Abstract
Integration-defective lentiviral vectors (IDLVs) have become an important alternative tool for gene therapy applications and basic research. Unfortunately, IDLVs show lower transgene expression as compared to their integrating counterparts. In this study, we aimed to improve the expression levels of IDLVs by inserting the IS2 element, which harbors SARs and HS4 sequences, into their LTRs (SE-IS2-IDLVs). Contrary to our expectations, the presence of the IS2 element did not abrogate epigenetic silencing by histone deacetylases. In addition, the IS2 element reduced episome levels in IDLV-transduced cells. Interestingly, despite these negative effects, SE-IS2-IDLVs outperformed SE-IDLVs in terms of percentage and expression levels of the transgene in several cell lines, including neurons, neuronal progenitor cells, and induced pluripotent stem cells. We estimated that the IS2 element enhances the transcriptional activity of IDLV LTR circles 6- to 7-fold. The final effect the IS2 element in IDLVs will greatly depend on the target cell and the balance between the negative versus the positive effects of the IS2 element in each cell type. The better performance of SE-IS2-IDLVs was not due to improved stability or differences in the proportions of 1-LTR versus 2-LTR circles but probably to a re-positioning of IS2-episomes into transcriptionally active regions.
Keywords: IDLV, HS4 insulator, gene therapy, scaffold or matrix attachment regions, lentiviral vector
Introduction
Lentiviral vectors (LVs) have proven to be highly successful in several gene therapy protocols over the last 20 years.1 Their success is partly explained by their ability to transduce dividing and non-dividing cells, including hematopoietic stem cells (HSCs),2, 3, 4 neurons,5, 6, 7 and T cells.8, 9, 10 However, concerns have been raised regarding the potential risk of inherent insertional mutagenesis caused by integrative LVs.11 The use of integration-defective LVs (IDLVs) is a logical option for minimizing insertional mutagenesis risk when target cells are quiescent. IDLVs are also an interesting alternative for transient expression in dividing cells.12, 13, 14, 15, 16, 17, 18, 19, 20 As with their integrative counterpart, the tropism of IDLV particles can be altered and adapted to target cells pseudotyped with different envelope proteins.21, 22
Current research in the field of retroviral vectors shows that LVs are often more effective than their IDLV counterparts in terms of gene expression23, 24 mainly due to the tendency of IDLVs to undergo epigenetic silencing as a result of nuclear chromatinization.25, 26 In 2013, Pelascini et al.27 showed that histone deacetylase (HDAC) activity is the principal cellular determinant underlying weak IDLV transcriptional activity. Different strategies have been used to enhance the transgene expression of episomal molecules in both viral- and non-viral-based systems. In non-viral episomal gene delivery systems, genomic elements based on scaffold or matrix attachment regions (SARs or S/MARs) are widely used to enhance transcription levels and to maintain long-term expression rates.28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 This is mainly due to the capacity of SARs elements to bind transcription factors such as special AT-rich binding protein 1 (SATB1), nuclear matrix protein 4 (Nmp4) and CCCTC-binding factor (CTCF), in addition to their capacity to promote nucleoprotein structural aggregation, histone acetyltransferase recruitment, and ATP-dependent chromatin remodeling complexes.42 As with the SAR elements, the inclusion of other cis-acting elements based on the 5′ cHS4 chicken hypersensitive site 4 (HS4) chicken β-globin insulator (cHS4) also increases non-viral episomal efficiency33 due, in part, to the interaction of the cHS4 element with the matrix via CTCF nuclear proteins.43 This nuclear factor avoids heterochromatin spreading in episomal DNA when bound to the cHS4 sequence.44, 45, 46, 47
Several research groups have achieved varying levels of success in their attempt to improve IDLV expression by inserting different fragments of the β-interferon SARs element into IDLVs.48, 49, 50 However, the use of SARs elements to improve IDLV transcription efficiency has only been studied in relation to immunoglobulin κ (Igκ) SARs sequences. Grandchamp et al.51 concluded that these elements had, in general, no effect on transduction efficiency and observed an improvement only in differentiated primary neural progenitor cells.
We have previously reported that the inclusion of the IS2 element (which combines a synthetic SAR [SAR2] and a 650-pb fragment of the chicken β-globin HS4 insulator) in LVs reduced biological viral titers but improved transgene expression and prevents epigenetic silencing in human embryonic stem cells (hESCs) and hematopoietic pluripotent cells (HSCs). However, these effects were cell type dependent, since no improvement in transgene expression could be observed on K562 cells and other immortalized cell lines.52 In the present study, we tested whether the IS2 element could improve IDLV gene expression in different cell types. Although the presence of the IS2 element did not abrogate epigenetic silencing, it did improve IDLV efficiency in several cell lines. Surprisingly, in spite of the improved expression levels, the inclusion of the IS2 element into IDLVs (SE-IS2-IDLVs) reduced 3–5 times the amount of episomal vector in transduced cells relative to those transduced with unmodified SE-IDLVs. We have estimated that the IS2 element enhances the transcriptional activity of SE-IS2-episomes 6- to 7-fold. The final effect of the IS2 element in IDLVs will greatly depend on the target cell and the balance between the negative versus the positive effects of the IS2 element in each cell type. We have also shown that the IS2 element does not improve the stability of IDLV episomes and that, although there is a change in the ratio of 1-LTR/2-LTR circles, this is not the mechanism involved in the increased transcriptional activity of the SE-IS2-IDLV episomes. Finally, a fluorescence in situ hybridization (FISH) analysis suggested that the improved behavior SE-IS2-IDLV episomes is probably due to a distinct nuclear re-positioning into transcriptionally active regions, as suggested by the aggregation of SE-IS2-IDLV episomes into DAPI-low regions.
