ABSTRACT
Mutation in minor spliceosome components is linked to the developmental disorder microcephalic osteodysplastic primordial dwarfism type 1 (MOPD1). Here, we inactivated the minor spliceosome in the developing mouse cortex (pallium) by ablating Rnu11, which encodes the crucial minor spliceosome small nuclear RNA (snRNA) U11. Rnu11 conditional knockout mice were born with microcephaly, which was caused by the death of self-amplifying radial glial cells (RGCs), while intermediate progenitor cells and neurons were produced. RNA sequencing suggested that this cell death was mediated by upregulation of p53 (Trp53 – Mouse Genome Informatics) and DNA damage, which were both observed specifically in U11-null RGCs. Moreover, U11 loss caused elevated minor intron retention in genes regulating the cell cycle, which was consistent with fewer RGCs in S-phase and cytokinesis, alongside prolonged metaphase in RGCs. In all, we found that self-amplifying RGCs are the cell type most sensitive to loss of minor splicing. Together, these findings provide a potential explanation of how disruption of minor splicing might cause microcephaly in MOPD1.
KEY WORDS: Cortical development, Microcephaly, Radial glial cells, Minor spliceosome, U11 snRNA, Cell cycle
Highlighted Article: Here, we report the first mammalian model to investigate the role of the minor spliceosome in cortical development and microcephaly – a conditional knockout mouse for Rnu11, the gene encoding the U11 small nuclear RNA.
INTRODUCTION
RNA splicing, the removal of noncoding introns from pre-mRNA transcripts, is an essential step in eukaryotic gene expression. This process is performed by one of two distinct spliceosomes, the major or the minor type (Patel and Steitz, 2003). The minor spliceosome contains four unique small nuclear RNAs (snRNAs) (U11, U12, U4atac and U6atac) and the U5 snRNA, which it shares with the major spliceosome (Tarn and Steitz, 1996a,b). Less than 0.5% of introns require the minor spliceosome for their removal; hence, they are called minor introns (Alioto, 2007). The importance of minor intron splicing in development is underscored by the discoveries that hypomorphic mutation in the U4atac snRNA causes the developmental disorders microcephalic osteodysplastic primordial dwarfism type 1 (MOPD1), Roifman syndrome and Lowry Wood syndrome (He et al., 2011; Merico et al., 2015; Farach et al., 2018). All three disorders, despite being caused by systemic loss of minor splicing, are characterized by microcephaly and dwarfism, which suggests a tissue-specific requirement for the minor spliceosome in development (He et al., 2011; Merico et al., 2015; Farach et al., 2018). Therefore, to understand the mechanism of disease pathogenesis underlying microcephaly in these syndromes, it is important to study the regulation of minor intron-containing genes (MIGs) in cortical development.
Proper cortical development requires a balance between amplification of the neural progenitor cells (NPCs) and neurogenesis. Overproduction of neurons at the expense of self-amplifying NPC divisions can deplete the NPC pool early in cortical development, resulting in microcephaly (Homem et al., 2015). The decision of an NPC to produce a neuron is regulated by cell cycle length, particularly the length of G1 and mitosis (Calegari and Huttner, 2003; Lange et al., 2009; Pilaz et al., 2009, 2016). Cell cycle defects that lengthen these phases would result in increased neuron production. Additionally, the presence of two different NPC types, radial glial cells (RGCs) and intermediate progenitor cells (IPCs), can regulate neurogenesis. RGCs can undergo symmetric proliferative divisions, which produce two RGCs, or asymmetric differentiative divisions, to produce one RGC and either one IPC or one neuron. In contrast, most IPCs undergo symmetric neurogenic divisions, to produce two neurons (Götz and Huttner, 2005). A small fraction undergoes symmetric proliferative division, producing two IPCs, prior to symmetric neurogenic division (Noctor et al., 2004). Ultimately, RGCs are required to maintain the NPC population, while IPCs primarily contribute to neuron production (Kowalczyk et al., 2009). In short, microcephaly can be caused by (1) death of the NPC or neuronal populations; (2) increased neuron production, resulting in depletion of the NPC pool; and/or (3) decreased IPC production, resulting in fewer neurons. It is unclear how these processes are affected by loss of minor splicing in MOPD1, Roifman syndrome and Lowry Wood syndrome. Here, we report the role of minor splicing in mammalian cortical development. For this, we leveraged a novel conditional knockout (cKO) mouse for the gene encoding the U11 snRNA, a crucial component of the minor spliceosome (Kolossova and Padgett, 1997).
RESULTS
Constitutive loss of U11 is embryonically lethal
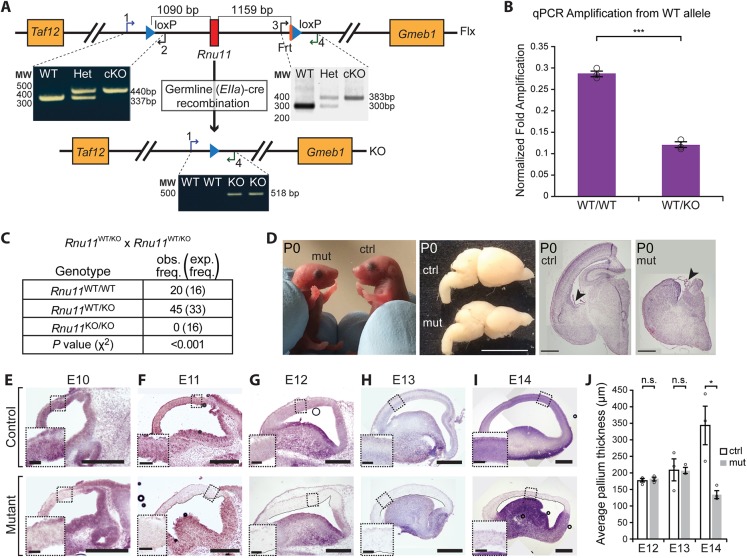
We generated the Rnu11 cKO mouse by engineering loxP sites 1090 bp upstream and 1159 bp downstream of the Rnu11 gene (Fig. 1A, Fig. S1). Successful targeting of the loxP sites was confirmed by long-range nested PCR in targeted embryonic stem (ES) cells (Fig. S1B,C) and further validated by the loss of the wild-type (WT) allele in Rnu11Flx/Flx mice (Fig. 1A). EIIa-Cre-mediated germline recombination generated Rnu11WT/KO mice, which showed the presence of the KO allele that was absent in Rnu11WT/WT genomic DNA (Fig. 1A). Quantitative PCR (qPCR) for the WT allele showed 50% reduction in Rnu11WT/KO mice compared with Rnu11WT/WT mice (Fig. 1B). Intercrossing Rnu11WT/KO mice did not yield Rnu11KO/KO mice (Fig. 1C), indicating embryonic lethality.
