Abstract
Candida blankii is a newly recognized human pathogen. Here we describe a case of bloodstream infection in a preterm neonate. The yeast was repeatedly isolated from blood, and its identity was confirmed by PCR sequencing of rDNA. Additionally, C. blankii DNA was detected directly in a blood sample. The isolates initially developed pink colonies on CHROMagar Candida which later turned into dark metallic blue similar to Candida tropicalis. Inaccurate identification by the VITEK 2 yeast identification system as Stephanoascus ciferrii and intrinsic resistance to fluconazole (MIC 12–16 μg/mL) underscore the need for its accurate identification for appropriate therapeutic management.
Keywords: Azole resistance, Candida blankii, molecular identification, neonate
Introduction
Two reports have highlighted the clinical significance of Candida blankii in human infections. The first report described its isolation from respiratory specimens of a patient with repeated exacerbations of cystic fibrosis [1]. The second patient, also with cystic fibrosis, yielded a germ-tube–negative Candida species along with Aspergillus [2]. This patient was receiving itraconazole when the sputum culture yielded this yeast. After bilateral lung transplantation, and despite liposomal amphotericin B prophylaxis, blood cultures grew a yeast which was subsequently identified as C. blankii. Here we describe second case of C. blankii fungaemia, diagnosed by repeated isolation of the yeast in blood cultures and detection of its DNA in serum sample by PCR, thus unequivocally establishing its aetiologic role as a bloodstream pathogen.
Case description
The patient was a 27-week-old preterm neonate. On day 8 of his birth, he presented a clinical picture simulating necrotizing enterocolitis associated with greenish gastric aspirate. The baby was maintained on total parenteral nutrition and was prescribed piperacillin/tazobactam and amphotericin B for the next 14 days, followed by 21 days of fluconazole prophylaxis. Because he continued to yield greenish gastric aspirate, an X-ray of the abdomen was performed, which revealed perforated necrotizing enterocolitis requiring surgical intervention. On day 40, ileostomy was performed. However, after surgery, he had multiple positive blood cultures for Staphylococcus epidermidis and then for Enterococcus faecalis. He was prescribed piperacillin/tazobactam and amphotericin B, and then vancomycin and liposomal amphotericin B (AmBisome). On day 76, antibiotic therapy was withdrawn, but his condition deteriorated and blood culture yielded Pseudomonas aeruginosa, which was treated with meropenem, and he continued to receive liposomal amphotericin B (AmBisome) empirically. Two days later, his blood culture grew E. faecalis, so vancomycin was prescribed for 21 days, which was replaced with teicoplanin as a result of renal toxicity. On day 117, before undergoing a second surgical procedure for prolapsed stoma, he developed septicaemia yielding S. epidermidis and a Candida species (Lab. No. Kw593/18) in blood cultures requiring addition of amikacin along with teicoplanin and liposomal amphotericin B (AmBisome) in the treatment regimen. The details of the disease progression and outcome are provided in Fig. 1. The isolate (Kw593/18) was referred to the Mycology Reference Laboratory for identification and antifungal susceptibility testing under routine patient care as a part of an ongoing study approved by ethical committee of the Ministry of Health, Kuwait. Three subsequent blood cultures on day 125 (Kw710/18), day 130 (Kw775/18) and day 133 (Kw807/18) yielded the same Candida species. Cultures of rectal swabs and endotracheal aspirates were negative for C. blankii and other Candida species.
Fig. 1.
Timeline and therapeutic course of Candida blankii fungaemia. AmBisome, liposomal amphotericin B; Amk, amikacin; Amp, ampicillin; AmB, amphotericin B; Cs, caspofungin; Ctx, cefotaxime; D, day; Fl, fluconazole; Mem, meropenem; NEC, necrotizing enterocolitis; NPO, nothing by mouth; Tec, teicoplanin; TPN, total parenteral nutrition; Tzp, piperacillin/tazobactam; Van, vancomycin.