Results
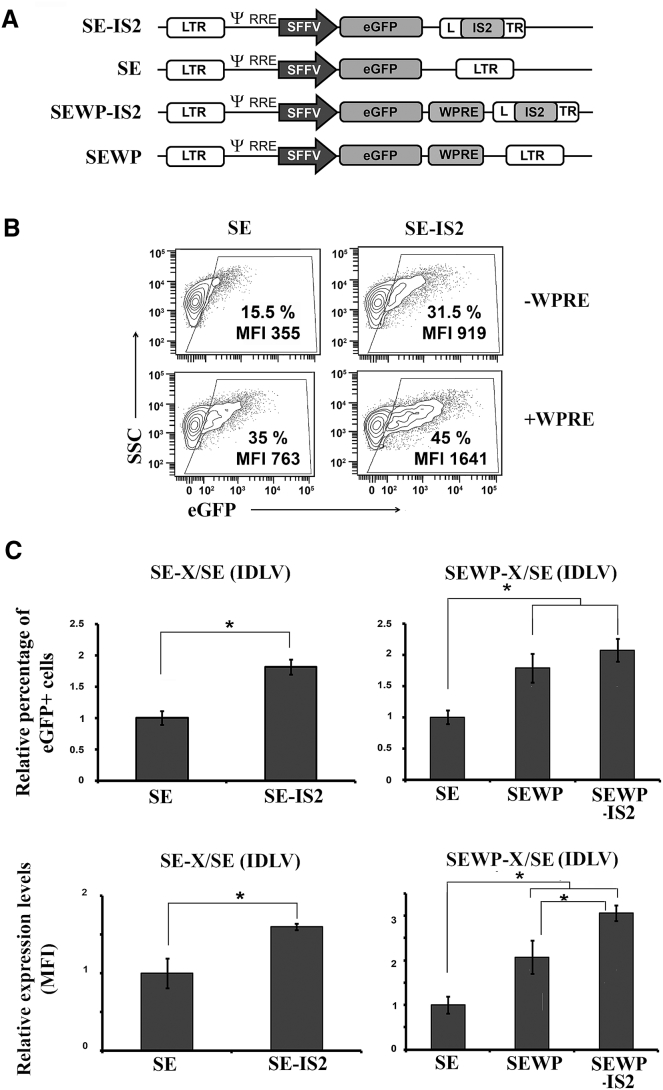
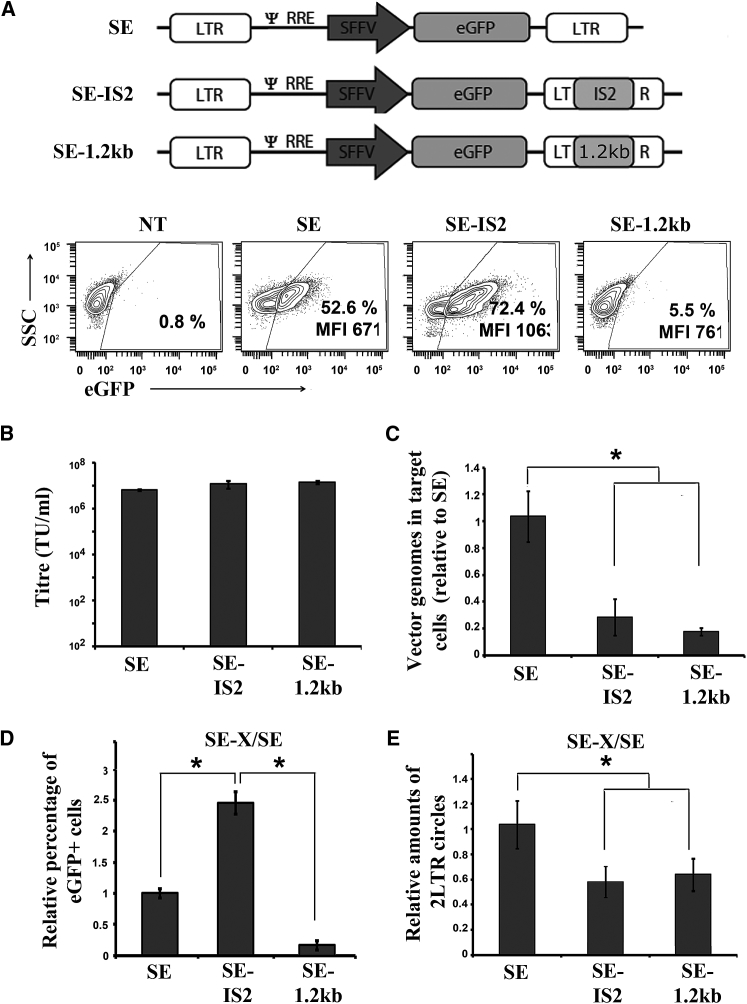
The Inclusion of the IS2 Insulator in the Long Terminal Repeat of IDLVs Improves Their Expression Levels in 293T Cells in an HDAC-Independent Manner
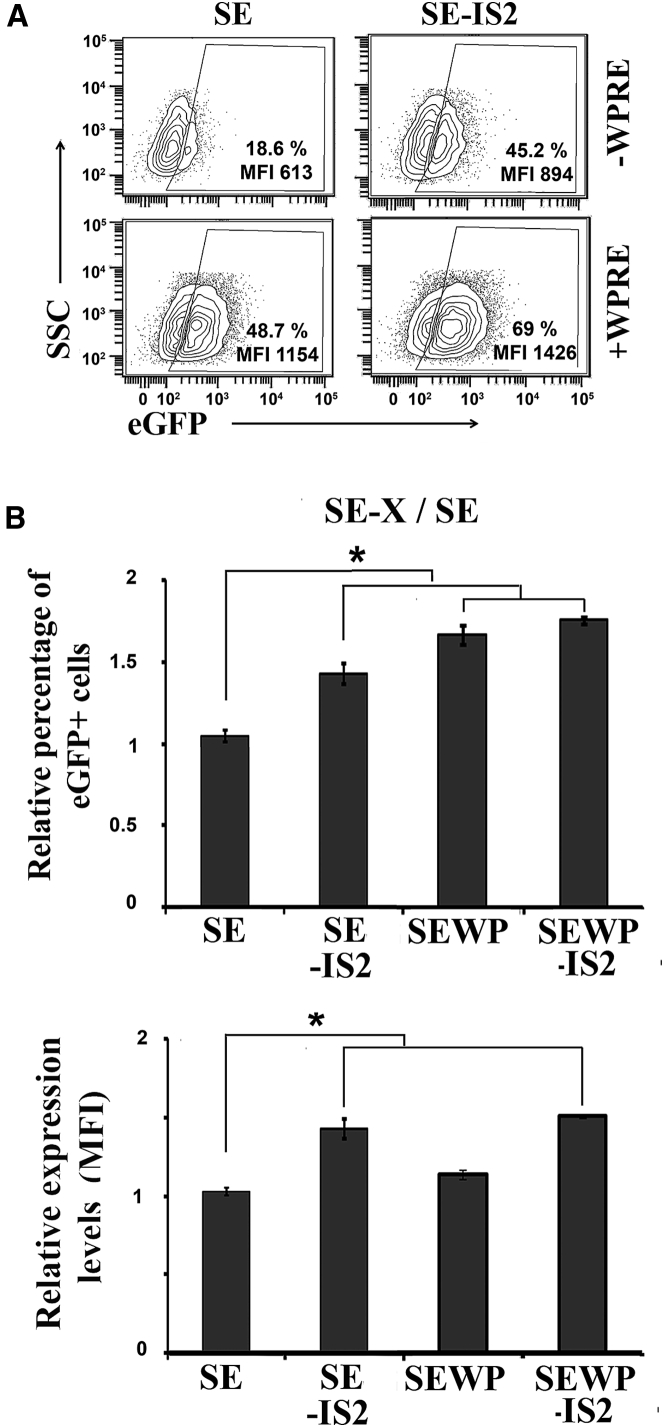
We generated IDLV particles from an SE lentiviral backbone containing or not the IS2 element52, 53, 54, 55 with and without WPRE (woodchuck hepatitis virus posttranscriptional regulatory element) (Figure 1A). We first analyzed the efficiency of different IDLVs in 293T cells. These cells were transduced with an equal MOI, estimated based on the Applied Biological Materials (ABM) Lentiviral qPCR Titer Kit (see Materials and Methods), and 3 days later, we analyzed the percentage of eGFP+ cells and the transgene expression levels (measured as mean fluorescence intensity [MFI] of the eGFP+ population). We found that the incorporation of the IS2 element into the IDLVs significantly increased the expression levels of eGFP in the absence and presence of the WPRE element (Figure 1B, MFI; Figure 1C, bottom graphs). We also found an increase in the percentage of GFP+ cells, which reached significance only in the absence of the WPRE element (Figure 1B, %; Figure 1C, upper graphs). We further corroborated that the effect of the IS2 element on IDLVs was maintained at higher MOIs (Figure S1).
Figure 1.
Inclusion of IS2 Element into IDLVs Enhances eGFP Expression Levels in 293T Cells
(A) Schematic representation of SE-IS2, SE, SEWP-IS2, and SEWP. eGFP, enhanced green florescence protein; SFFV, spleen focus forming virus promoter; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element. (B) Representative plots showing eGFP expression profiles of 293T cells transduced with the different IDLVs. An MOI of 0.3 was used to maintain the percentage of eGFP+ cells below 50%. The eGFP+ population gates were set to 0.2%–0.7% of eGFP+ cells in the untransduced population and subtracted from the % obtained under the different vectors and conditions for the analysis. The percentages (%) and expression levels (MFI) of the eGFP+ population are shown in each plot. (C) Graphs showing relative % of GFP+ cells (top graphs) and relative expression levels (MFI, bottom graphs) in 293T cells of SE-IS2-IDLVs and SE-IDLVs in the absence (left graphs) or presence (right graphs) of the WPRE element. Values represent means ± SEM of at least four separate experiments (*p < 0.05).
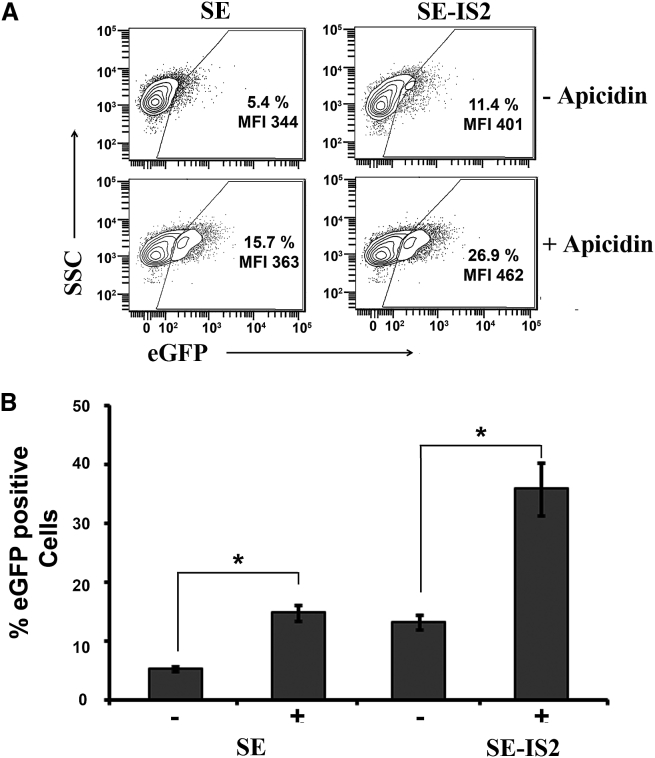
Prevention of histone deacetylation, the main factor underlying weak IDLV transcriptional activity, could explain the higher SE-IS2-IDLVs expression levels.27 In order to study this possibility, we analyzed SE-IDLV and SE-IS2-IDLV GFP expression levels in the presence and absence of apicidin, an HDAC inhibitor (HDACi). As can be observed in Figure 2, the addition of apicidin enhanced the eGFP expression to a similar degree in cells transduced with SE-IDLVs and in those transduced with SE-IS2-IDLVs (2.90-fold and 2.35-fold, respectively). These findings suggest that IS2-mediated enhancement is caused by an HDAC-independent mechanism.
Figure 2.
Apicidin Enhances Gene Expression of IDLV Transduced Cells Independently of the Presence of the IS2 Element
(A) Representative plots showing eGFP expression profiles of 293T cells transduced with the SE or SE-IS2 at MOI = 0.2, in the absence or presence of 0.4 μM apicidin. (B) Graphs showing % of GFP+ cells transduced with the SE-IS2-IDLVs and SE-IDLVs in the absence (−) or presence (+) of 0.4 μM apicidin. Values represent means ± SEM of at least four separate experiments (*p < 0.05).
The Insertion of the IS2 Element into the IDLV Backbone Does Not Affect RNA Packaging into Vector Particles but Reduces the Amount of IDLV Episomes in the Target Cells
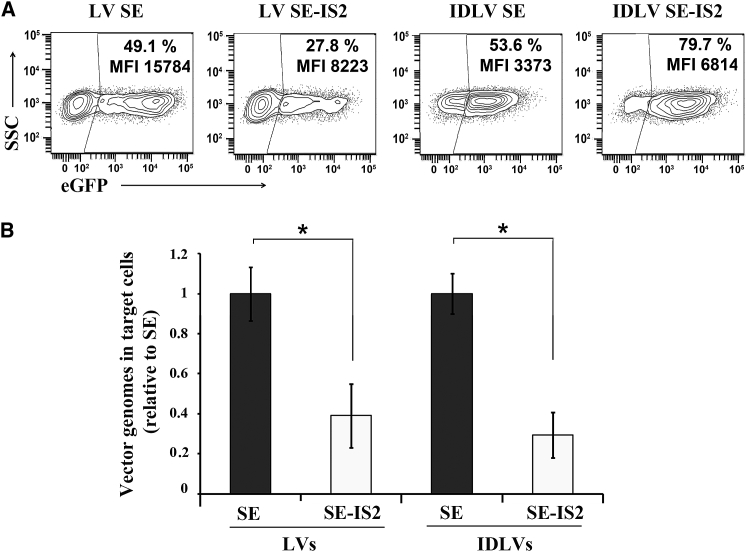
It has been described previously that insertions of large fragments into the 3′ long terminal repeat (LTR) of LVs reduce their efficacy. These insertions do not affect viral particle production but reduce reverse transcription efficacy in target cells.53, 55, 56 We therefore analyzed the effect of IS2 (1.2 kb long) on LV and IDLV transgene expression, vector production, and reverse transcription products in 293T target cells. The vector production efficacy was measured from the vector supernatants using ABM’s Lentiviral qPCR Titer Kit. This kit calculates the transduction units (TU) per mL based on an equation that convert genome copies per mL (GC/mL) values to TU/mL (see Materials and Methods for details). The value obtained using this formula is generally very close to the amount of effective particles per mL,57 although this can vary depending on the vector backbone. The relative amounts of reverse transcription products of the different vectors were quantified by qPCR using the U3Fw/PBSRev primers pair (see Figure S2). As expected, we did not observe any effect of the IS2 on vector production of LVs or IDLVs (data not shown). Also in agreement with previous observations,52, 53, 54, 55 the insertion of IS2 into the LTR had a negative effect on the expression levels of integrative LVs (Figure 3A, left plots) that correlated with a reduction of the amount of reverse transcribed products in target cells (Figure 3B, left bars). Interestingly, although the insertion IS2 also caused a similar reduction of IDLV reverse transcription products (Figure 3B, right bars), we observed a significant improvement in both the percentage and the expression levels (Figure 3A, right plots).