Fig. 1.
U11 loss in the developing mouse neocortex causes severe microcephaly. (A) Schematic of the Rnu11 floxed (Flx) allele with positions of the loxP sites (blue triangles), with agarose gel image showing PCR results detecting the upstream (left) and downstream (right) loxP sites. Below is a schematic of the Rnu11 knockout (KO) allele, confirmed by PCR. See also Fig. S1 and Table S7. (B) Results of qPCR detecting the WT allele. See also Table S7. (C) Table showing genotype frequency of pups produced from crosses of Rnu11WT/KO mice. (D) Images of P0 Rnu11WT/Flx::Emx1-Cre+/− (control, ctrl) and Rnu11Flx/Flx::Emx1-Cre+/− (mutant, mut) pups, with lateral view of P0 ctrl and mut brains. Scale bar: 5 mm. The two right composite images show coronal sections of P0 ctrl and mut brains stained with Hematoxylin and Eosin. Arrowheads indicate the choroid plexis. Scale bars: 500 µm. (E-I) ISH for U11 expression (purple signal) in composite images of sagittal sections of the telencephalon from ctrl (top row) and mut (bottom row) E10 (E), E11 (F), E12 (G), E13 (H) and E14 (I) embryos. Scale bars: 500 µm. Insets show higher magnification views of the boxed regions (scale bars: 200 µm). (J) Bar graphs showing the average thickness of the E12, E13 and E14 ctrl and mut pallium. Quantification data are presented as mean±s.e.m. For details of statistical methods, see Table S8. n.s., not significant; *P<0.05; ***P<0.001.
Ablation of U11 in the pallium causes microcephaly
We employed Emx1-Cre mice to specifically ablate Rnu11 in the pallium (Gorski et al., 2002). Emx1-Cre expression begins at embryonic day (E) 9.5 in the neuroepithelium of the pallium, prior to the production of RGCs, IPCs or neurons at E10-E11 (Gorski et al., 2002). Ablation of U11 in the pallium resulted in profound microcephaly at birth in Rnu11Flx/Flx::Emx1-Cre+/− mutant mice, owing to collapse of the cortex and absence of the hippocampus (Fig. 1D). To understand how this microcephaly precipitated, we sought to determine the kinetics of U11 snRNA loss after Emx1-Cre-mediated Rnu11 ablation. In situ hybridization (ISH) for U11 snRNA revealed a reduction in U11 signal (purple) in the E10 mutant pallium, relative to the control (Rnu11WT/Flx::Emx1-Cre+/−) (Fig. 1E). This U11-null domain was expanded in the E11 mutant pallium and, by E12, the majority of cells in the Emx1-Cre domain lacked U11 expression (Fig. 1F,G). We observed scattered U11+ cells at this point (Fig. 1G), which could represent either inefficient Emx1-Cre recombination or migrating interneurons produced in the ventral telencephalon, where Emx1-Cre is not active (Gorski et al., 2002; Bartolini et al., 2013). There were no observable morphological differences between mutant and littermate control pallia at E10, E11 and E12 (Fig. 1E-G). At E13, we observed persistence of the U11-null domain alongside collapse of the lateral ventricle in the mutant telencephalon, without a significant change in pallial thickness (Fig. 1H,J). At E14, the thickness of the mutant pallium was significantly reduced relative to the control (Fig. 1I-J).
Cell death contributes to microcephaly in the U11 cKO mice
We first performed fluorescent ISH (FISH) for U11 to identify the U11-null domain, followed by terminal dUTP nick-end labeling (TUNEL) to detect apoptotic cells. Results showed sporadic loss of U11 snRNA in the E11 mutant pallium without a significant increase in TUNEL+ cells (Fig. 2A). Onset of cell death was observed at E12, with a statistically significant increase in TUNEL+ cells in the mutant mice compared with littermate controls (Fig. 2B), which continued at E13 and E14 (Fig. 2C,D). Because TUNEL marks cells in late stages of apoptosis, we sought to verify the timing of cell death in the mutant pallium using a marker of early apoptosis (Kyrylkova et al., 2012). For this, we employed immunofluorescence (IF) for cleaved caspase 3 (CC3; Casp3 – Mouse Genome Informatics) (Faleiro et al., 1997) at E11 and E12, which confirmed that the onset of cell death was occurring at E12 (Fig. 2E,F).
Fig. 2.
