenced gene expression and function, implying that GNA13 may act as a tumor suppressor gene [38], [39]. Our results were similar to a previous study; however, our prognostic value results were limited by the relatively small sample size and retrospective nature of our study.
The lymphoma-related gene panel included gene mutations that might have an important role in treatment options. These mutations, currently targeted by precision therapies, may serve as alternative targeted therapies (Figure 7, Supplementary Table 5). In lymphoma, especially DLBCL, commonly used small molecular inhibitors include Bruton's tyrosine kinase inhibitor (ibrutinib), PI3K inhibitor (copanlisib, buparlisib), histone deacetylase inhibitor (vorinostat, belinostat), PIM kinase inhibitor (SGI-1776), protein kinase C inhibitor (sotrastaurin), and mTOR inhibitor (temsirolimus and everolimus) [40], [41], [42], [43].
Figure 7.
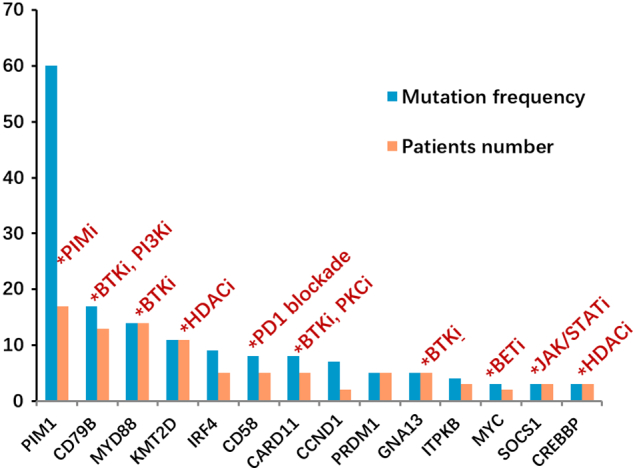
Gene mutation with potential targeted therapy. The top 14 mutation genes in PCNSL cohort and the potential targets highlighted by indicating the appropriate class of molecules, which could potentially be used as therapy.
Bruton's tyrosine kinase (BTK) is a kinase linking BCR signaling to NF-κB activity and inhibits chronic activation of the BCR pathway in DLBCL [44]. Ibrutinib is a selective, covalent, and irreversible BTK inhibitor with high activity in other lymphoid malignancies such as mantle cell lymphoma or chronic lymphocytic leukemia [45], [46], [47]. Recently, a phase I/II clinical trial of 80 patients with R/R DLBCL [48] showed that the response rate of ibrutinib in ABC type was 37%, while MYD88/CD79B double mutation patients responded significantly better to ibrutinib. A MYD88 or CD79B mutation alone did not affect the sensitivity of patients to ibrutinib, which highlights the importance of the clinical utility of targeted NGS to patients. Because PCNSL has a special structure and its treatment largely depends on drugs crossing the blood-brain barrier as well as whole brain radiation, its outcome is significantly inferior to that of systemic DLBCL [49]. However, it was recently shown that it closely resembles the ABC subtype of DLBCL, characterized by specific somatic mutations [11], [29], [31], [37]. In our cohort, CD79B and MYD88 mutations were found in the majority of cases, with a higher ratio compared with systemic ABC DLBCL. For these reasons, strategies that target BCR and MYD88 signaling might be promising for the treatment of PCNSL. In addition to the independent mutations of MYD88 and CD79B, the MYD88/CD79B double mutation occurred in 8 of 22 patients, who might be more receptive to ibrutinib treatment, although this requires confirmation in future studies.
MYD88 is a common somatic mutation target in lymphomas, and L265P is a hot mutation locus. A previous study reported that 29% of cases had mutations in MYD88 (L265P) in ABC-type DLBCL [50], whereas they were rare in GCB type and PMBL. MYD88 is a crucial linking protein in the BCR/TLR signaling pathway, and current phase I/II clinical trials for patients with relapsed or refractory Waldenstrom's macroglobulinemia have been initiated (NCT02092909). Other promising strategies for the targeted therapy of MYD88-mutated DLBCLs include targeting the TAK1 and IRAK4 mediator proteins of MYD88 signaling and homodimerization [51], [52]. However, growing evidence has indicated that cross talk between the BCR and TLR signaling components may contribute to lymphomagenesis, indicating a dual blocking strategy for the treatment [53]. In our study, 14 patients (63.64%) had a MYD88 mutation, 59.09% had a MYD88 (L265P) mutation, and 36.36% had a double MYD88/CD78B mutation, indicating that all of these genetic alterations may play an important role in the pathogenesis of PCNSL and might provide a therapeutic target.