Figure 3.
The Insertion of the IS2 Element into the LV Backbone Reduces Their Efficiency to Generate Vector Genomes in Target Cells
(A) Representative plots showing eGFP expression profiles of 293T cells transduced with integrative LVs (left plots) and IDLVs (right plots) with (SE-IS2) or without (SE) the IS2 element. All the experiments were carried out using 0.7 viral particle/cells. (B) Graph showing the amounts of reverse-transcribed products (vector genomes) in 293T cells transduced with LVs (left bars) and IDLVs (right bars) with (SE-IS2) or without (SE) the IS2 element at 72 hr after transduction. Values represent means ± SEM of at least four separate experiments (*p < 0.05).
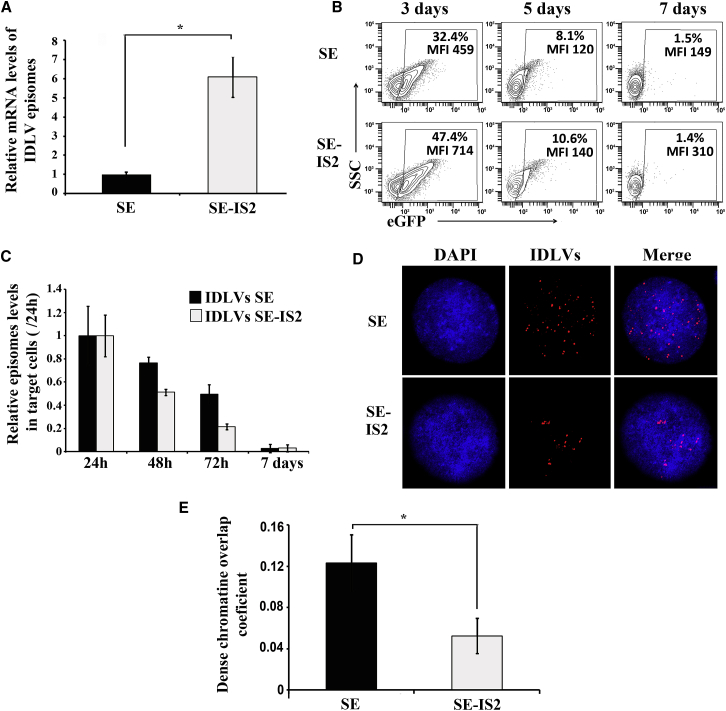
IDLV Episomes Harboring the IS2 Element Express Higher mRNA Levels and Have a Distinct Nuclear Localization
The IS2 is a chimeric DNA element containing a synthetic SAR (SAR2) and a 650-pb fragment of the chicken β-globin HS4 insulator.52 As mentioned in the introduction, these elements could enhance transcription or increase episomal stability28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 41 that can also lead to an improved transgene expression. In order to study the mechanisms involved in the IS2 effect, we first analyzed the increment in transcriptional efficacy of SE-IS2 episomes in comparison with SE episomes. To do that, we measured eGFP mRNA expression levels of 293T cells transduced with equivalent MOIs of SE-IDLVs and SE-IS2-IDLVs at 72 hr post-transduction and normalized to the amount of vector genomes (Figure 4A). This analysis showed that the SE-IS2 episomes express 6–7 times more mRNA than SE-episomes.
Figure 4.
The IS2 Element Does Not Affect the Stability of IDLV Episomes but Enhances Their Transcriptional Efficacy and Relocates Them into DAPI-Low Nuclear Domains
(A) Graph showing the relative amount of eGFP mRNA expression level in 293T cells transduced with IDLV-SE and IDLV-SE-IS2 normalized to the amount of vector genomes in each target cell. (B) Representative plots showing eGFP expression profiles of 293T cells transduced with SE (top) and SE-IS2 (bottom) IDLVs at MOI = 0.5 and analyzed at days 3, 5, and 7 post-transduction. The percentages (%) and expression levels (MFI) of the eGFP+ population are shown in each plot. (C) Relative amount of SE-IS2-IDLVs episomes related to SE-IDLVs at different time points post-transduction. (D) Representative confocal images showing nuclear distribution of SE- and SE-IS2-IDLV episomes. The cells were transduced at MOI = 10 in order to obtain several episomes inside the cells. The cells were subsequently fixed, methanol permeabilized, and incubated with Alexa Fluor 555 IDLV-labeled probes. (E) Graph showing the chromatin/IDLV colocalization in 293T cells transduced with SE- and SE-IS2-IDLVs. Values represent the Mander’s overlap coefficient (MOC; see Materials and Methods for details) between DAPI (DNA staining blue) and Alexa Fluor 555 (IDLV staining, in red). Values represent means ± SEM of at least four separate experiments (*p < 0.05).
We next analyzed whether the enhanced transcriptional activity of SE-IS2 episomes was due to an effect on their longtime stability. We therefore studied the eGFP expression levels (Figure 4B) and the relative amount of SE-IS2 episomes (Figure 4C) related to SE episomes at different time points post-transduction (24 hr to 7 days). Our data showed that the IS2 element does not influence the stability of the IDLV expression (Figure 4B) or the stability of IDLV episomes (Figure 4C).
We finally analyzed whether the presence of IS2 in the IDLV episomes could influence their nuclear localization to transcriptionally active sites since SAR elements bind several factors that promote the aggregation of nucleoproteins, HDAC recruitment, and chromatin remodeling complexes that enhance transcription.42, 44, 45, 46, 47 We performed a FISH analysis on 293T cells transduced with SE-IDLVs and SE-IS2-IDLVs at an MOI of 10 using SE plasmid as probe. As we can observe in Figures 4D and S3, the SE-IDLV episomes are uniformly distributed inside the nuclei, while the SE-IS2-IDLV episomes follow a more aggregated pattern. An analysis of the DAPI/probe co-localization indicated that the episomes harboring IS2 localized preferentially into regions with lower DAPI signal compared to the SE-IDLV episomes (Figure 4E). These data indicated that the better behavior of SE-IS2-IDLV episomes is probably due, in part, to a distinct nuclear re-positioning compared to SE-IDLV episomes.
Insertion of the IS2 Element into the IDLV Reduces the Formation of Lower Expressing 2-LTR Circles
Large LTR inserts such as the IS2 element can also increase homologous LTR recombination favoring 1-LTR circle formation that are superior to 2-LTR circles in terms of transgenic expression,56, 58 and this could be another potential mechanism behind the improved transcriptional activity of SE-IS2-IDLVs. In order to study this possibility, we generated a control IDLV (SE-1.2kb) harboring an equivalent insertion (1.2 kb) of irrelevant DNA at the same LTR location (Figure 5A, top) and compared it effect on IDLVs behavior. As observed previously with the IS2 element, the insertion of the 1.2-kb fragment did not affect vector titer, estimated based on ABM’s Lentiviral qPCR Titer Kit (see Materials and Methods) (Figure 5B) and reduced 4–5 times the amount of reverse-transcribed products in 293T target cells (Figure 5C). Additionally, as expected by their increased LTR size, the amount of 2-LTR circles (calculated by qPCR using the q2-LTRfw/q2-LTRrev primers [see Figure S2]) related to total episomes (calculated by qPCR using the ΔU3fw/PBSrev primers [see Figure S2]) was reduced similarly on SE-IS2 and SE-1.2kb IDLVs (Figure 5E), suggesting a similar increase on 1-LTR circles in both IDLVs. However, contrary to the SE-IS2-IDLVs, the transgene expression of SE-1.2kb IDLVs was 2.5 lower than SE-IDLVs (Figure 5A, plots, and Figure 5D), which correlates with the observed reduction on IDLVs episomes (Figure 5C). These data indicate that the decrease of the 2-LTR circles is not the main mechanism involved on the increased transcriptional activity of the SE-IS2-IDLV episomes, since the 293T cells transduced with the SE-1.2kb IDLVs showed a similar 2-LTR decrease but no effect on transcription efficacy.
Figure 5.
Insertion of the IS2 Element or a Control 1.2-kb Fragment into the IDLVs Reduce the Total Number of Episomes and the Formation of Lower Expressing 2-LTR Circles
(A) Top, schematic representation of SE, SE-IS2, and SE-1.2kb eGFP; SFFV (spleen focus-forming virus) promoter. Bottom, representative plots showing eGFP expression profiles of 293T cells transduced with the different IDLVs at MOI = 0.7. The percentages (%) and expression levels (MFI) of the eGFP+ population are shown in each plot. (B) Graph showing the titer of the different IDLV supernatants using the ABM’s Lentiviral qPCR Titer Kit (see Materials and Methods for details). (C) Relative amounts of vector genomes in 293T cells transduced with the different IDLVs at 72 hr post-transduction and normalized to the levels observed in SE IDLVs (see Materials and Methods for details). (D) Graph showing the percentage of GFP positive cells in 293T cells transduced with the different IDLVs and normalized to the levels observed in SE-IDLVs. (E) Graph showing the relative amounts of 2-LTR circles relative to total viral genomes in 293T target cells at 72 hr post-transduction and normalized to the levels observed in SE-IDLVs (see Materials and Methods for details). The values represent means ± SEM of at least four separate experiments (*p < 0.05). The data were reported to an equal amount of cells, estimated by β-albumin housekeeping gene.