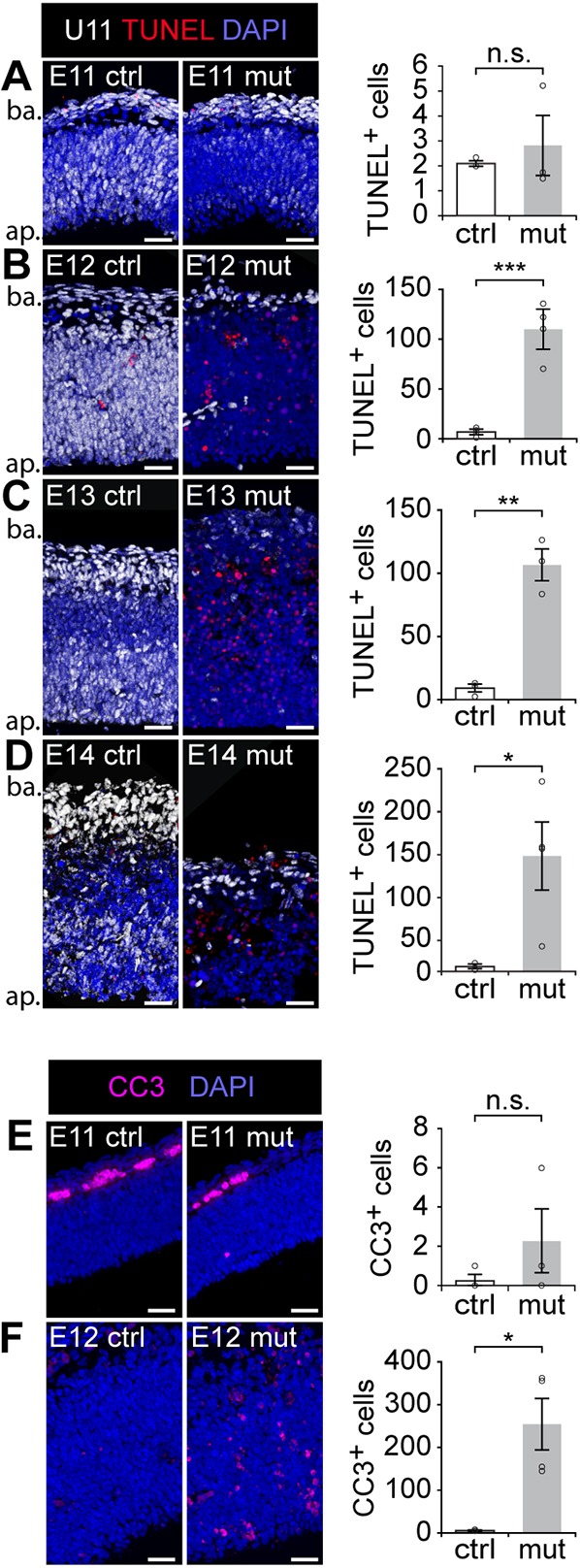
Cell death contributes to microcephaly in the U11 cKO mice. (A-E) Fluorescent ISH (FISH) signal for U11 (white) combined with TUNEL (red) on control (ctrl, left) and mutant (mut, middle) pallium sections from E11 (A), E12 (B), E13 (C) and E14 (D) embryos, with quantification of TUNEL+ cells (right). Ap, apical; ba, basal. (E,F) IF for cleaved caspase 3 (CC3) in ctrl (left) and mut (middle) pallium sections from E11 (E) and E12 (F) embryos, with quantification of CC3+ cells (right). Scale bars: 30 µm. Data are presented as mean±s.e.m. For details of statistical methods, see Table S8. n.s., not significant; *P<0.05; **P<0.01; ***P<0.001.
Microcephaly is predominantly caused by loss of NPCs
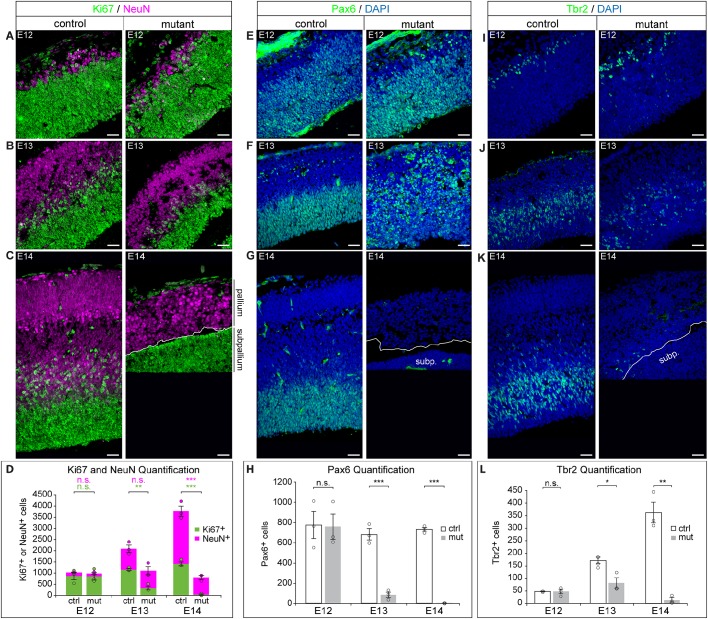
To identify the cell type being lost in the mutant pallium, we investigated the numbers of proliferating NPCs (Ki67+; Mki67+) versus neurons (NeuN+; Rbfox3+) by IF (Wolf et al., 1996; Scholzen and Gerdes, 2000). We found no change in the number of proliferating NPCs in the E12 mutant pallium compared with the control (Fig. 3A,D). However, at E13 and E14, the number of Ki67+ NPCs was significantly reduced in the mutant pallium compared with the control (Fig. 3B-D). Unlike NPCs, we did not observe a significant difference in the number of neurons between the control and mutant at E12 or E13 (Fig. 3A,B,D). To confirm that the neurons present in the E12 and E13 mutant pallium were in fact U11 null, we performed U11 FISH in conjunction with IF for Ki67 and NeuN, which showed the presence of both U11-null NPCs and U11-null neurons (Fig. S2A). Finally, at E14, there were significantly fewer neurons in the mutant compared with the control, which was consistent with the depletion of the NPC population at E13 (Fig. 3B-D). To further investigate whether neurons were dying in the U11-null pallium, we employed IF for Tuj1 (Tubb3) (which marks both immature and mature neurons) (Zhang and Jiao, 2015) and CC3, and quantified the percentage of CC3+ dying cells that were Tuj1+ neurons. In the E12 mutant pallium, the vast majority (93.4±2.1%) of the CC3+ cells were pyknotic, indicative of late-stage apoptotic cells (Fig. S2C). By E13, virtually all CC3+ cells were pyknotic (data not shown). Because pyknotic cells in the late stages of apoptosis are unlikely to retain Tuj1 expression, we only considered nonpyknotic CC3+ cells for the analysis. This quantification revealed that 4.44±2.42% of nonpyknotic CC3+ cells in the E12 mutant pallium were Tuj1+ neurons, which was not significantly higher than that observed in the control (0%; Fig. S2C). Together, these data suggested that most of the dying cells were likely the U11-null NPCs, not the U11-null neurons.