Results
All bloodstream isolates were recovered in BACTEC Peds Plus/F culture vials after 2 to 3 days of incubation at 37°C and were identified by VITEK 2 yeast identification system as Stephanoascus ciferrii with 89% probability and remained unidentified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (VITEK MS; bioMérieux, Marcy l’Etoile, France). The isolates formed typical yeastlike cream-coloured colonies with a smooth surface and entire margins on Sabouraud dextrose agar and initially developed pink colonies (Fig. 2(A,a)) on CHROMagar Candida [3], which subsequently developed into dark metallic blue similar to Candida tropicalis (Fig. 2(A,b). Slide culture on cornmeal agar showed clusters of budding yeast cells with pseudohyphae (Fig. 2(B)). The isolate showed good growth up to 42°C. The MIC values during antifungal susceptibility testing of the four isolates by Etest were as follows: fluconazole, 12–16 μg/mL; voriconazole, 0.19–0.38 μg/mL; itraconazole, 0.75 μg/mL; posaconazole, 0.5–0.75 μg/mL; amphotericin B, 0.125 μg/mL; caspofungin, 0.25–0.5 μg/mL; micafungin, 0.125 μg/mL and anidulafungin, 0.19 μg/mL (Fig. 3(A) and (B)). Although no approved susceptibility breakpoints for C. blankii are yet available, our susceptibility results to antifungal agents are comparable with a previous report [2]. Because the isolate showed reduced susceptibility to fluconazole by Etest (≥12 μg/mL) and VITEK 2 identified it as S. ciferrii with only 89% probability, molecular identification was carried out. PCR sequencing of internally transcribed spacer (ITS) region of rDNA was performed for species-specific identification [4], [5]. The isolate (Kw593/18) was unequivocally identified as C. blankii because the ITS region of rDNA (GenBank accession no. LT993736) exhibited 100% sequence identity with the corresponding sequence from type strain (CBS1898) as well as several other (CBS6427, CBS7205, CBS1989 and CBS6788) strains, and only one nucleotide difference with a bloodstream isolate (HCFMUSP01) of C. blankii obtained from a cystic fibrosis patient in Brazil [2]. The ITS region of rDNA sequences from C. blankii CBS1898, C. blankii CBS6427 and several other closely related species [6], as well as sequences from other well-known pathogenic Candida spp. available from GenBank were retrieved. Multiple sequence alignments were performed with Clustal Omega, and the phylogenetic tree was constructed with MEGA 6.1 software using the neighbour-joining method with the Kimura two-parameter model. The robustness of tree branches was assessed by bootstrap analysis with 1000 replicates. The dendrogram (Fig. 4) showed that our isolate was identical with C. blankii CBS1898 and C. blankii CBS6427, which belong to the Stephanoascus clade, while most pathogenic Candida spp. (such as C. albicans, C. tropicalis and C. parapsilosis) belong to the CTG clade. The DNA from the blood sample yielding C. blankii Kw593/18 was also extracted as described previously; the ITS-1 region was amplified by PCR using ITS1 and ITS2 primers and sequenced by using ITS1FS and ITS2 primers [4]. The DNA sequence data of the ITS-1 region matched completely with the corresponding sequence from C. blankii Kw593/18. Despite combination antifungal therapy with amphotericin B and caspofungin for 5 days, the patient died on day 138 of life (Fig. 1).
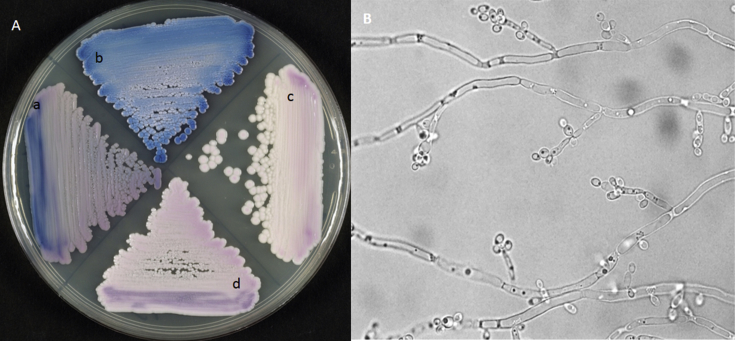
Fig. 2.
(A) CHROMagar Candida showing colony characteristics of the following Candida species after 48 hours' incubation at 37°C: C. blankii (a), C. tropicalis (b), C. auris (c) and C. haemulonii (d). (B) Slide culture of C. blankii on cornmeal agar showing clusters of budding yeast cells with pseudohyphae. Original magnification, ×1000.