The Final Effect of the IS2 Element Depends on the Target Cell and Vector Backbone
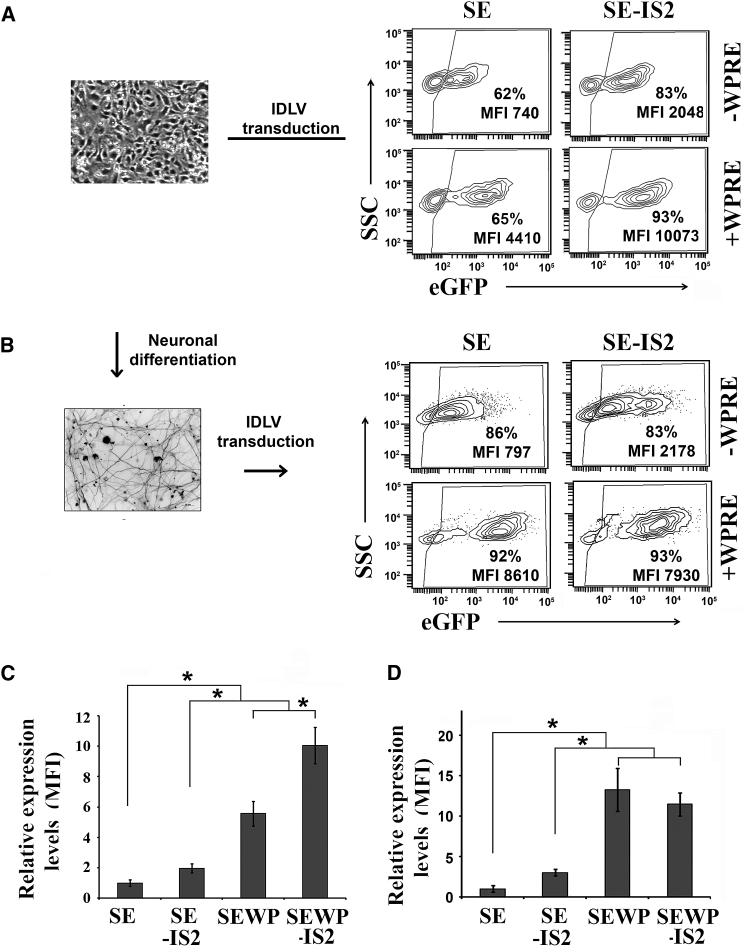
We next studied the potential applications of IS2-IDLVs in different target cells of interest with regard to gene therapy and/or basic research, such as neural progenitor cells (NPCs), neuronal cells (NCs), induced pluripotent stem cells (iPSCs), mesenchymal stromal cells (MSCs), human oral mucosa (HOM), human skin fibroblasts (HSF), and T cells. The different cell types were transduced with equal amounts of SE-IDLVs/SE-IS2-IDLV (NPCs, MOI = 3; iPSCs, MOI = 10; HSFs and HOMs, MOI = 10; T cells, MOI = 5; hMSCs, MOI = 5) and, 3 days later, we analyzed transgene expression levels in terms of MFI and percentage of eGFP+ cells. The incorporation of the IS2 element resulted in a marked increase in GFP expression levels in NPCs regardless of the presence or absence of the WPRE element (MFI SE = 740 versus SE-IS2 = 2,048, and SEWP = 4,410 versus SEWP-IS2 = 10,073) (Figures 6A and 6C). However, in differentiated neurons (Figures 6B and 6D; Figure S4), the inclusion of IS2 enhanced expression levels only in WPRE-negative IDLVs (MFI SE = 797 versus SE-IS2 = 2,178), a positive effect that was masked by the inclusion of the WPRE sequence (Figure 6B, bottom; Figure 6D, right bars). As previously shown in undifferentiated neurons, the incorporation of IS2 into the IDLV backbone significantly improved performance in iPSCs either in the absence (Figure 7A, top plots; MFI SE-IS2 = 894 versus MFI SE = 613) or in the presence of WPRE (Figure 7A, bottom plots; MFI SEWP-IS2 = 1,426 versus MFI SEWP = 1,154; Figure 7B, right graph). Contrary to NPCs and iPSCs, we could not find any significant positive effect of IS2-IDLVs in terms of global effects on eGFP+ expression pattern in the different T cell subpopulations (Figure S5), HOM, HSF, or hMSCs (Figure S6 and data not shown).
Figure 6.
The IS2 Sequence Improves Transgene Expression in Neural Progenitor Cells and Differentiated Neuron-like Cells
(A) Representative image (left) and plots (right) showing eGFP expression profiles of undifferentiated neural stem cells transduced at MOI = 3 with SE, SE-IS2, SEWP, and SEWP-IS2-IDLVs and analyzed 72 hr post-transduction. The percentage (%) of the eGFP+ cells and the eGFP expression levels (MFI) are shown in each plot. (B) Representative image (left) and plots (right) showing eGFP expression profiles of differentiated neuron-like cells (positive for β-tubulin, see Materials and Methods for details) transduced at MOI = 3 with SE-, SE-IS2-, SEWP-, and SEWP-IS2-IDLVs and analyzed 72 hr post-transduction. The percentage (%) of the eGFP+ cells and the eGFP expression levels (MFI) are shown in each plot. (C) Graph showing the relative expression levels of eGFP (MFI) in NPCs transduced with the different IDLVs and normalized to the levels observed in SE-IDLVs. (D) Graph showing the relative expression levels of eGFP (MFI) in differentiated NPCs (neuron-like cells) transduced with the different IDLVs and normalized to the levels observed in SE-IDLVs. Values represent means ± SEM of at least four separate experiments (*p < 0.05).
Figure 7.
The IS2 Sequence Improves Transgene Expression in iPSCs
(A) Representative plots showing eGFP expression profiles of iPSCs transduced with the SE, SE-IS2, SEWP, and SEWP-IS2 IDLVs at MOI = 10. The percentage (%) of the eGFP+ population and the relative transgene expression levels (MFI) are shown in each plot. (B) Graphs showing the relative percentage of eGFP+ cells (top) and the relative transgene expression levels (bottom) in iPSCs transduced with different IDLVs and normalized to the levels obtained with SE-IDLVs. The values represent means ± SEM of at least four separate experiments (*p < 0.05).
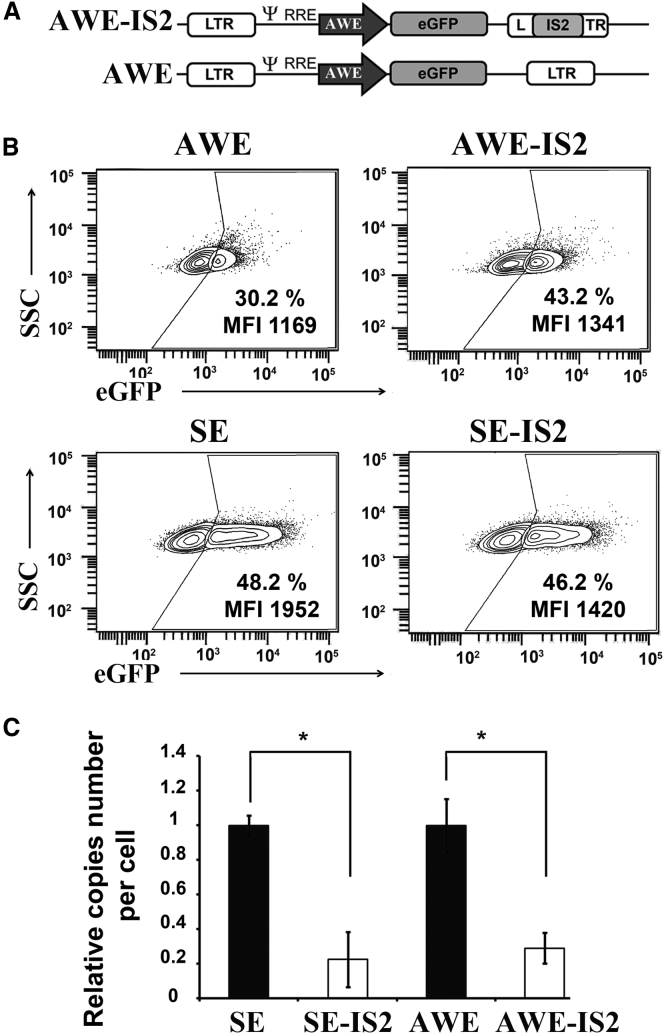
We finally analyzed whether the IS2 element had a similar effect when inserted in a different IDLV backbone expressing the transgene under a physiological promoter. We used an LV backbone previously published by our group (AWE)59 that express eGFP through the WAS (Wiskott-Aldrich syndrome) gene promoter only in hematopoietic cells60 and generated the IS2-AWE (Figure 8, top). Since the WAS promoter is hematopoietic specific, we tested the behavior of AWE-IDLVs versus AWE-IS2-IDLVs in Jurkat cells (an immortalized line of human T lymphocytes). As can be observed in Figure 8, the results were similar to that obtained with the SE-IDLVs. Indeed, although the cells transduced with AWE-IS2-IDLVs had 3–4 times lower episome numbers on target cells, compared to those transduced with AWE-IDLVs (Figure 8, graph), they expressed similar or slightly higher levels of eGFP (Figure 8, plots).
Figure 8.
Performance of the IS2-IDLVs Expressing the Transgene through the Human WAS Promoter
(A) Scheme of the IDLVs harboring the human WAS promoter. (B) Representative plots showing eGFP expression profiles of Jurkat cells transduced with AWE and AWE-IS2 (top) and the SE- and SEIS2 (bottom)-IDLVs at MOI = 0.5. The percentage (%) of the eGFP+ population and the expression levels (MFI) are shown in each plot. (C) Graphs showing the relative vector DNA genomes in IDLV-transduced cells at 72 hr after transduction. Values represent means ± SEM of at least four separate experiments (*p < 0.05).
Discussion
The success of gene therapy greatly depends on the availability of appropriate gene transfer vectors for the selected strategy. The broad application of gene therapy to different diseases has been possible thanks to the development of a wide range of vectors with different properties.1 In general, integrative vectors are the vectors of choice when stable gene expression in actively dividing cells is required, whereas non-integrative vectors are preferred for stable expression in non-dividing cells or when transient expression is sufficient or desirable. Integrative LVs have several properties that make them an attractive tool for gene delivery, such as the ability to deliver inserts of up to 12 kb, the active translocation to intact nuclei, and the possibility of using different envelopes that enable efficient gene delivery in almost all the cell lines analyzed. These characteristics have led scientists to extend the range of potential applications by developing IDLVs,61 which maintain several properties of LVs while expressing the transgene without integrating their genome into the host chromosome. IDLVs extend the applicability of LVs, which are safer when stable expression is required in non-dividing cells (neurons, hepatocytes) and can also achieve transient expression in actively dividing cells.62, 63 However, the expression levels and titers of IDLVs are generally lower than those of LVs,64, 65, 66 which has limited the applications of this technology.