Fig. 3.
U11 loss results in depletion of RGCs. (A-D) IF for Ki67 (green) and NeuN (magenta) on sagittal sections of the control (left) and mutant (right) pallium across E12 (A), E13 (B) and E14 (C), with quantification (D). (E-H) IF for Pax6 (green) on sagittal sections of the control (left) and mutant (right) pallium across E12 (E), E13 (F) and E14 (G), with quantification (H). (I-L) IF for Tbr2 (green) on sagittal sections of the control (left) and mutant (right) pallium across E12 (I), E13 (J) and E14 (K), with quantification (L). Scale bars: 30 µm. Data are presented as mean±s.e.m. For details of statistical methods, see Table S8. subp., subpallium. n.s., not significant; *P<0.05; **P<0.01; ***P<0.001. See also Fig. S2.
Self-amplifying RGCs require the minor spliceosome for survival
We next sought to identify the type of NPC being lost in the U11-null pallium. Using the nuclear RGC marker Pax6, we did not observe a significant difference in the number of RGCs between the control and mutant at E12 (Fig. 3E,H) (Götz et al., 1998). At E13, there was a significant reduction in the number of RGCs with nuclear Pax6 signal in the mutant compared with the control (Fig. 3F,H). However, widespread cytoplasmic Pax6 expression, which was rarely seen in control sections, was observed in the mutant (Fig. 3F). This cytoplasmic staining was primarily observed around condensed nuclei, suggesting that the cytoplasmic Pax6 expression was occurring in dying cells. This was confirmed by colocalization of the cytoplasmic Pax6 expression with CC3 (Fig. S2B-B‴). Therefore, only cells with nuclear Pax6 expression were quantified. At E14, there were few RGCs in the mutant, which was consistent with the small number of Ki67+ cells at this timepoint (Fig. 3D,G,H). Next, we investigated the number of IPCs in the U11-null pallium using the marker Tbr2 (Eomes) (Englund et al., 2005), which revealed no significant difference in the E12 mutant compared with the control (Fig. 3I,L). At E13, we observed a significant reduction in the IPC population in the mutant relative to the control (Fig. 3J,L), which continued at E14 (Fig. 3K,L).
Both the RGC and IPC populations were significantly reduced in the mutant pallium at E13, but not at E12, relative to the control (Fig. 3D,H). However, the trends of RGC and IPC loss across mutant pallium development were not similar: although there was a significant decline in the number of RGCs in the E13 mutant pallium compared with the E12 mutant pallium, there was no significant change in the number of IPCs between these timepoints in the mutant (Fig. S2D,E). A significant decline in IPC number was only observed between E13 and E14 in the mutant pallium (Fig. S2E). Therefore, the decline in RGC number occurred earlier in the mutant pallium than the decline in IPCs, suggesting that IPCs were being produced in the U11-null pallium.
The observation that the E13 control and mutant pallia both contained similar numbers of neurons suggested that neuron production was also occurring in the E12 mutant pallium (Fig. 3B,D). To study neuron production in the mutant pallium, we pulsed pregnant dames carrying E12 embryos with the thymidine analog 5-bromo-2′-deoxyruidine (BrdU), to mark S-phase NPCs, followed by harvest at E13 (Takahashi et al., 1995). After removing pyknotic BrdU+ cells, quantification showed that a significant decrease in NeuN+/BrdU+ cells as a percentage of all NeuN+ cells in the E13 mutant pallium compared with the control. In contrast, there was no significant difference in the percentage of nonpyknotic BrdU+ cells that were NeuN+ between the E13 control (15.7±2.17%) and mutant (15.5±3.5%; Fig. S2F) pallia. The presence of BrdU+ neurons in the mutant indicated that U11-null NPCs produced neurons between E12 and E13. Together, these findings suggested that the majority of cells dying between E12 and E13 were self-amplifying RGCs.
DNA damage and p53 upregulation occur in U11-null RGCs
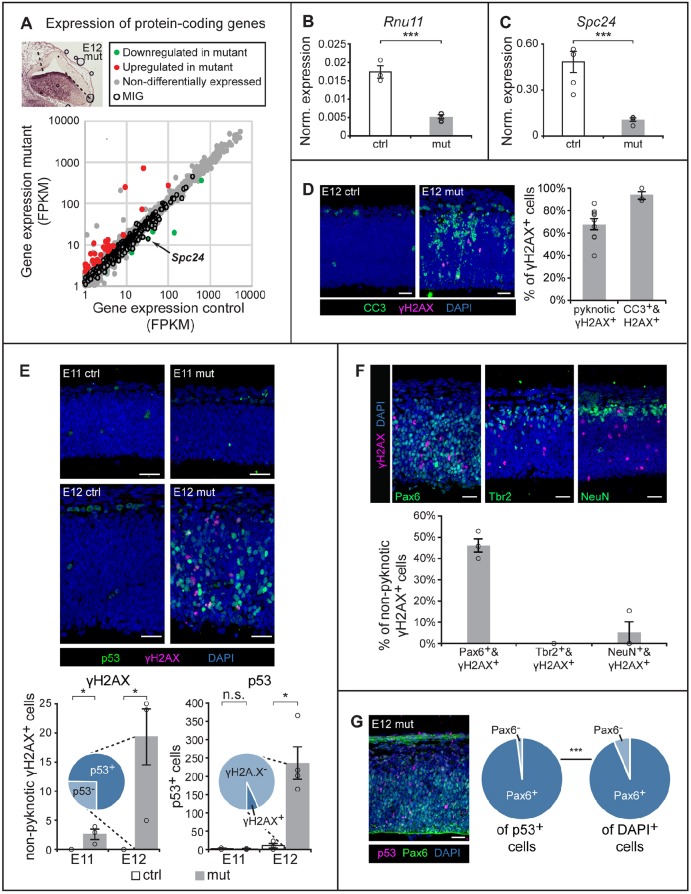
To understand the molecular defects underlying the loss of self-amplifying RGCs, we performed RNA sequencing (RNAseq) on control (n=5) and mutant (n=5) pallia from E12 embryos (Fig. 4A, inset). At this timepoint, U11 loss occurs throughout the entire Emx1-Cre domain without major histological changes, such as shifts in NPC number (Figs 1G and 3D). Confirmation of the pallial dissection was reflected by the expression of Emx1 in the control [19.1 fragments per kilobase per million mapped reads (FPKM)] and mutant (20.3 FPKM). Expression of Dbx1, which marks the pallial-subpallial boundary, and Lhx6, which is expressed in the medial ganglionic eminence (Grant et al., 2012), was below 1 FPKM in both the control and mutant samples (Table S1). Together, these results confirm the integrity of our dissection. Expression of Rnu11 was reduced by 59.2% in the mutant compared with the control, which was further confirmed by quantitative reverse transcriptase-PCR (qRT-PCR) (Fig. 