Recently, small molecule inhibitors of PIM kinase, serine/threonine kinase inhibitors competitively combined with ATP [54], have been studied and developed for DLBCL. The oncogene PIM1 is a member of the PIM family (including PIM1, PIM2, and PIM3) and is mainly involved in regulation of the cell cycle, transcription and translation of proteins, promotion of apoptosis, regulation of cellular metabolism, and mediating drug resistance [54], [55], [56]. Therefore, it is closely related to the development of tumors. Small molecule PIM1 inhibitors have been investigated in phase I/II clinical trials for prostate cancer and relapsed/refractory NHLs (NCT00848601). In our study, PIM1 had the highest frequency of mutation. Different from MYD88 gene mutations, there was a large heterogeneity in the mutations, with no clear mutation hot spots. In our study, 51 different loci mutations were found, and these were dominated by missense mutations. Interestingly, studies showed that the sensitivity of PIM kinase inhibitors in DLBCL was not related to the expression of PIM kinases [57], suggesting that the presence of modified PIM1 mutations may not necessarily affect PIM1 inhibitor treatment. In addition, in ABC-DLBCL in vitro experiments, PIM1 mutations were associated with endogenous ibrutinib (BTK inhibitor) resistance. Mutations of PIM1 showed stable protein expression and enhanced NF-κB signaling transduction. In addition, a combination of pan-PIM inhibitor and ibrutinib was more effective than ibrutinib monotherapy [58]. Therefore, sequencing analysis of PIM1 mutation status and loci might help screen eligible candidates suitable for PIM inhibitors and avoid drug resistance to improve patient treatment efficacy.
The PI3K signaling pathway is involved in cellular processes such as proliferation and survival. Chronic activation of the BCR signaling pathway in DLBCL activates PI3K, which indicates the existence of cross talk between NF-κB and PI3K pathways [44], [59]. Studies showed that CD79B mutant ABC cell lines were more sensitive to PI3K inhibitors than CD79B wild-type models, demonstrating that PI3K inhibition may reduce NF-κB activity in DLBCL cell lines [59]. Similarly, in GCB cell lines, the PTEN gene regulated the response to PI3K inhibitors, caused by mutation, deletion, or unknown molecular mechanisms [60]. Recently, the oral pan-PI3K inhibitor buparlisib was tested in a phase 2 clinical trial. The preliminary results showed that the overall response rate in DLBCL was 12% [61]. Another study showed that buparlisib had excellent brain penetration regardless of the influence of the blood-brain barrier, complete oral bioavailability, and efficient intracranial target inhibition at clinically achievable plasma concentrations [62]. These studies provide information that will aid the development of targeted therapeutic strategies for PCNSL patients.
In addition, protein kinase C (PKC) is a downstream target of multiple signaling pathways, including BCR and NF-κB. In B cells, PKCβ is thought to be the predominant PKC isoform that mediates BCR/NF-κB activation, maybe in part through the phosphorylation of CARD11 [63], [64]. Studies showed that mutations of CARD11 and TNFAIP3 resulted in the decreased activity of ibrutinib and sotrastaurin (PKC inhibitor) and that the CD79A/B mutation was associated with the sensitivity of sotrastaurin [65], [66]. Another study tested sotrastaurin in combination with the mTOR inhibitor everolimus in patients with CD79-mutant or ABC subtype of DLBCL (NCT01854606). In this study, mutated CD79B (59.09%), CARD11 (22.73%), CD79A (9.09%), and TNFAIP3 (4.55%) might have influenced the treatment effectiveness of PCNSL patients.