The relatively low expression levels of IDLVs have been linked to epigenetic silencing through cellular defense mechanisms that apply heterochromatin marks to episomal viral sequences. This cellular response, which is not restricted to IDLV systems, affects different vector genomes such as herpes simplex viruses and adenoviruses.25, 26, 67, 68 The episomal viral DNA is “chromatinized” and acquires nucleosome-like properties immediately after entry into the nucleus.26, 68 In particular, IDLV genomes have been previously reported to undergo heterochromatinization through histone deacetylation, a process that can be reversed using HDACi such as sodium butyrate27 and valproic acid.24 For basic research purposes and some clinical applications, HDACi could be used to improve IDLV efficiency; however, in most gene therapy settings, the use of HDACi is not desirable due to potential severe side effects, such as the development of malignancies. We have previously described an improved IS2 insulator (which combines a synthetic SAR element [SAR2] and a 650-pb fragment of the chicken β-globin HS4 insulator) that avoid silencing and enhance LV expression in hESCs and HSCs.52 We therefore hypothesized that the inclusion of the IS2 element in IDLVs could improve their behavior by avoiding epigenetic transcriptional silencing through HS4 activity and by improving transcription efficiency through SAR activity. Contrary to expectations, the presence of the IS2 element did not abrogate epigenetic silencing by HDACs, although it did improve the transcriptional efficiency of episomal IDLVs.
Up to now, most attempts to improve IDLVs have focused on improving the stability of DNA episomal circles either through transient cell cycle arrest50 or by the inclusion of SAR elements alone48, 49, 50 or combined with replication origin (http://www.vivebiotech.com/technology). However, efficient stable transgene expression in highly dividing cells using IDLVs can be difficult to achieve. As an alternative, we have focused on improving the transient transgene expression levels of IDLVs through insertion of the IS2 element. Although the presence of SAR sequences in this element could also affect expression stability, we did not find this type of effect, which is in line with the observations of Kymäläinen et al.,50 who did not observe any differences in episomal establishment in IDLVs containing a SAR sequence. However, these data differ from other studies that show that the insertion of a full 1.2-kb or minimal 155-pb fragment of the β-interferon SAR elements in IDLVs provided a sustained transgene expression.48, 49 These contradictory findings could be explained by the differences in SAR elements used in the different IDLVs. Verghese et al.49 and Xu et al.48 used the full β-interferon SARs element and a 155-bp fragment, respectively. On the other hand, Kymäläinen et al.50 used a truncated form of 0.7 kb, while the IS2 element contained a synthetic SARs element consisting of 4 SAR recognition signatures (MRS).48 It is therefore possible that both the 1.2-kb β-interferon SAR element and the 155-bp fragment are required for episomal maintenance.
We inserted the IS2 element into the lentiviral backbone 3′ LTR in order to be duplicated during the reverse transcription process. This procedure enhances the effect of SAR sequences (the more sequences the better) and/or increases homologous recombination in order to promote the formation of 1-LTR circles, which is reported to be 2- to 4-fold more effective for expression than 2-LTR circles.69 To differentiate between these two effects, we constructed a 1.2 kb IDLV lentiviral backbone that has the same insertion in the LTR as IS2-IDLV but contains an irrelevant sequence. The 1-LTR form is the result of homologous recombination between the LTRs, while 2-LTR circles are the result of non-homologous end joining (NHEJ),70, 71 meaning that longer LTRs are expected to render higher levels of 1-LTRs. Our analysis showed that the inclusion of both the IS2 element and the irrelevant sequence 1.2 kb did not affect vector titer (estimated based on ABM’s Lentiviral qPCR Titer Kit; see Materials and Methods) but reduced the amount of episomes on transduced cells and also the amount of 2-LTR circles related to total episomes. While IS2-IDLVs episomes showed an increased eGFP transgene expression as compared to unmodified IDLVs, the 1.2kb IDLV episomes showed no effect. These data suggest that, as described previously,56 the insertion of the IS2 element into the LTRs reduced the reverse transcription process, rendering these IDLVs less efficient in generating IDLV episomes in target cells. Additionally, they also indicated that the enhanced expression of IS2-IDLV episomes cannot be explained by the increment in 1-LTR forms, since this effect is not present in the SE-1.2kb IDLVs.
The improved behavior of IDLVs harboring the IS2 element could be due to an improved transcriptional activity of the IDLV episomes or to improved mRNA stability and/or expression of IS2-containing transcripts. However, the insertion of IS2 into integrative LVs reduced their expression levels in 293T cells, and they also contained the IS2 element in their mRNAs. We can therefore conclude that the better performance of IS2-containing episomes must be due to an effect related to transcription and not to mRNA stability and/or other effects on the mRNA. We therefore focused our attention on trying to understand the potential mechanism involved in this enhanced transcriptional activity of SE-IS2-IDLVs. By normalizing the transcription levels to the relative amount of total vector DNA genomes, we estimated that the transcriptional activity of IS2 episomes is 6- to 7-fold higher than unmodified SE episomes. These positive effects of the IS2 sequence counter-balance the negative effect on episome generation in target cells. Therefore, the final effect the IS2 element in IDLVs will greatly depend on the target cell and the balance between the negative (less efficacy of episome generation) versus the positive (enhanced transcription) effects of the IS2 element in each cell type. Interestingly, the IS2 element still has a similar activity when inserted into a different LV backbone expressing the transgene through the WAS promoter, suggesting that the effect could be independent of the promoter used.
The elements contained in the IS2 (HS4 and SAR sequences) function as DNA anchor points for the chromatin scaffold and organize the chromatin into structural domains that separate different transcriptional units from each other and provide a platform for the assembly of the factors involved in transcription regulation.42, 72 Several studies indicate that these elements are located in proximity to expressed genes at the 5′ end or near transcription start sites.72 Several pieces of evidence suggest that these elements may poise the DNA for transcription by allowing interaction with ubiquitous tissue-specific transcription factors, such as special SATB-1,73, 74 NMP4,75 and CTCF;76 these, in turn, recruit regulatory proteins such topoisomerases and ATP-dependent chromatin remodeling complexes to mediate a more expression-permissive state.77 These nuclear domains involved in transcription and replication stain poorly with DAPI due to a lower DNA context.78 In this direction, our FISH analysis showed that, while the SE-IDLV episomes were uniformly distributed throughout the nuclei, the SE-IS2-IDLV episomes followed a more aggregated pattern into DAPI-low regions. These observations indicate that the improved behavior of SE-IS2-IDLV episomes is probably in part due to a preferential nuclear re-positioning into transcriptionally active regions.
In summary, we have designed a new LV backbone that improves the expression levels of IDLVs through a 6- to 7-fold increase in the transcriptional activity of IDLV episomes due to the inclusion of the IS2 element at the LTR. However, as the inclusion of this element also negatively affects the amount of episomal vectors produced in target cells, the final effect on different cell types varies. We did observe an objective increase in transgene expression only in certain cell types, such as 293T cells, NPCs, neurons, and iPSCs. Additional improvements such as the reduction of IS2 element size and/or the design of new backbones harboring this element in different locations could further enhance the effectiveness of IDLVs as a vector system.
Materials and Methods
Cell Lines
293T cells (CRL11268; American Type Culture Collection [ATCC], Rockville, MD) were cultured in DMEM (Invitrogen, Edinburgh, Scotland), supplemented with 10% fetal bovine serum (FBS; Invitrogen). The Jurkat T lymphocyte line (TIB-152) (ATCC, Manassas, VA) was grown in RPMI medium supplemented with 10% FBS and Penicillin-Streptomycin Solution (Biowest). Human neural progenitor cells (Gibco human neural stem cells [NSCs], H9 hESC-derived, cat. no. N7800) were maintained in StemPro NSC serum-free medium (SFM) (KnockOut DMEM/F-12, StemPro NSC SFM supplement, basic FGF recombinant protein, and EGF recombinant protein; Gibco, Thermo Fisher Scientific). Plates were previously coated with poly-L-Ornithine 10 μg/mL (Sigma-Aldrich, St. Louis, MO; https://www.sigmaaldrich.com) and laminin 20 μg/mL (Thermo Fisher Scientific, cat. no. 23017-015). The human iPSC line PBMC1-iPS4F1 were grown as previously described by Montes et al.79 All of the human cells used in this study were donated by healthy individuals after informed consent according to the Institutional Guidelines.
Neural Cell Differentiation In Vitro
When the human neural progenitor cells were 90% confluent, the complete growth medium was replaced by the neural differentiation media (NSC SFM Media without basic fibroblast growth factor [FGF] recombinant protein or epidermal growth factor [EGF] recombinant protein) and maintained at 37°C in 5% CO2 20%O2. The medium was changed every 3–4 days for 28 days. The mature neural-like cells were characterized by immunocytochemistry (see Supplemental Information).
Lentiviral Vector Constructs
The IS2 element, which combine the HS4-650 fragment and the SAR2, were designed and synthesized as previously described.52 SE,80 SEWP,81 AWE,59 and SEIS252 plasmids have been previously described. The SEWP-IS2 and AWE-IS2 were generated by the insertion of the IS2 element into the Bbs1 site of the 3′ LTR of the SEWP and AWE plasmids, respectively. The SE-1.2kb plasmid was generated by inserting a 1.2-kb fragment from SEN (SFFV-eGFP-Nluc) plasmid (https://www.genscript.com and our unpublished data) into the SE plasmid.
Viral Production and Titration
LVs and IDLVs were generated by transient transfection of 293T cells using the different vector plasmid together with the packaging plasmid pCMVDR8.91 (http://www.addgene.org/Didier_Trono) for LVs and pCMVDRD8.74 (kindly provided by Howe S.J. [UCL Great Ormond Street Institute of Child Health, University College London, London, UK]) for IDLVs, as well as the p-MD-G plasmid encoding the vesicular stomatitis virus (VSV-G) envelope gene (http://www.addgene.org/Didier_Trono). Transfection was performed with the aid of LipoD293 (SignaGen Laboratories, Ijamsville, MD, USA) according to the manufacturer’s instructions, and the supernatants were harvested at 48 hr and 72 hr after transfection. Viral titers were determined by the estimation of TU/mL using ABM’s Lentiviral qPCR Titer Kit to convert the amount of viral copies per mL (GC/mL) to TU/mL as indicated by the manufacturer.