4B, Table S1). The incomplete loss of U11 expression in the mutant likely reflects (1) contamination of non-Emx1-lineage cells of the meninges and blood vessels (Fig. 1G) (Siegenthaler and Pleasure, 2011); (2) contamination from non-Emx1-lineage cells migrating into the pallium (Fig. 1G) (Bartolini et al., 2013); and/or (3) incomplete recombination by Emx1-Cre. Regardless, the mutant samples showed a clear reduction in U11 expression compared with the control, which was in agreement with the ISH analysis (Fig. 1G). Next, we interrogated the expression of all protein-coding genes with ≥1 FPKM in the control or mutant. Overall, there was an increase in the number of genes upregulated [74; ≥2-fold change (FC), P≤0.01] relative to those downregulated (11; ≥2-FC, P≤0.01) in the mutant (Fig. 4A, Table S1). DAVID analysis (Huang et al., 2009) performed on the downregulated protein-coding genes revealed no significant enrichments for gene ontology (GO) terms by Benjamini score. However, the upregulated protein-coding genes were significantly enriched for the GO term ‘intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator’ (Table S2), implicating this pathway in the cell death observed at this timepoint (Fig. 2).
Fig. 4.
U11 loss triggers DNA damage and p53 upregulation in RGCs. (A) Scatterplot depicting the expression of protein-coding genes ≥1 FPKM in the control (x-axis) and mutant (y-axis) E12 pallium. Inset shows the E12 mutant pallium, with dashed lines marking the dissected region. (B,C) Normalized expression, determined by qRT-PCR on E12 control and mutant pallium, for Rnu11 (B) and Spc24 (C). (D) IF for CC3 (green) and γH2AX (magenta) in the E12 control (ctrl) and mutant (mut) pallium, with quantification. (E) IF for γH2AX (magenta) and p53 (green) in E11 and E12 ctrl and mut sagittal pallial sections, with quantification. Inset pie charts show the percentage of γH2AX+ cells that upregulated p53 (p53+) (left) and the percentage of p53+ cells that were γH2AX+ (right). (F) IF for γH2AX (magenta) and Pax6 (green), Tbr2 (green) or NeuN (green), on sagittal sections of the E12 mut pallium, with quantification. (G) IF for p53 (magenta) and Pax6 (green) in the E12 mut pallium, with pie charts showing the percentage of Pax6+ cells of the p53+ population (left) and of all DAPI+ cells (right). Scale bars: 30 µm. Quantification data are presented as mean±s.e.m. For details of statistical methods, see Table S8. n.s., not significant; *P<0.05; ***P<0.001.
To test whether DNA damage and p53 (Trp53) activation were occurring in the E12 mutant, we employed IF with the DNA damage marker γH2AX (γH2AFX) (Sharma et al., 2012), along with antibody against p53. We observed that the majority of γH2AX+ cells were pyknotic (54.1±3.5%), suggesting that they were dying cells (Fig. 4D). To verify this, we employed the early-apoptotic marker CC3, which revealed that 93.2±3.5% of γH2AX+ cells were CC3+ (Fig. 4D). Because most cells undergoing the final stages of apoptosis do not express nuclear markers, we removed all pyknotic γH2AX+ cells from the quantification strategy. Using this approach, we observed significantly more nonpyknotic γH2AX+ cells scattered throughout the E12 U11-null pallium relative to the control (Fig. 4E). Similarly, we observed significantly more cells upregulating p53 throughout the E12 U11-null pallium compared with the control (Fig. 4E). Given the GO term enriched for by the upregulated protein-coding genes, and reports that p53 stabilization occurs in response to DNA damage (Cheng and Chen, 2010), we expected that most γH2AX+ cells would also upregulate p53. Indeed, 74.7±11.9% of γH2AX+ cells were also upregulating p53 (Fig. 4E, left chart). In contrast, only 6.7±2.0% of p53-upregulating cells were also γH2AX+ (Fig. 4E, right chart). At E11, we observed significantly more γH2AX+ cells in the mutant pallium (2.67±0.76) compared with the control (0). However, we did not observe a significant difference in the number of p53-upregulating cells in the control and mutant, suggesting that the precipitation of DNA damage begins at E11, while cell death occurs at E12 (Fig. 4E).
Because the majority of the cells dying between E12 and E13 were self-amplifying RGCs, we next investigated whether RGCs were the cell type accumulating DNA damage and upregulating p53. We found that 46.3±3.2% of γH2AX+ cells were Pax6+ RGCs, while virtually none were NeuN+ neurons (5.1±5.1%) or Tbr2+ IPCs (0%; Fig. 4F). To test whether RGCs were the primary cell type upregulating p53, we also performed IF for p53 and Pax6. This revealed that the vast majority of p53-upregulating cells were Pax6+ RGCs (98.1±0.85%), a significant enrichment compared with the percentage of Pax6+ RGCs found in the mutant pallium (93.61±0.56%; Fig. 4G). In all, we found that U11-null RGCs are the primary cell type both accumulating DNA damage and upregulating p53, which most likely contribute to their widespread death between E12 and E13.