Targeting epigenetic changes is also being increasingly explored in the realm of precision therapy, notably using the inhibitors of DNA methyltransferase and histone deacetylase (HDAC) [67]. This study included KMT2D, CREBBP, EP300, MEF2B, and EZH2, which are within our lymphoma panel. Previous studies have shown that frequent inactivating mutations of CREBPP and EP300 in DLBCL provide a basis for targeted therapy via HDAC inhibitors with the aim of reestablishing acetylation levels [68]. More specific epigenetic inhibitors are also being investigated: for example, HDAC inhibitors or a combination of BCL2 and histone methyltransferase inhibitors was considered a possible therapeutic option in the absence of both KMT2D and BCL2 deregulation [69]. In our cohort, CREBBP and EP300 mutations were identified in 13.64% and 9.09% of PCNSL patients. KMT2D, accounting for 50.0% of our PCNSL patients, was one of the most frequently mutated genes in our cohort and had a higher frequency compared with de novo DLBCLs (approximately 30%) [38], [70], [71]. This indicates that KMT2D has an irreplaceable role in the tumorigenesis and development of PCNSL. Targeted epigenetic alterations have been developed through genome-wide sequencing, exons sequencing, epigenomes, and transcriptomes and may become a new therapeutic option.
PIM1 is highly expressed in several types of carcinomas and is related to prognosis; however, little is known about its role in PCNSL. Therefore, we focused on PIM1 expression and its prognostic value in PCNSL. Our results showed that PIM1 was highly expressed in 71.93% (41/57) of PCNSL patients, which was higher than in lymphadenitis (30%). The expression of PIM1 was correlated with the risk score of patients (P = .023), suggesting that it might be involved in the occurrence and development of PCNSL. However, there was no significant correlation between PIM1 and PCNSL patient age, gender, or LDH level. Furthermore, patients with high PIM1 expression had a poorer OS compared with patients with low PIM1 expression. Our findings are similar to those of Shuai [72], Guo [73], and His [74] reporting different kinds of carcinomas, suggesting that PIM1 may act as a prognostic factor for PCNSL patients.
Myeloid differentiation factor 88 (MYD88), which encodes a soluble adapter protein, is activated in the early genetic response of bone marrow cells to differentiation and growth inhibitory stimuli and also functions as a transcription factor to transduce most Toll-like receptors and cohesion proteins, including IL-1 and IL-18 [75], [76], [77]. In human ABC DLBCL cell lines, MYD88 forms complex receptor-associated kinase 4 with IL-1 receptor-associated kinase 1 and IL-1 to promote the activation of NF-κB and Janus kinase signal transducers and activators of transcription 3 (JAK-STAT3) signaling, resulting in the survival of lymphoma cells [76]. However, few studies have investigated the relationship between MYD88 and PCNSL and the clinical significance. Our results showed that MYD88 was highly expressed in 70.18% (40/57) of PCNSL patients, which was higher than in proliferative lymph node patients (15%). The expression of PIM1 was correlated with the risk score and LDH level of patients, which suggested that MYD88 may be involved in the occurrence and development of PCNSL. We also found that patients with high MYD88 expression had a poorer OS compared with those with low MYD88 expression. Moreover, multivariate analysis showed that high MYD88 expression was an independent prognostic factor for PCNSL patient overall survival. Choi et al. [78] reported that high MYD88 expression was related to poor DFS in DLBCL patients. Chen [79] discovered that high MYD88 expression in breast cancer patients was associated with a poor prognosis. Our results were in accord with these findings, suggesting that MYD88 may predict treatment efficacy and prognosis in patients with PCNSL.
NGS is increasingly accessible in current academic research but has not been fully utilized in conventional clinical settings. Our study demonstrated that by using a limited set of genes, we could track popular hot mutations and identify new and rare mutations to provide new potential treatment targets. Targeted NGS allows extensive depth and breadth of sequence studies, which play a fundamental role in the study of tumor subtype and heterogeneity. We used FFPE tumor samples, which are also the predominant form of acquired lymphoma specimens. Hopefully, the use of liquid biopsy in lymphoma diagnosis and early recurrence monitoring will become more common. Under the guidance of new technologies and concepts, we hope to promote and create new genomic research and targeted precision therapy for PCNSL patients.
Conclusion
Regarding the mutational status of PCNSL patients using NGS, we identified highly recurrent genetic lesions in components of the NF-κB pathway, CD79B and KMT2D. Mutations of CD79B and GNA13 may be related to the prognosis of PCNSL patients. Furthermore, PIM1 and MYD88 were highly expressed in the PCNSL cohort and were associated with poor survival. The high expression of MYD88 may be an independent prognostic factor affecting OS in PCNSL patients.
Conflict of Interest
The authors declare no conflicts of interest.