Cell Transduction
Concentrated as well as unconcentrated LV and IDLV supernatants were used to transduce different cell lines. Cells were washed with Dulbecco’s PBS (1×) (Biowest), counted, and plated on 48-well plates and incubated for 5 hr with different LV and IDLV particles at different MOIs (from 0.3 to 10). Media were changed after 5 hr. NPCs and 293T cells were washed with Dulbecco’s PBS (1×) (Biowest), dissociated with TrypLE (Gibco), and plated on 24-well plates in the presence of the fresh viral particles at different TU/cells. The cell line PBMC1-iPS4F1 was incubated for 5 hr on the day of passage with a concentrated virus in the presence of 8 μg/mL Polybrene and 10 μM Y-27632 (Sigma-Aldrich).
Flow Cytometry
At different times (72 hr to 7 days) after transduction, the different cell types were harvested and washed twice with fluorescence-activated cell sorting (FACS) buffer (PBS containing 2 mM EDTA and 2% FBS), acquired on a FACS Canto II flow cytometer and analyzed using FACS Diva software (BD Biosciences). eGFP expression was detected in the fluorescein isothiocyanate (FITC) channel in a flow cytometer (BD FACScanto).
Profile of Extrachromosomal Forms of IDLV DNA
The 2-LTR:total IDLV DNA ratio in transduced cells was determined by real-time PCR using different primer pairs (Table 1) that enable us to discriminate 2-LTR from total IDLVs DNA. As an internal control, we also used primers for the human albumin locus (hAlb). DNA of transduced cells was extracted 72 hr after transduction using a QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany; https://www.qiagen.com:443/us). Real-time PCRs were performed using the QuantiTect SYBRGreen PCR kit (QIAGEN) on a Stratagene MX3005P System (Agilent Technologies, Santa Clara, CA; https://www.agilent.com). The PCRs were performed using the following run program: 10’ at 95°C for denaturation, 40 cycles of 15 min at 95°C, 60 min at 60°C, and 72°C for 60’’ followed by the melting curve. PCR data were analyzed according to the comparative CT method.82
Table 1.
Pairs of Primers Used to Establish the 2-LTR Relative to the Total IDLV DNA Ratio by Real-Time PCR and Estimate the cDNA Amount of eGFP
| Primer Pair | Orientation | Sequence | Amplified Forms |
|---|---|---|---|
| q2-LTR | Fw | 5′-GCCTCAATAAAGCTTGCCTTG-3′ | 2-LTR circles |
| Rv | 5′-TGGGAGTGAATTAGCCCTTCCA-3′ | ||
| ΔU3 | Fw | 5′-GATCTGCTTTTTGCTTGTACT-3′ | total viral DNA |
| PBS | Rv | 5′-GAGTCCTGCGTCGAGAGAGC-3′ | |
| qhAlb | Fw | 5′-GCTGTCATCTCTTGTGGGCTGT-3′ | genomic normalization |
| Rv | 5′-ACTCATGGGAGCTGCTGGTTC-3′ | ||
| GFP | Fw | 5′-GCCCGACAACCACTACCT-3′ | GFP cDNA |
| Rv | 5′-CGTCCATGCCGAGAGTGA-3′ | ||
| GAPDH | Fw | 5′-ATGGGGAAGGTGAAGGTCG-3′ | GAPDH cDNA |
| Rv | 5′-GGGGTCATTGATGGCAACAATA-3′ |
The q2-LTR pair of primer flanks the 2-LTR circle junction. The second pair of primers amplify all forms of reverse transcribed products (bottom). The qhAlb is used as a control for genomic normalization. GFP cDNA primers pair amplifies the eGFP cDNA. GAPDH primers were used for cDNA normalization. FW, forward; Rv, reverse.
mRNA Analysis by RT-qPCR
Total RNA was obtained using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA samples were reverse transcribed using the Superscript first-strand system (Invitrogen), and qPCRs were performed using the QuantiTect SYBRGreen PCR kit (QIAGEN) on a Stratagene MX3005P system (Agilent Technologies, Santa Clara, CA; https://www.agilent.com). The primers used are listed in Table 1.
FISH
FISH based on the use of an orange 8 (Alexa Fluor 555) fluorescent probe was employed to localize IDLVs in cells as previously described.83 In brief, to generate the IDLV-FISH probe, DNA from the vector plasmid was directly labeled by Nick Translation according to the manufacturer’s specifications (Invitrogen, Edinburgh, Scotland). Fixed cells with Carnoy’s fixative were hybridized overnight at 37°C with the SE probe. After post-hybridization washes, the cells were counterstained with DAPI in anti-fade solution (Molecular Probes). Images were acquired on a Zeiss LSM 710 confocal microscope (Carl Zeiss, Jena, Germany; https://www.zeiss.com). The extent of colocalization was performed using Mander’s overlap coefficient (MOC)84 to quantify the degree.
Statistical Analysis
All data are represented as means ± SEM. Statistical analysis was performed using GraphPad Prism software (GraphPad Software, La Jolla, CA; https://www.graphpad.com) by applying the unpaired two-tailed t test. Statistical significance was defined as a p value < 0.05.
Author Contributions
K.B. and F.M. wrote the manuscript and designed the experiments. A.G.-G., S.R.-P., and S.S.-H. performed the experiments on 293T cells. A.B.C.-G. and P.A. performed the experiments on hMSC cells. J.C.-A. and R.F.-V. performed the experiments skin fibroblast cells. M.T.-M., S.S.-H., and R.M. performed the experiments on T cells. K.B., S.S.-H., L.S., and J.L.G.-P. performed the experiments on NPCs. S.S.-H., R.M., and P.J.R. performed the experiments on iPSCs. K.B., F.M., A.G.-G., and S.S.-H. analyzed the data. All the authors reviewed the manuscript.
Conflicts of Interest
The authors have applied for the patent “IS2 element to improved integration-defective lentiviral vectors. Aplication number: EP18382613.0” Applicant: Fundación Progreso y Salud. This patent application does not alter the authors’ adherence to the policies of Molecular Therapy — Nucleic Acids on sharing data and materials. Therefore, all material presented in this manuscript will be freely available, though not for commercial purposes, to the scientific community.
Acknowledgments
This study is financed by the ISCIII Health Research Fund (Spain) and the European Regional Development Fund (FEDER) through research grants PI12/01097 and PI15/02015 (F.M.) and CD09/0020, PI15/00794, and CPII15/00032 (P.A.); the ISCIII Cellular Therapy Network (TerCel, RD12/0019/0006) (F.M.); a Juan de la Cierva fellowship (JCI_2012_12666; to R.M.); the CICE and CS of the Junta de Andalucía FEDER/European Cohesion Fund (FSE) for Andalucía 2007-2013 through research grants P09-CTS-04532, PI-57069, PI-0001/2009, and PAIDI-Bio-326 (F.M.) and PI-0160/2012 and PI-0014-2016 (K.B.); the Ministerio de Economía, Industria y competitividad through research grant SAF2017-89745-R (J.L.G.-P.); a RYC contract. (RYC-2015-18382; to P/J.R.); an FPU fellowship (FPU16/05467; to M.T.-M.); a PEJ contract (PEJ-2014-A-46314; to A.B.C.-G. and PEJ-2014-A-17105 S.S.-H.); the European Research Council (ERC-Consolidator ERC-STG-2012-233764); and a private donation from Ms. Francisca Serrano (Trading y Bolsa para Torpes, Granada, Spain).We also wish to thank Michael O’Shea for proofreading the article. J.L.G.-P’s lab is supported by MINECO-FEDER (SAF2017-89745-R).
Footnotes
Supplemental Information includes Supplemental Materials and Methods and six figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.08.007.
Contributor Information
Francisco Martin, Email: francisco.martin@genyo.es.
Karim Benabdellah, Email: karim.benabdel@genyo.es.