U11 loss results in elevated minor intron retention in MIGs that regulate nucleotide binding, protein transport and the cell cycle
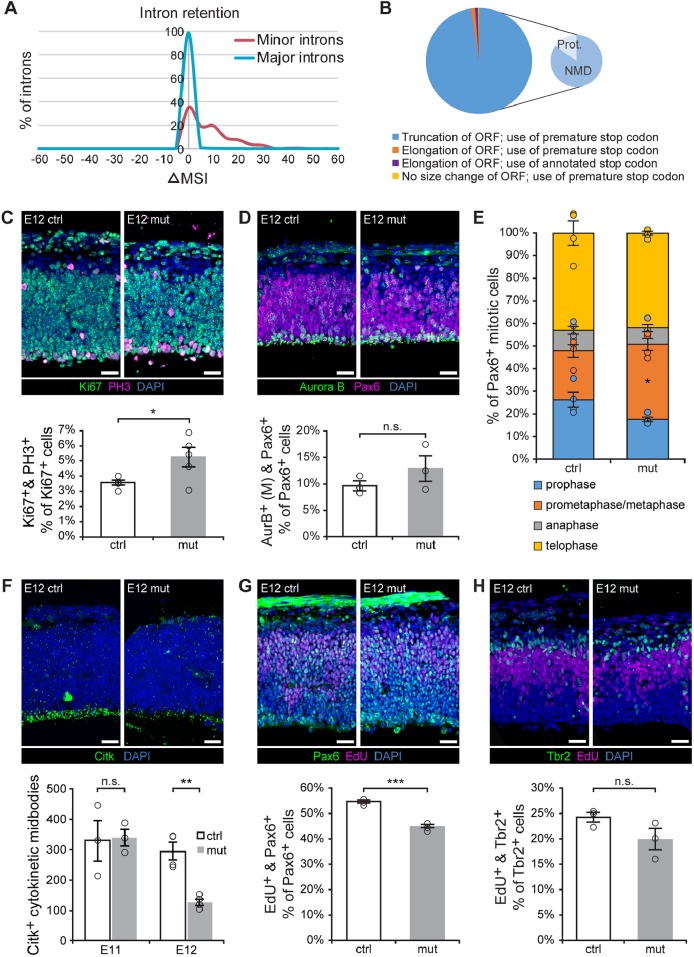
Because U11 is an essential component of the minor spliceosome, the expected result of U11 loss is disruption of minor intron splicing. However, none of the genes upregulated in the E12 mutant pallium that enriched for p53-mediated apoptosis contained minor introns, indicating that their upregulation is a secondary effect of U11 loss. Therefore, we next investigated the expression of the minor spliceosome targets, i.e. the MIGs, and the splicing of their minor introns. Overall, the vast majority of MIGs were not differentially expressed (Fig. 4A). Only one MIG, Spc24, was reduced by >50% in the mutant (Fig. 4A, arrow), which was independently confirmed by qRT-PCR (Fig. 4C). To assess minor intron splicing, we employed the strategy previously described by (Madan et al., 2015), which utilizes a mis-splicing index (MSI) to reflect intron retention. Using this strategy, we identified 299 (of 545) minor introns and 11,187 (of 54,500 randomized major introns) that passed the filtering criteria, indicating that they show some level of intron retention. We then calculated the difference in MSI for each intron between the mutant and the control (mutant MSI – control MSI; ΔMSI) and generated frequency plots (Fig. 5A, Table S3). These revealed a rightward ΔMSI shift for the 299 minor introns, whereas the 11,187 randomized major introns followed a normal distribution (Table S3). This indicated that loss of U11 inactivates the minor spliceosome, resulting in elevated retention of minor, but not of major, introns.
Fig. 5.
U11-null RGCs exhibit S-phase, metaphase and cytokinesis defects. (A) Frequency plot of the change in mis-splicing index (ΔMSI, x-axis) of major (blue) and minor (red) introns. (B) Pie chart showing the effect of minor intron retention on the open reading frame (ORF), for the 186 minor introns with significantly elevated retention, across all annotated MIG transcripts requiring minor intron splicing. The truncation category is further broken down into the percentage of transcripts predicted to be degraded by nonsense-mediated decay (NMD) or to produce protein (prot.). (C) IF for Ki67 (green) and PH3 (magenta) in sagittal sections of the E12 control (ctrl) and mutant (mut) pallium, with quantification. (D) IF for Aurora B (green, AurB) and Pax6 (magenta) in sagittal sections from E12 ctrl and mut pallium, with quantification of mitosis-specific Aurora B staining (M). (E) Quantification of the percentage of Aurora B+ cells in different mitotic phases for E12 ctrl and mut pallium. (F) IF for Citk (green) in sagittal sections of ctrl and mut E12 pallium, quantification of Citk+ cytokinetic midbodies along the ventricular edge in the E11 and E12 control and mutant pallium. (G) IF for Pax6 (green) and EdU (magenta) on E12 ctrl and mut sagittal pallial sections, with quantification. (H) IF for Tbr2 (green) and EdU (magenta) on sagittal sections of E12 ctrl and mut pallium, with quantification. Scale bars: 30 µm. Data are presented as mean±s.e.m. For details of statistical methods, see Table S8. n.s., not significant, *P<0.05, **P<0.01, ***P<0.001. See also Figs S3 and S4, and Tables S3 and S4.