Acknowledgement
This study was supported by National Natural Science Foundation of China (No. 81570200) and Natural Science Foundation of Hunan Province (No. 14JJ2042). And we thank for the Burning Rock Biomedical Company (Guangzhou, China) for providing the next-generation platform.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2018.08.012.
Contributor Information
Jianhua Zhou, Email: zhoujh15@163.com.
Meizuo Zhong, Email: z402524@csu.edu.cn.
Appendix A. Supplementary data
Supplementary Table 1. Lymphoma-Related 64-Gene Panel
Supplementary Table 2. Identification of Gene Mutations Among 22 PCNSL Patients
Supplementary Table 3. Signaling Pathway Distribution Among Mutation Genes
Supplementary Table 4. Analysis of Gene Mutation Status with Prognostic Indicators
Supplementary Table 5. Genes Mutation with Potential Targeted Therapy
Supplementary Table 6. Raw Data for Next-Generation Sequencing in 22 PCNSL Patients
References
- 1.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol. 2012;14(Suppl. 5):v1–49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patrick LB, Mohile NA. Advances in primary central nervous system lymphoma. Curr Oncol Rep. 2015;17(12):60. doi: 10.1007/s11912-015-0483-8. [DOI] [PubMed] [Google Scholar]
- 3.Rubenstein JL, Gupta NK, Mannis GN, Lamarre AK, Treseler P. How I treat CNS lymphomas. Blood. 2013;122(14):2318–2330. doi: 10.1182/blood-2013-06-453084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuurmans M, Bromberg JE, Doorduijn J. Primary central nervous system lymphoma in the elderly: a multicentre retrospective analysis. Br J Haematol. 2010;151(2):179–184. doi: 10.1111/j.1365-2141.2010.08328.x. [DOI] [PubMed] [Google Scholar]
- 5.Carnevale J, Rubenstein JL. The challenge of primary central nervous system lymphoma. Hematol Oncol Clin North Am. 2016;30(6):1293–1316. doi: 10.1016/j.hoc.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitz N. Treatment of primary CNS lymphoma. Blood. 2015;125(9):1360–1361. doi: 10.1182/blood-2015-01-622498. [DOI] [PubMed] [Google Scholar]
- 7.Sethi TK, Reddy NM. Treatment of newly diagnosed primary central nervous system lymphoma: current and emerging therapies. Leuk Lymphoma. 2018:1–13. doi: 10.1080/10428194.2018.1466296. [DOI] [PubMed] [Google Scholar]
- 8.Norden AD, Drappatz J, Wen PY, Claus EB. Survival among patients with primary central nervous system lymphoma, 1973-2004. J Neurooncol. 2011;101(3):487–493. doi: 10.1007/s11060-010-0269-7. [DOI] [PubMed] [Google Scholar]
- 9.Perkins A, Liu G. Primary brain tumors in adults: diagnosis and treatment. Am Fam Physician. 2016;93(3):211–217. [PubMed] [Google Scholar]
- 10.Montesinos-Rongen M, Schmitz R, Brunn A. Mutations of CARD11 but not TNFAIP3 may activate the NF-kappaB pathway in primary CNS lymphoma. Acta Neuropathol. 2010;120(4):529–535. doi: 10.1007/s00401-010-0709-7. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura T, Tateishi K, Niwa T. Recurrent mutations of CD79B and MYD88 are the hallmark of primary central nervous system lymphomas. Neuropathol Appl Neurobiol. 2016;42(3):279–290. doi: 10.1111/nan.12259. [DOI] [PubMed] [Google Scholar]
- 12.Compagno M, Lim WK, Grunn A. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung CO, Kim SC, Karnan S. Genomic profiling combined with gene expression profiling in primary central nervous system lymphoma. Blood. 2011;117(4):1291–1300. doi: 10.1182/blood-2010-07-297861. [DOI] [PubMed] [Google Scholar]
- 14.Braggio E, McPhail ER, Macon W. Primary central nervous system lymphomas: a validation study of array-based comparative genomic hybridization in formalin-fixed paraffin-embedded tumor specimens. Clin Cancer Res. 2011;17(13):4245–4253. doi: 10.1158/1078-0432.CCR-11-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turco MC, Romano MF, Petrella A, Bisogni R, Tassone P, Venuta S. NF-kappaB/Rel-mediated regulation of apoptosis in hematologic malignancies and normal hematopoietic progenitors. Leukemia. 2004;18(1):11–17. doi: 10.1038/sj.leu.2403171. [DOI] [PubMed] [Google Scholar]