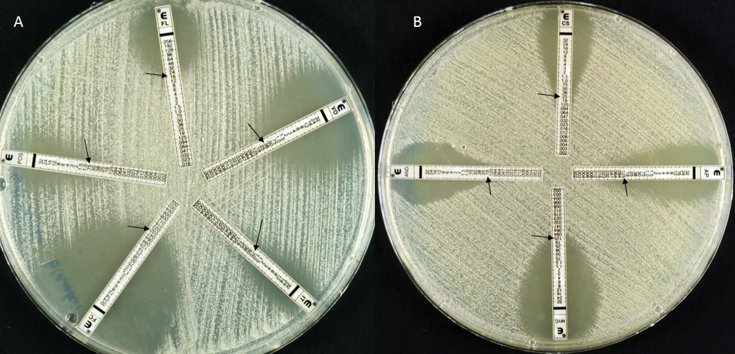
Fig. 3.
(A) Etest susceptibility performed on RPMI 1640 medium showing minimum inhibitory concentrations. (A) Fluconazole (FL), 16 μg/mL, voriconazole (VO), 0.38 μg/mL, itraconazole (IT), 0.75 μg/mL, posaconazole (POS), 0.75 μg/mL, 5-flucytosine (FC), 0.032 μg/mL. (B) Amphotericin B (AP), 0.125 μg/mL, micafungin (MYC), 0.125, caspofungin (CS), 0.25 μg/mL, anidulafungin (AND), 0.19 μg/mL.
Fig. 4.
Neighbour-joining phylogenetic tree based on internally transcribed spacer regions of rDNA sequence data for Candida blankii Kw593/18 from Kuwait together with reference strains of several other closely related as well as many well-known pathogenic Candida spp. Numbers on node branches are bootstrap frequencies.
Discussion
Our report unequivocally establishes the aetiologic role of C. blankii in human infections by repeatedly isolating it from blood specimens of a preterm neonate and also by demonstrating its DNA in blood. PCR sequencing of DNA isolated from blood showed 100% identity with the sequence of the bloodstream isolate. C. blankii was first described from blood of a mink in 1968 by Buckley and van Uden [7]. Two other studies have also described the isolation of this yeast from clinical specimens, including bloodstream of a fungaemia patient [1], [2].
Therapeutic experience with C. blankii fungaemia is limited to only one previous case [2]. The patient experienced clearing the yeast from blood after 3 days of micafungin therapy and was discharged after 14 days of maintenance therapy with no postinfection complication. Our patient, who received antifungal prophylaxis with amphotericin B (14 days) and fluconazole (21 days) and then was treated empirically with amphotericin B, followed by combination therapy of amphotericin B and caspofungin (5 days), died of infection probably due to polymicrobial septicaemia caused by enteric pathogens possibly originating from the leaky gut together with C. blankii.
C. blankii is not a regular component of human skin/mucosal microbiota and is less virulent than C. albicans [2]. Because of identification difficulties, it is probable that some isolates previously identified as S. ciferrii by VITEK 2 were actually C. blankii. Stephanoascus ciferrii has rarely been implicated in human infections [8], [9], [10]. Moreover, like C. blankii, it also shows reduced susceptibility to antifungal agents in previous reports [11], [12]. Candida species exhibiting reduced susceptibility to fluconazole have been implicated in breakthrough infections in patients receiving prophylaxis with this drug for prolonged periods [13]. A limitation of our study is that we determined the MIC values by Etest and not by a reference microdilution method. Although the Etest generally shows good reproducibility and correlation with the broth microdilution method, occasionally the MIC values obtained by the two methods may also show subtle variations [14].
In conclusion, this report highlights the emergence of this newly recognized azole-resistant Candida species in human infections warranting greater understanding of its epidemiology and pathogenic potential.
Acknowledgements
The authors acknowledge cooperation received from the healthcare staff of the neonatal intensive care unit, and excellent technical support provided by M. Asadzadeh, S. Vayalil and O. Al-Musallam of Mycology Reference Laboratory.
Conflict of Interest
None declared.