To identify the specific minor introns with elevated retention in the mutant, we employed Student's t-tests to compare the MSI of each minor intron between the control and mutant samples. Of the 299 minor introns, we identified 186 minor introns, found in 178 MIGs, with significantly elevated minor intron retention in the mutant (Table S3). For three of these MIGs (Coa3, Parp1 and Pten), this was confirmed by qRT-PCR (Fig. S3A). Minor intron retention has been predicted to result in the introduction of premature stop codons, subsequently targeting MIG transcripts for degradation by the nonsense-mediated decay (NMD) pathway (Patel et al., 2002). To test whether retention of these 186 minor introns would indeed introduce a premature stop codon, we curated a list of all annotated isoforms of these 178 MIGs and extracted all the isoforms that would require minor intron splicing for their production, resulting in a list of 330 isoforms (Table S4). Bioinformatics analysis showed that minor intron retention in 99.1% of these isoforms would introduce a stop codon prior to the annotated stop codon (Fig. 5B, Table S4). Because stop codons that are found >50 nucleotides (nt) upstream of an exon-exon junction are predicted to trigger the NMD pathway (Nagy and Maquat, 1998), we also interrogated the location of the premature stop codons in these 330 MIG transcripts in relation to their exon-exon junctions. Again, we found that the majority (83.8%) of the premature stop codons would be predicted to activate NMD, resulting in transcript degradation (Fig. 5B, Table S4). For the remaining 53 MIG transcripts, minor intron retention would introduce a stop codon in the 3′ end of the transcript, potentially allowing these transcripts to escape NMD and be translated, resulting in the production of truncated proteins (Fig. 5B, Table S4).
Regardless of whether a MIG transcript would be degraded via NMD or would produce an aberrant protein, the functions executed by these MIGs would likely be compromised. To identify which biological processes would be affected in the mutant, we submitted the 178 MIGs with elevated minor intron retention to DAVID (Huang et al., 2009). Functions significantly enriched for by these MIGs included both broad GO terms, such as nucleotide binding (36 genes) and protein transport (19 genes), and more specific GO terms, such as cell cycle (18 genes) and centrosome (13 genes). The enrichment of these more specific functions suggested that aberrant minor intron splicing might result in cell cycle defects (Fig. S3B, Table S5). Moreover, the only downregulated MIG in the mutant pallium, Spc24, is involved in kinetochore assembly (Fig. 4A,C, Table S1) (McCleland et al., 2004). In all, disruption of minor intron splicing likely affected cell cycle in the E12 mutant pallium.
U11-null RGCs experience S-phase, prometaphase/metaphase and cytokinesis defects
Among the 21 cell cycle-regulating MIGs we identified with significantly elevated minor intron retention, there were two noteworthy enrichments: nine MIGs that regulate mitosis and cytokinesis, and six MIGs that regulate S-phase and DNA replication (Table S5). Therefore, we expected that mitosis/cytokinesis and S-phase would likely be affected in the mutant pallium.
To study mitosis in the U11-null pallium, we performed IF for Ki67 and phospho-histone H3 (PH3), a marker of mitosis (Gurley et al., 1978), and quantified PH3+ mitotic cells as a percentage of the entire Ki67+ cycling cell population, to identify the mitotic fraction of the NPC population (Fig. 5C). This quantification revealed a significantly larger mitotic fraction in the E12 mutant pallium relative to the control (Fig. 5C). To investigate whether a specific stage within mitosis was affected in the U11-null E12 pallium, we employed the mitotic marker Aurora B. The staining pattern of Aurora B corresponds to specific phases within mitosis (Sun et al., 2008) (Fig. S4A); by quantifying cells within these specific phases of mitosis, we were able to identify the fraction of mitotic cells in prophase, prometaphase+metaphase, anaphase and telophase. Aurora B staining was combined with IF for either Pax6 or Tbr2, to assess cell type-specific defects. This staining paradigm revealed a significantly higher percentage of mitotic Aurora B+/Pax6+ RGCs in prometaphase+metaphase in the E12 mutant pallium than in the control, whereas prophase, anaphase and telophase were not significantly different (Fig. 5D,E). In mitotic Tbr2+ IPCs, we did not observe any significant differences in the fractions in prophase, prometaphase+metaphase, anaphase or telophase (Fig. S4B,C). To investigate cytokinesis in the E12 mutant pallium, we performed IF for citron kinase (Citk; Cit), which marks the cytokinetic midbody (Paramasivam et al., 2007). At E12, there was a significant reduction in Citk+ cytokinetic midbodies lining the ventricular edge in the U11-null pallium (Fig. 5F). This reduction was not observed at E11, when the number of Citk+ cytokinetic midbodies across the U11-null pallium was similar to that in the control (Fig. 5F, Fig. S4D). These data suggest that U11 loss causes both prolonged prometaphase/metaphase, specifically in RGCs, and cytokinesis defects.
Next, we explored how minor intron retention in the S phase-regulating MIGs impacted S phase kinetics in the mutant pallium. For this, we pulsed pregnant females with the thymidine analog 5-ethynyl-2-deoxyuridine (EdU; Chehrehasa et al., 2009), followed by harvest 1 h later. EdU detection revealed a significantly lower percentage of EdU+/Pax6+ RGCs in the mutant compared with the control at E12 (Fig. 5G), while there was no significant difference in the percentage of EdU+/Tbr2+ IPCs between the E12 control and mutant pallium (Fig. 5H). In addition, we observed no change in the percentage of EdU+/Pax6+ RGCs in the E11 mutant pallium compared with the control (Fig. S4E). Thus, U11 loss in RGCs causes S-phase defects at E12, whereas U11-null IPCs are spared.
Together, these findings indicate that U11 loss disrupts the splicing of MIGs that are involved in mitosis, cytokinesis and S-phase, resulting in defects in these cell cycle phases. To determine whether other cell cycle phases were affected in the U11-null pallium, we employed the markers cyclin B1 (which is cytoplasmic in G2 phase) and Aurora B (which is localized to condensing chromosomes in cells in G2) (Clute and Pines, 1999; Crosio et al., 2002) (Fig. S4A). Neither quantification of cyclin B1+ G2 cells, nor quantification of the percentage of G2 Aurora B+ RGCs or IPCs revealed a significant difference between the E12 mutant and control pallium (Fig. S4B,F,G), further underscoring that U11 loss causes three significant and specific cell cycle defects, affecting (1) prometaphase/metaphase, (2) cytokinesis and (3) S-phase.