- 16.Turturro F. Constitutive NF-κB activation underlines major mechanism of drug resistance in relapsed refractory diffuse large B cell lymphoma. Biomed Res Int. 2015;2015:484537. doi: 10.1155/2015/484537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q, Fu W, Jiang H. Clinicopathological implications of nuclear factor κB signal pathway activation in diffuse large B-cell lymphoma. Hum Pathol. 2015;46(4):524–531. doi: 10.1016/j.humpath.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 18.Yu L, Li L, Medeiros LJ, Young KH. NF-κB signaling pathway and its potential as a target for therapy in lymphoid neoplasms. Blood Rev. 2017;31(2):77–92. doi: 10.1016/j.blre.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X, Meng B, Iqbal J. Activation of the STAT3 signaling pathway is associated with poor survival in diffuse large B-cell lymphoma treated with R-CHOP. J Clin Oncol. 2013;31(36):4520–4528. doi: 10.1200/JCO.2012.45.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson P, Klein-Hitpass L, Grabellus F. Recurrent mutations in NF-κB pathway components, KMT2D, and NOTCH1/2 in ocular adnexal MALT-type marginal zone lymphomas. Oncotarget. 2016;7(38):62627–62639. doi: 10.18632/oncotarget.11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y, Zhong M, Liu Y, Wang L, Tang Y. Expression of TIM-3 and LAG-3 in extranodal NK/T cell lymphoma, nasal type. Histol Histopathol. 2018;33(3):307–315. doi: 10.14670/HH-11-931. [DOI] [PubMed] [Google Scholar]
- 22.Fan W, Fan SS, Feng J, Xiao D, Fan S, Luo J. Elevated expression of HSP10 protein inhibits apoptosis and associates with poor prognosis of astrocytoma. PLoS One. 2017;12(10) doi: 10.1371/journal.pone.0185563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA, Banks E, Poplin R. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreri AJ, Blay JY, Reni M. Prognostic scoring system for primary CNS lymphomas: the International Extranodal Lymphoma Study Group experience. J Clin Oncol. 2003;21(2):266–272. doi: 10.1200/JCO.2003.09.139. [DOI] [PubMed] [Google Scholar]
- 27.Visco C, Li Y, Xu-Monette ZY. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26(9):2103–2113. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright GW, Wilson WH, Staudt LM. Genetics of diffuse large B-cell lymphoma. N Engl J Med. 2018;379(5):493–494. doi: 10.1056/NEJMc1806191. [DOI] [PubMed] [Google Scholar]
- 29.Bruno A, Boisselier B, Labreche K. Mutational analysis of primary central nervous system lymphoma. Oncotarget. 2014;5(13):5065–5075. doi: 10.18632/oncotarget.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vater I, Montesinos-Rongen M, Schlesner M. The mutational pattern of primary lymphoma of the central nervous system determined by whole-exome sequencing. Leukemia. 2015;29(3):677–685. doi: 10.1038/leu.2014.264. [DOI] [PubMed] [Google Scholar]
- 31.Braggio E, Van Wier S, Ojha J. Genome-wide analysis uncovers novel recurrent alterations in primary central nervous system lymphomas. Clin Cancer Res. 2015;21(17):3986–3994. doi: 10.1158/1078-0432.CCR-14-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okosun J, Bödör C, Wang J. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Grubor V, Love CL. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith DR, Quinlan AR, Peckham HE. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 2008;18(10):1638–1642. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubois S, Viailly PJ, Mareschal S. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: a LYSA study. Clin Cancer Res. 2016;22(12):2919–2928. doi: 10.1158/1078-0432.CCR-15-2305. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence MS, Stojanov P, Mermel CH. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukumura K, Kawazu M, Kojima S. Genomic characterization of primary central nervous system lymphoma. Acta Neuropathol. 2016;131(6):865–875. doi: 10.1007/s00401-016-1536-2. [DOI] [PubMed] [Google Scholar]
- 38.Morin RD, Mendez-Lago M, Mungall AJ. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morin RD, Mungall K, Pleasance E. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhodes J, Landsburg DJ. Small-molecule inhibitors for the treatment of diffuse large B cell lymphoma. Curr Hematol Malig Rep. 2018 doi: 10.1007/s11899-018-0467-5. [DOI] [PubMed] [Google Scholar]