Supplemental Information
References
- 1.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 2.Ungari S., Montepeloso A., Morena F., Cocchiarella F., Recchia A., Martino S., Gentner B., Naldini L., Biffi A. Design of a regulated lentiviral vector for hematopoietic stem cell gene therapy of globoid cell leukodystrophy. Mol. Ther. Methods Clin. Dev. 2015;2:15038. doi: 10.1038/mtm.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biffi A., Aubourg P., Cartier N. Gene therapy for leukodystrophies. Hum. Mol. Genet. 2011;20(R1):R42–R53. doi: 10.1093/hmg/ddr142. [DOI] [PubMed] [Google Scholar]
- 4.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 5.Kato S., Kobayashi K., Kobayashi K. Improved transduction efficiency of a lentiviral vector for neuron-specific retrograde gene transfer by optimizing the junction of fusion envelope glycoprotein. J. Neurosci. Methods. 2014;227:151–158. doi: 10.1016/j.jneumeth.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Allodi I., Mecollari V., González-Pérez F., Eggers R., Hoyng S., Verhaagen J., Navarro X., Udina E. Schwann cells transduced with a lentiviral vector encoding Fgf-2 promote motor neuron regeneration following sciatic nerve injury. Glia. 2014;62:1736–1746. doi: 10.1002/glia.22712. [DOI] [PubMed] [Google Scholar]
- 7.Towne C., Aebischer P. Lentiviral and adeno-associated vector-based therapy for motor neuron disease through RNAi. Methods Mol. Biol. 2009;555:87–108. doi: 10.1007/978-1-60327-295-7_7. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Q., Uhlig K.M., Muth A., Kimpel J., Lévy C., Münch R.C., Seifried J., Pfeiffer A., Trkola A., Coulibaly C. Exclusive Transduction of Human CD4+ T Cells upon Systemic Delivery of CD4-Targeted Lentiviral Vectors. J. Immunol. 2015;195:2493–2501. doi: 10.4049/jimmunol.1500956. [DOI] [PubMed] [Google Scholar]
- 9.Amirache F., Lévy C., Costa C., Mangeot P.E., Torbett B.E., Wang C.X., Nègre D., Cosset F.L., Verhoeyen E. Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123:1422–1424. doi: 10.1182/blood-2013-11-540641. [DOI] [PubMed] [Google Scholar]
- 10.Humbert J.M., Frecha C., Amirache Bouafia F., N’Guyen T.H., Boni S., Cosset F.L., Verhoeyen E., Halary F. Measles virus glycoprotein-pseudotyped lentiviral vectors are highly superior to vesicular stomatitis virus G pseudotypes for genetic modification of monocyte-derived dendritic cells. J. Virol. 2012;86:5192–5203. doi: 10.1128/JVI.06283-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cesana D., Ranzani M., Volpin M., Bartholomae C., Duros C., Artus A., Merella S., Benedicenti F., Sergi Sergi L., Sanvito F. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol. Ther. 2014;22:774–785. doi: 10.1038/mt.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lombardo A., Cesana D., Genovese P., Di Stefano B., Provasi E., Colombo D.F., Neri M., Magnani Z., Cantore A., Lo Riso P. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 13.Lombardo A., Genovese P., Beausejour C.M., Colleoni S., Lee Y.L., Kim K.A., Ando D., Urnov F.D., Galli C., Gregory P.D. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 14.Negri D.R., Rossi A., Blasi M., Michelini Z., Leone P., Chiantore M.V., Baroncelli S., Perretta G., Cimarelli A., Klotman M.E., Cara A. Simian immunodeficiency virus-Vpx for improving integrase defective lentiviral vector-based vaccines. Retrovirology. 2012;9:69. doi: 10.1186/1742-4690-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negri D.R., Michelini Z., Bona R., Blasi M., Filati P., Leone P., Rossi A., Franco M., Cara A. Integrase-defective lentiviral-vector-based vaccine: a new vector for induction of T cell immunity. Expert Opin. Biol. Ther. 2011;11:739–750. doi: 10.1517/14712598.2011.571670. [DOI] [PubMed] [Google Scholar]
- 16.Michelini Z., Negri D.R., Baroncelli S., Spada M., Leone P., Bona R., Klotman M.E., Cara A. Development and use of SIV-based Integrase defective lentiviral vector for immunization. Vaccine. 2009;27:4622–4629. doi: 10.1016/j.vaccine.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoban M.D., Cost G.J., Mendel M.C., Romero Z., Kaufman M.L., Joglekar A.V., Ho M., Lumaquin D., Gray D., Lill G.R. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125:2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez-Guerrero A., Sanchez-Hernandez S., Galvani G., Pinedo-Gomez J., Martin-Guerra R., Sanchez-Gilabert A., Aguilar-González A., Cobo M., Gregory P., Holmes M. Comparison of Zinc Finger Nucleases Versus CRISPR-Specific Nucleases for Genome Editing of the Wiskott-Aldrich Syndrome Locus. Hum. Gene Ther. 2018;29:366–380. doi: 10.1089/hum.2017.047. [DOI] [PubMed] [Google Scholar]
- 19.Ortinski P.I., O’Donovan B., Dong X., Kantor B. Integrase-Deficient Lentiviral Vector as an All-in-One Platform for Highly Efficient CRISPR/Cas9-Mediated Gene Editing. Mol. Ther. Methods Clin. Dev. 2017;5:153–164. doi: 10.1016/j.omtm.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coluccio A., Miselli F., Lombardo A., Marconi A., Malagoli Tagliazucchi G., Gonçalves M.A., Pincelli C., Maruggi G., Del Rio M., Naldini L. Targeted gene addition in human epithelial stem cells by zinc-finger nuclease-mediated homologous recombination. Mol. Ther. 2013;21:1695–1704. doi: 10.1038/mt.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steffen I., Simmons G. Pseudotyping Viral Vectors With Emerging Virus Envelope Proteins. Curr. Gene Ther. 2016;16:47–55. doi: 10.2174/1566523216666160119093948. [DOI] [PubMed] [Google Scholar]
- 22.Cronin J., Zhang X.Y., Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joglekar A.V., Hollis R.P., Kuftinec G., Senadheera S., Chan R., Kohn D.B. Integrase-defective lentiviral vectors as a delivery platform for targeted modification of adenosine deaminase locus. Mol. Ther. 2013;21:1705–1717. doi: 10.1038/mt.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joglekar A.V., Stein L., Ho M., Hoban M.D., Hollis R.P., Kohn D.B. Dissecting the mechanism of histone deacetylase inhibitors to enhance the activity of zinc finger nucleases delivered by integrase-defective lentiviral vectors. Hum. Gene Ther. 2014;25:599–608. doi: 10.1089/hum.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takacs M., Banati F., Koroknai A., Segesdi J., Salamon D., Wolf H., Niller H.H., Minarovits J. Epigenetic regulation of latent Epstein-Barr virus promoters. Biochim. Biophys. Acta. 2010;1799:228–235. doi: 10.1016/j.bbagrm.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Knipe D.M., Lieberman P.M., Jung J.U., McBride A.A., Morris K.V., Ott M., Margolis D., Nieto A., Nevels M., Parks R.J., Kristie T.M. Snapshots: chromatin control of viral infection. Virology. 2013;435:141–156. doi: 10.1016/j.virol.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelascini L.P., Maggio I., Liu J., Holkers M., Cathomen T., Gonçalves M.A. Histone deacetylase inhibition rescues gene knockout levels achieved with integrase-defective lentiviral vectors encoding zinc-finger nucleases. Hum. Gene Ther. Methods. 2013;24:399–411. doi: 10.1089/hgtb.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizutani A., Kikkawa E., Matsuno A., Shigenari A., Okinaga H., Murakami M., Ishida H., Tanaka M., Inoko H. Modified S/MAR episomal vectors for stably expressing fluorescent protein-tagged transgenes with small cell-to-cell fluctuations. Anal. Biochem. 2013;443:113–116. doi: 10.1016/j.ab.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Haase R., Magnusson T., Su B., Kopp F., Wagner E., Lipps H., Baiker A., Ogris M. Generation of a tumor- and tissue-specific episomal non-viral vector system. BMC Biotechnol. 2013;13:49. doi: 10.1186/1472-6750-13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Argyros O., Wong S.P., Gowers K., Harbottle R.P. Genetic modification of cancer cells using non-viral, episomal S/MAR vectors for in vivo tumour modelling. PLoS ONE. 2012;7:e47920. doi: 10.1371/journal.pone.0047920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong S.P., Argyros O., Coutelle C., Harbottle R.P. Non-viral S/MAR vectors replicate episomally in vivo when provided with a selective advantage. Gene Ther. 2011;18:82–87. doi: 10.1038/gt.2010.116. [DOI] [PubMed] [Google Scholar]
- 32.Argyros O., Wong S.P., Niceta M., Waddington S.N., Howe S.J., Coutelle C., Miller A.D., Harbottle R.P. Persistent episomal transgene expression in liver following delivery of a scaffold/matrix attachment region containing non-viral vector. Gene Ther. 2008;15:1593–1605. doi: 10.1038/gt.2008.113. [DOI] [PubMed] [Google Scholar]
- 33.Hagedorn C., Antoniou M.N., Lipps H.J. Genomic cis-acting Sequences Improve Expression and Establishment of a Nonviral Vector. Mol. Ther. Nucleic Acids. 2013;2:e118. doi: 10.1038/mtna.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagedorn C., Wong S.P., Harbottle R., Lipps H.J. Scaffold/matrix attached region-based nonviral episomal vectors. Hum. Gene Ther. 2011;22:915–923. doi: 10.1089/hum.2011.084. [DOI] [PubMed] [Google Scholar]
- 35.Sgourou A., Routledge S., Spathas D., Athanassiadou A., Antoniou M.N. Physiological levels of HBB transgene expression from S/MAR element-based replicating episomal vectors. J. Biotechnol. 2009;143:85–94. doi: 10.1016/j.jbiotec.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 36.Giannakopoulos A., Stavrou E.F., Zarkadis I., Zoumbos N., Thrasher A.J., Athanassiadou A. The functional role of S/MARs in episomal vectors as defined by the stress-induced destabilization profile of the vector sequences. J. Mol. Biol. 2009;387:1239–1249. doi: 10.1016/j.jmb.2009.02.043. [DOI] [PubMed] [Google Scholar]
- 37.Lufino M.M., Manservigi R., Wade-Martins R. An S/MAR-based infectious episomal genomic DNA expression vector provides long-term regulated functional complementation of LDLR deficiency. Nucleic Acids Res. 2007;35:e98. doi: 10.1093/nar/gkm570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao C.P., Guo X., Chen S.J., Li C.Z., Yang Y., Zhang J.H., Chen S.N., Jia Y.L., Wang T.Y. Matrix attachment region combinations increase transgene expression in transfected Chinese hamster ovary cells. Sci. Rep. 2017;7:42805. doi: 10.1038/srep42805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T.Y., Yang R., Qin C., Wang L., Yang X.J. Enhanced expression of transgene in CHO cells using matrix attachment region. Cell Biol. Int. 2008;32:1279–1283. doi: 10.1016/j.cellbi.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Girod P.A., Nguyen D.Q., Calabrese D., Puttini S., Grandjean M., Martinet D., Regamey A., Saugy D., Beckmann J.S., Bucher P., Mermod N. Genome-wide prediction of matrix attachment regions that increase gene expression in mammalian cells. Nat. Methods. 2007;4:747–753. doi: 10.1038/nmeth1076. [DOI] [PubMed] [Google Scholar]
- 41.Girod P.A., Zahn-Zabal M., Mermod N. Use of the chicken lysozyme 5′ matrix attachment region to generate high producer CHO cell lines. Biotechnol. Bioeng. 2005;91:1–11. doi: 10.1002/bit.20563. [DOI] [PubMed] [Google Scholar]