References
- 1.Zaragoza S., Galanternik L., Vazquez M., Teper A A., Cordoba S., Finquelievich J. Candida blankii: new agent in cystic fibrosis airways? J Cyst Fibros. 2015;14(Suppl. 1):S140. [Google Scholar]
- 2.de Almeida J.N., Jr., Campos S.V., Thomaz D.Y., Thomaz L., de Almeida R.K.G., Del Negro G.M.B. Candida blankii: an emergent opportunistic yeast with reduced susceptibility to antifungals. Emerg Microbes Infect. 2018;7:24. doi: 10.1038/s41426-017-0015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan Z.U., Ahmad S., Al-Sweih N., Joseph L., Alfouzan F., Asadzadeh M. Increasing prevalence, molecular characterization and antifungal drug susceptibility of serial Candida auris isolates in Kuwait. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan Z.U., Ahmad S., Hagen F., Fell J.W., Kowshik T., Chandy R. Cryptococcus randhawai sp. nov., a novel anamorphic basidiomycetous yeast isolated from tree trunk hollow of Ficus religiosa (peepal tree) from New Delhi, India. Antonie Van Leeuwenhoek. 2010;97:253–259. doi: 10.1007/s10482-009-9406-8. [DOI] [PubMed] [Google Scholar]
- 5.Schoch C.L., Seifert K.A., Huhndorf S., Robert V., Spouge J.L., Levesque C.A. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prasad G.S., Mayilraj S., Sood N., Singh V., Biswas K., Lal B. Candida digboiensis sp. nov., a novel anamorphic yeast species from an acidic tar sludge–contaminated oilfield. Int J Syst Evol Microbiol. 2005;55:967–972. doi: 10.1099/ijs.0.63313-0. [DOI] [PubMed] [Google Scholar]
- 7.Buckley H.R., van Uden N. Five new Candida species. Mycopathol Mycol Appl. 1968;36:257–266. doi: 10.1007/BF02050372. [DOI] [PubMed] [Google Scholar]
- 8.Kurtzman C.P., Christie J., Robnett C.J. Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen.nov. and 14 new species combinations. FEMS Yeast Res. 2007;7:141–151. doi: 10.1111/j.1567-1364.2006.00157.x. [DOI] [PubMed] [Google Scholar]
- 9.Demiray T., Hafizoglu T., Koroglu M., Ozbek A., Altindis M. The first case of Stephanoascus ciferrii infection in a newborn and review of literature. Nobel Medicus. 2015;11:97–100. [Google Scholar]
- 10.Villnueva-Lozano H., Trevino-Rangel R.D.J., Hernandez-Balbola C.L., Gonzalez G.M., Martinez-Resendez M.F. An unusual case of Candida ciferrii fungemia in an immunocompromised patient with Crohn's and Mycobacterium bovis disease. J Infect Dev Ctries. 2016;10:1156–1158. doi: 10.3855/jidc.8228. [DOI] [PubMed] [Google Scholar]
- 11.Agın H., Ayhan Y., Devrim I., Gülfidan G., Tulumoglu S., Kayserili E. Fluconazole-, amphotericin-B–, caspofungin-, and anidulafungin-resistant Candida ciferrii: an unknown cause of systemic mycosis in a child. Mycopathologia. 2011;172:237–239. doi: 10.1007/s11046-011-9418-6. [DOI] [PubMed] [Google Scholar]
- 12.Arendrup M.C., Boekhout T., Akova M., Meis J.F., Cornely O.A., Lortholary O., European Society of Clinical Microbiology and Infectious Diseases Fungal Infection Study Group; European Confederation of Medical Mycology ESCMID and ECMM joint clinical guidelines for the diagnosis and management of rare invasive yeast infections. Clin Microbiol Infect. 2014;20(Suppl. 3):76–98. doi: 10.1111/1469-0691.12360. [DOI] [PubMed] [Google Scholar]
- 13.Cuervo G., Garcia-Vidal C., Nucci M., Puchades F., Fernández-Ruiz M., Obed M. Breakthrough candidaemia in the era of broad-spectrum antifungal therapies. Clin Microbiol Infect. 2016;22:181–188. doi: 10.1016/j.cmi.2015.09.029. [DOI] [PubMed] [Google Scholar]
- 14.Cuenca-Estrella M., Gomez-Lopez A., Mellado E., Rodriguez-Tudela J.L. Correlation between the procedure for antifungal susceptibility testing for Candida spp. of the European Committee on Antibiotic Susceptibility Testing (EUCAST) and four commercial techniques. Clin Microbiol Infect. 2005;11:486–492. doi: 10.1111/j.1469-0691.2005.01166.x. [DOI] [PubMed] [Google Scholar]