- 41.Dunleavy K, Erdmann T, Lenz G. Targeting the B-cell receptor pathway in diffuse large B-cell lymphoma. Cancer Treat Rev. 2018;65:41–46. doi: 10.1016/j.ctrv.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Bohers E, Mareschal S, Bertrand P. Activating somatic mutations in diffuse large B-cell lymphomas: lessons from next generation sequencing and key elements in the precision medicine era. Leuk Lymphoma. 2015;56(5):1213–1222. doi: 10.3109/10428194.2014.941836. [DOI] [PubMed] [Google Scholar]
- 43.Dubois S, Jardin F. The role of next-generation sequencing in understanding the genomic basis of diffuse large B cell lymphoma and advancing targeted therapies. Expert Rev Hematol. 2016;9(3):255–269. doi: 10.1586/17474086.2016.1130616. [DOI] [PubMed] [Google Scholar]
- 44.Davis RE, Ngo VN, Lenz G. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byrd JC, Furman RR, Coutre SE. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang ML, Rule S, Martin P. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369(6):507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burger JA, Tedeschi A, Barr PM. Ibrutinib as initial therapy for patients with chronic lymphocytic leukemia. N Engl J Med. 2015;373(25):2425–2437. doi: 10.1056/NEJMoa1509388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson WH, Young RM, Schmitz R. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Batchelor TT. Primary central nervous system lymphoma. Hematology Am Soc Hematol Educ Program. 2016;2016(1):379–385. doi: 10.1182/asheducation-2016.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ngo VN, Young RM, Schmitz R. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loiarro M, Volpe E, Ruggiero V. Mutational analysis identifies residues crucial for homodimerization of myeloid differentiation factor 88 (MyD88) and for its function in immune cells. J Biol Chem. 2013;288(42):30210–30222. doi: 10.1074/jbc.M113.490946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ansell SM, Hodge LS, Secreto FJ. Activation of TAK1 by MYD88 L265P drives malignant B-cell growth in non-Hodgkin lymphoma. Blood Cancer J. 2014;4 doi: 10.1038/bcj.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dufner A, Schamel WW. B cell antigen receptor-induced activation of an IRAK4-dependent signaling pathway revealed by a MALT1-IRAK4 double knockout mouse model. Cell Commun Signal. 2011;9(1):6. doi: 10.1186/1478-811X-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merkel AL, Meggers E, Ocker M. PIM1 kinase as a target for cancer therapy. Expert Opin Investig Drugs. 2012;21(4):425–436. doi: 10.1517/13543784.2012.668527. [DOI] [PubMed] [Google Scholar]
- 55.An N, Cen B, Cai H, Song JH, Kraft A, Kang Y. Pim1 kinase regulates c-Kit gene translation. Exp Hematol Oncol. 2016;5:31. doi: 10.1186/s40164-016-0060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiménez-García MP, Lucena-Cacace A, Robles-Frías MJ, Narlik-Grassow M, Blanco-Aparicio C, Carnero A. The role of PIM1/PIM2 kinases in tumors of the male reproductive system. Sci Rep. 2016;6:38079. doi: 10.1038/srep38079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brault L, Menter T, Obermann EC. PIM kinases are progression markers and emerging therapeutic targets in diffuse large B-cell lymphoma. Br J Cancer. 2012;107(3):491–500. doi: 10.1038/bjc.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuo HP, Ezell SA, Hsieh S. The role of PIM1 in the ibrutinib-resistant ABC subtype of diffuse large B-cell lymphoma. Am J Cancer Res. 2016;6(11):2489–2501. [PMC free article] [PubMed] [Google Scholar]
- 59.Kloo B, Nagel D, Pfeifer M. Critical role of PI3K signaling for NF-kappaB–dependent survival in a subset of activated B-cell–like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A. 2011;108(1):272–277. doi: 10.1073/pnas.1008969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfeifer M, Grau M, Lenze D. PTEN loss defines a PI3K/AKT pathway-dependent germinal center subtype of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(30):12420–12425. doi: 10.1073/pnas.1305656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bedard PL, Tabernero J, Janku F. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin Cancer Res. 2015;21(4):730–738. doi: 10.1158/1078-0432.CCR-14-1814. [DOI] [PubMed] [Google Scholar]