- 42.Wang T.Y., Han Z.M., Chai Y.R., Zhang J.H. A mini review of MAR-binding proteins. Mol. Biol. Rep. 2010;37:3553–3560. doi: 10.1007/s11033-010-0003-8. [DOI] [PubMed] [Google Scholar]
- 43.Yusufzai T.M., Felsenfeld G. The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA. 2004;101:8620–8624. doi: 10.1073/pnas.0402938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zielke K., Full F., Teufert N., Schmidt M., Müller-Fleckenstein I., Alberter B., Ensser A. The insulator protein CTCF binding sites in the orf73/LANA promoter region of herpesvirus saimiri are involved in conferring episomal stability in latently infected human T cells. J. Virol. 2012;86:1862–1873. doi: 10.1128/JVI.06295-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holohan E.E., Kwong C., Adryan B., Bartkuhn M., Herold M., Renkawitz R., Russell S., White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohan M., Bartkuhn M., Herold M., Philippen A., Heinl N., Bardenhagen I., Leers J., White R.A., Renkawitz-Pohl R., Saumweber H., Renkawitz R. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herold M., Bartkuhn M., Renkawitz R. CTCF: insights into insulator function during development. Development. 2012;139:1045–1057. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 48.Xu Z., Chen F., Zhang L., Lu J., Xu P., Liu G., Xie X., Mu W., Wang Y., Liu D. Non-integrating lentiviral vectors based on the minimal S/MAR sequence retain transgene expression in dividing cells. Sci. China Life Sci. 2016;59:1024–1033. doi: 10.1007/s11427-016-0067-0. [DOI] [PubMed] [Google Scholar]
- 49.Verghese S.C., Goloviznina N.A., Skinner A.M., Lipps H.J., Kurre P. S/MAR sequence confers long-term mitotic stability on non-integrating lentiviral vector episomes without selection. Nucleic Acids Res. 2014;42:e53. doi: 10.1093/nar/gku082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kymäläinen H., Appelt J.U., Giordano F.A., Davies A.F., Ogilvie C.M., Ahmed S.G., Laufs S., Schmidt M., Bode J., Yáñez-Muñoz R.J., Dickson G. Long-term episomal transgene expression from mitotically stable integration-deficient lentiviral vectors. Hum. Gene Ther. 2014;25:428–442. doi: 10.1089/hum.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grandchamp N., Henriot D., Philippe S., Amar L., Ursulet S., Serguera C., Mallet J., Sarkis C. Influence of insulators on transgene expression from integrating and non-integrating lentiviral vectors. Genet. Vaccines Ther. 2011;9:1. doi: 10.1186/1479-0556-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benabdellah K., Gutierrez-Guerrero A., Cobo M., Muñoz P., Martín F. A chimeric HS4-SAR insulator (IS2) that prevents silencing and enhances expression of lentiviral vectors in pluripotent stem cells. PLoS ONE. 2014;9:e84268. doi: 10.1371/journal.pone.0084268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramezani A., Hawley T.S., Hawley R.G. Performance- and safety-enhanced lentiviral vectors containing the human interferon-beta scaffold attachment region and the chicken beta-globin insulator. Blood. 2003;101:4717–4724. doi: 10.1182/blood-2002-09-2991. [DOI] [PubMed] [Google Scholar]
- 54.Arumugam P.I., Scholes J., Perelman N., Xia P., Yee J.K., Malik P. Improved human beta-globin expression from self-inactivating lentiviral vectors carrying the chicken hypersensitive site-4 (cHS4) insulator element. Mol. Ther. 2007;15:1863–1871. doi: 10.1038/sj.mt.6300259. [DOI] [PubMed] [Google Scholar]
- 55.Arumugam P.I., Urbinati F., Velu C.S., Higashimoto T., Grimes H.L., Malik P. The 3′ region of the chicken hypersensitive site-4 insulator has properties similar to its core and is required for full insulator activity. PLoS ONE. 2009;4:e6995. doi: 10.1371/journal.pone.0006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urbinati F., Arumugam P., Higashimoto T., Perumbeti A., Mitts K., Xia P., Malik P. Mechanism of reduction in titers from lentivirus vectors carrying large inserts in the 3′LTR. Mol. Ther. 2009;17:1527–1536. doi: 10.1038/mt.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J.Y., Lee H.H. A new chemical complex can rapidly concentrate lentivirus and significantly enhance gene transduction. Cytotechnology. 2018;70:193–201. doi: 10.1007/s10616-017-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw A.M., Joseph G.L., Jasti A.C., Sastry-Dent L., Witting S., Cornetta K. Differences in vector-genome processing and illegitimate integration of non-integrating lentiviral vectors. Gene Ther. 2017;24:12–20. doi: 10.1038/gt.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frecha C., Toscano M.G., Costa C., Saez-Lara M.J., Cosset F.L., Verhoeyen E., Martin F. Improved lentiviral vectors for Wiskott-Aldrich syndrome gene therapy mimic endogenous expression profiles throughout haematopoiesis. Gene Ther. 2008;15:930–941. doi: 10.1038/gt.2008.20. [DOI] [PubMed] [Google Scholar]
- 60.Muñoz P., Toscano M.G., Real P.J., Benabdellah K., Cobo M., Bueno C., Ramos-Mejía V., Menendez P., Anderson P., Martín F. Specific marking of hESCs-derived hematopoietic lineage by WAS-promoter driven lentiviral vectors. PLoS ONE. 2012;7:e39091. doi: 10.1371/journal.pone.0039091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wanisch K., Yáñez-Muñoz R.J. Integration-deficient lentiviral vectors: a slow coming of age. Mol. Ther. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanoun N., Gayral M., Pointreau A., Buscail L., Cordelier P. Initial Characterization of Integrase-Defective Lentiviral Vectors for Pancreatic Cancer Gene Therapy. Hum. Gene Ther. 2016;27:184–192. doi: 10.1089/hum.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sürün D., Schwäble J., Tomasovic A., Ehling R., Stein S., Kurrle N., von Melchner H., Schnütgen F. High Efficiency Gene Correction in Hematopoietic Cells by Donor-Template-Free CRISPR/Cas9 Genome Editing. Mol. Ther. Nucleic Acids. 2018;10:1–8. doi: 10.1016/j.omtn.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naldini L., Blömer U., Gage F.H., Trono D., Verma I.M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loewen N., Leske D.A., Chen Y., Teo W.L., Saenz D.T., Peretz M., Holmes J.M., Poeschla E.M. Comparison of wild-type and class I integrase mutant-FIV vectors in retina demonstrates sustained expression of integrated transgenes in retinal pigment epithelium. J. Gene Med. 2003;5:1009–1017. doi: 10.1002/jgm.447. [DOI] [PubMed] [Google Scholar]
- 66.Park F., Kay M.A. Modified HIV-1 based lentiviral vectors have an effect on viral transduction efficiency and gene expression in vitro and in vivo. Mol. Ther. 2001;4:164–173. doi: 10.1006/mthe.2001.0450. [DOI] [PubMed] [Google Scholar]
- 67.Lieberman P.M. Chromatin organization and virus gene expression. J. Cell. Physiol. 2008;216:295–302. doi: 10.1002/jcp.21421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knipe D.M. Nuclear sensing of viral DNA, epigenetic regulation of herpes simplex virus infection, and innate immunity. Virology. 2015;479-480:153–159. doi: 10.1016/j.virol.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cara A., Cereseto A., Lori F., Reitz M.S., Jr. HIV-1 protein expression from synthetic circles of DNA mimicking the extrachromosomal forms of viral DNA. J. Biol. Chem. 1996;271:5393–5397. doi: 10.1074/jbc.271.10.5393. [DOI] [PubMed] [Google Scholar]
- 70.Kilzer J.M., Stracker T., Beitzel B., Meek K., Weitzman M., Bushman F.D. Roles of host cell factors in circularization of retroviral dna. Virology. 2003;314:460–467. doi: 10.1016/s0042-6822(03)00455-0. [DOI] [PubMed] [Google Scholar]
- 71.Avettand-Fènoël V., Hocqueloux L., Ghosn J., Cheret A., Frange P., Melard A., Viard J.P., Rouzioux C. Total HIV-1 DNA, a Marker of Viral Reservoir Dynamics with Clinical Implications. Clin. Microbiol. Rev. 2016;29:859–880. doi: 10.1128/CMR.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keaton M.A., Taylor C.M., Layer R.M., Dutta A. Nuclear scaffold attachment sites within ENCODE regions associate with actively transcribed genes. PLoS ONE. 2011;6:e17912. doi: 10.1371/journal.pone.0017912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cai S.X., Liu A.R., He H.L., Chen Q.H., Yang Y., Guo F.M., Huang Y.Z., Liu L., Qiu H.B. Stable genetic alterations of β-catenin and ROR2 regulate the Wnt pathway, affect the fate of MSCs. J. Cell. Physiol. 2014;229:791–800. doi: 10.1002/jcp.24500. [DOI] [PubMed] [Google Scholar]
- 74.Kohwi-Shigematsu T., Maass K., Bode J. A thymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry. 1997;36:12005–12010. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- 75.Feister H.A., Torrungruang K., Thunyakitpisal P., Parker G.E., Rhodes S.J., Bidwell J.P. NP/NMP4 transcription factors have distinct osteoblast nuclear matrix subdomains. J. Cell. Biochem. 2000;79:506–517. [PubMed] [Google Scholar]
- 76.Dunn K.L., Zhao H., Davie J.R. The insulator binding protein CTCF associates with the nuclear matrix. Exp. Cell Res. 2003;288:218–223. doi: 10.1016/s0014-4827(03)00185-x. [DOI] [PubMed] [Google Scholar]
- 77.Zhang P., Torres K., Liu X., Liu C.G., Pollock R.E. An Overview of Chromatin-Regulating Proteins in Cells. Curr. Protein Pept. Sci. 2016;17:401–410. doi: 10.2174/1389203717666160122120310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deutsch M.J., Ott E., Papior P., Schepers A. The latent origin of replication of Epstein-Barr virus directs viral genomes to active regions of the nucleus. J. Virol. 2010;84:2533–2546. doi: 10.1128/JVI.01909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montes R., Romero T., Cabrera S., Ayllon V., Lopez-Escamez J.A., Ramos-Mejia V., Real P.J. Generation and characterization of the human iPSC line PBMC1-iPS4F1 from adult peripheral blood mononuclear cells. Stem Cell Res. (Amst.) 2015;15:614–617. doi: 10.1016/j.scr.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 80.Martín F., Toscano M.G., Blundell M., Frecha C., Srivastava G.K., Santamaría M., Thrasher A.J., Molina I.J. Lentiviral vectors transcriptionally targeted to hematopoietic cells by WASP gene proximal promoter sequences. Gene Ther. 2005;12:715–723. doi: 10.1038/sj.gt.3302457. [DOI] [PubMed] [Google Scholar]
- 81.Demaison C., Parsley K., Brouns G., Scherr M., Battmer K., Kinnon C., Grez M., Thrasher A.J. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 82.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 83.Torres R., Garcia A., Jimenez M., Rodriguez S., Ramirez J.C. An integration-defective lentivirus-based resource for site-specific targeting of an edited safe-harbour locus in the human genome. Gene Ther. 2014;21:343–352. doi: 10.1038/gt.2014.1. [DOI] [PubMed] [Google Scholar]
- 84.Manders R.J., Koopman R., Sluijsmans W.E., van den Berg R., Verbeek K., Saris W.H., Wagenmakers A.J., van Loon L.J. Co-ingestion of a protein hydrolysate with or without additional leucine effectively reduces postprandial blood glucose excursions in Type 2 diabetic men. J. Nutr. 2006;136:1294–1299. doi: 10.1093/jn/136.5.1294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.