- 62.de Gooijer MC, Zhang P, LCM B. Buparlisib is a brain penetrable pan-PI3K inhibitor. Sci Rep. 2018;8(1):10784. doi: 10.1038/s41598-018-29062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsumoto R, Wang D, Blonska M. Phosphorylation of CARMA1 plays a critical role in T cell receptor–mediated NF-kappaB activation. Immunity. 2005;23(6):575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 64.Sommer K, Guo B, Pomerantz JL. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 2005;23(6):561–574. doi: 10.1016/j.immuni.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 65.Naylor TL, Tang H, Ratsch BA. Protein kinase C inhibitor sotrastaurin selectively inhibits the growth of CD79 mutant diffuse large B-cell lymphomas. Cancer Res. 2011;71(7):2643–2653. doi: 10.1158/0008-5472.CAN-10-2525. [DOI] [PubMed] [Google Scholar]
- 66.Zheng X, Ding N, Song Y, Feng L, Zhu J. Different sensitivity of germinal center B cell-like diffuse large B cell lymphoma cells towards ibrutinib treatment. Cancer Cell Int. 2014;14(1):32. doi: 10.1186/1475-2867-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clozel T, Yang S, Elstrom RL. Mechanism-based epigenetic chemosensitization therapy of diffuse large B-cell lymphoma. Cancer Discov. 2013;3(9):1002–1019. doi: 10.1158/2159-8290.CD-13-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andersen CL, Asmar F, Klausen T, Hasselbalch H, Grønbæk K. Somatic mutations of the CREBBP and EP300 genes affect response to histone deacetylase inhibition in malignant DLBCL clones. Leuk Res Rep. 2012;2(1):1–3. doi: 10.1016/j.lrr.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ortega-Molina A, Boss IW, Canela A. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pasqualucci L, Dominguez-Sola D, Chiarenza A. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasqualucci L, Trifonov V, Fabbri G. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mou S, Wang G, Ding D. Expression and function of PIM kinases in osteosarcoma. Int J Oncol. 2016;49(5):2116–2126. doi: 10.3892/ijo.2016.3708. [DOI] [PubMed] [Google Scholar]
- 73.Guo S, Mao X, Chen J. Overexpression of Pim-1 in bladder cancer. J Exp Clin Cancer Res. 2010;29:161. doi: 10.1186/1756-9966-29-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsi ED, Jung SH, Lai R. Ki67 and PIM1 expression predict outcome in mantle cell lymphoma treated with high dose therapy, stem cell transplantation and rituximab: a Cancer and Leukemia Group B 59909 correlative science study. Leuk Lymphoma. 2008;49(11):2081–2090. doi: 10.1080/10428190802419640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rossi D. Role of MYD88 in lymphoplasmacytic lymphoma diagnosis and pathogenesis. Hematology Am Soc Hematol Educ Program. 2014;2014(1):113–118. doi: 10.1182/asheducation-2014.1.113. [DOI] [PubMed] [Google Scholar]
- 76.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 77.Miyazaki K. Treatment of diffuse large B-cell lymphoma. J Clin Exp Hematop. 2016;56(2):79–88. doi: 10.3960/jslrt.56.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choi JW, Kim Y, Lee JH, Kim YS. MYD88 expression and L265P mutation in diffuse large B-cell lymphoma. Hum Pathol. 2013;44(7):1375–1381. doi: 10.1016/j.humpath.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 79.Chen X, Zhao F, Zhang H, Zhu Y, Wu K, Tan G. Significance of TLR4/MyD88 expression in breast cancer. Int J Clin Exp Pathol. 2015;8(6):7034–7039. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Lymphoma-Related 64-Gene Panel
Supplementary Table 2. Identification of Gene Mutations Among 22 PCNSL Patients
Supplementary Table 3. Signaling Pathway Distribution Among Mutation Genes
Supplementary Table 4. Analysis of Gene Mutation Status with Prognostic Indicators
Supplementary Table 5. Genes Mutation with Potential Targeted Therapy
Supplementary Table 6. Raw Data for Next-Generation Sequencing in 22 PCNSL Patients