p53 upregulation occurs predominantly in G1 phase
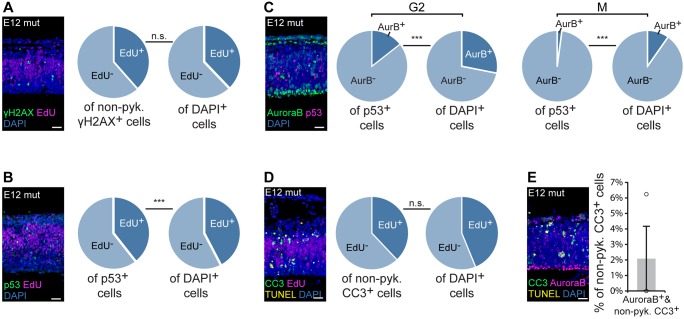
Because the MIGs with significantly elevated minor intron retention enriched for cell cycle functions, we expected that the observed cell cycle defects were the primary cause of RGC death, whereas the observed DNA damage and p53 upregulation were secondary effects. To determine whether DNA damage and p53 upregulation were occurring in NPCs in S-phase and mitosis/cytokinesis, we combined IF for either γH2AX or p53 with different cell cycle markers. To determine whether cells accruing DNA damage were in S-phase, we employed the aforementioned EdU pulse strategy. IF for γH2AX combined with EdU detection revealed that 38.5±9.3% of γH2AX+ cells were also EdU+, similar to the total percentage of EdU+ cells in the mutant pallium (37.8±1.0%; Fig. 6A). We did not observe any γH2AX+/PH3+ cells in the E12 mutant pallium, indicating that cells with DNA damage were not in mitosis (Fig. S5).
Fig. 6.
p53 upregulation and apoptosis predominantly occur in either cells in G1 or postmitotic cells. (A) IF for γH2AX (green) and EdU (magenta) in sagittal sections of E12 mutant (mut) pallium, with pie charts showing the percentage of EdU+ cells of the nonpyknotic (non-pyk.) γH2AX+ and DAPI+ populations. See also Fig. S5. (B) IF for p53 (green) and EdU (magenta) in sagittal sections of E12 mut pallium, with pie charts showing the percentage of EdU+ cells of the p53+ and DAPI+ populations. (C) IF for Aurora B (green, AurB) and p53 (magenta) in sagittal sections of the E12 mut pallium, with pie charts showing the percentage of G2 Aurora B+ cells of the p53+ and DAPI+ populations (left), and the percentage of mitotic (M) Aurora B+ cells of the p53+ and DAPI+ populations (right). (D) IF for CC3 (green), EdU (magenta) and TUNEL (yellow) in sagittal sections of E12 mut pallium, with pie charts showing the percentage of EdU+ cells of the nonpyknotic CC3+ cell population. (E) IF for CC3 (green), Aurora B (magenta) and TUNEL (yellow) in sagittal sections of the E12 mut pallium, with quantification. Scale bars: 30 µm. Quantification data are presented as mean±s.e.m. For details of statistical methods, see Table S8. n.s., not significant, ***P<0.001.
To investigate convergence between p53 upregulation and cell cycle phase, we employed the aforementioned EdU pulse paradigm along with IF for Aurora B. In the E12 mutant pallium, approximately one third (32.5±4.4%) of p53-upregulating cells were also EdU+, a significantly lower percentage than the total percentage of EdU+ cells present in the E12 mutant pallium (42.3±1.2%; Fig. 6B). We then leveraged Aurora B to interrogate p53 upregulation in G2 and mitosis. Quantification of cells with G2-specific Aurora B staining in the E12 mutant pallium revealed that 14.4±2.1% of p53-upregulating cells were in G2 phase, a significantly lower percentage than would be expected based on the total percentage of G2 cells in the mutant pallium (26.5±1.4%; Fig. 6C). Similar quantification of mitotic Aurora B+ cells showed that 2.4±0.3% of p53-upregulating cells were mitotic, which was significantly lower than expected from the total percentage of mitotic cells in the mutant pallium (8.5±0.5%; Fig. 6C). Thus, approximately half of p53-upregulating cells were not labeled by these S-, G2- and M-phase markers, suggesting that these p53-upregulating cells were in G1 phase.
Given that we observed a relationship between p53 upregulation and cell cycle phase, we sought to determine whether apoptosis in U11-null cells was also linked to cell cycle phase. Because pyknotic cells in later stages of apoptosis would not express cell cycle markers, we employed IF for CC3, to mark cells in early-to-late apoptosis, and TUNEL, to mark cells in late stages of apoptosis. For our analysis, we excluded pyknotic TUNEL+ cells, so that we only considered cells in early stages of apoptosis. First, to investigate S-phase cells, we employed the aforementioned EdU pulse paradigm. Of the CC3+ cells in the E12 mutant pallium, 38.0±19.1% were also EdU+, which was expected based on the total percentage of EdU+ cells in the mutant pallium (42.2±1.2%; Fig. 6D). To assess G2 and mitotic cells, we employed IF for Aurora B. In the E12 mutant pallium, we observed virtually no CC3+/Aurora B+ cells (2.1±2.1%; Fig. 6E), indicative of minimal death of cells in G2 and M phases. Together, these findings indicate a relationship between apoptosis and cell cycle that mirrors that of p53 upregulation and cell cycle: few dying cells are in G2 or M phase, and less than half are in S phase. Therefore, the majority of dying cells in the E12 mutant pallium are most likely in G1 phase or are neurons.
DISCUSSION
Patients with MOPD1, Roifman syndrome or Lowry Wood syndrome are thought to escape embryonic lethality owing to impairment, but not complete inactivation, of minor spliceosome activity (He et al., 2011; Merico et al., 2015; Farach et al., 2018). Despite the systemic impairment of minor splicing, the specific phenotypes observed in the brain and limbs of patients indicate tissue/cell-type specific sensitivity to minor spliceosome activity (He et al., 2011; Merico et al., 2015; Farach et al., 2018). Even within the brain, progenitor cells of the cortex are the most sensitive to inhibited minor splicing, as evidenced by the consistent set of cortical abnormalities observed in MOPD1 (Merico et al., 2015). However, investigation into the requirement of minor splicing for cortical development, and whether this requirement is cell type-specific within the cortex has, until now, been unexplored.
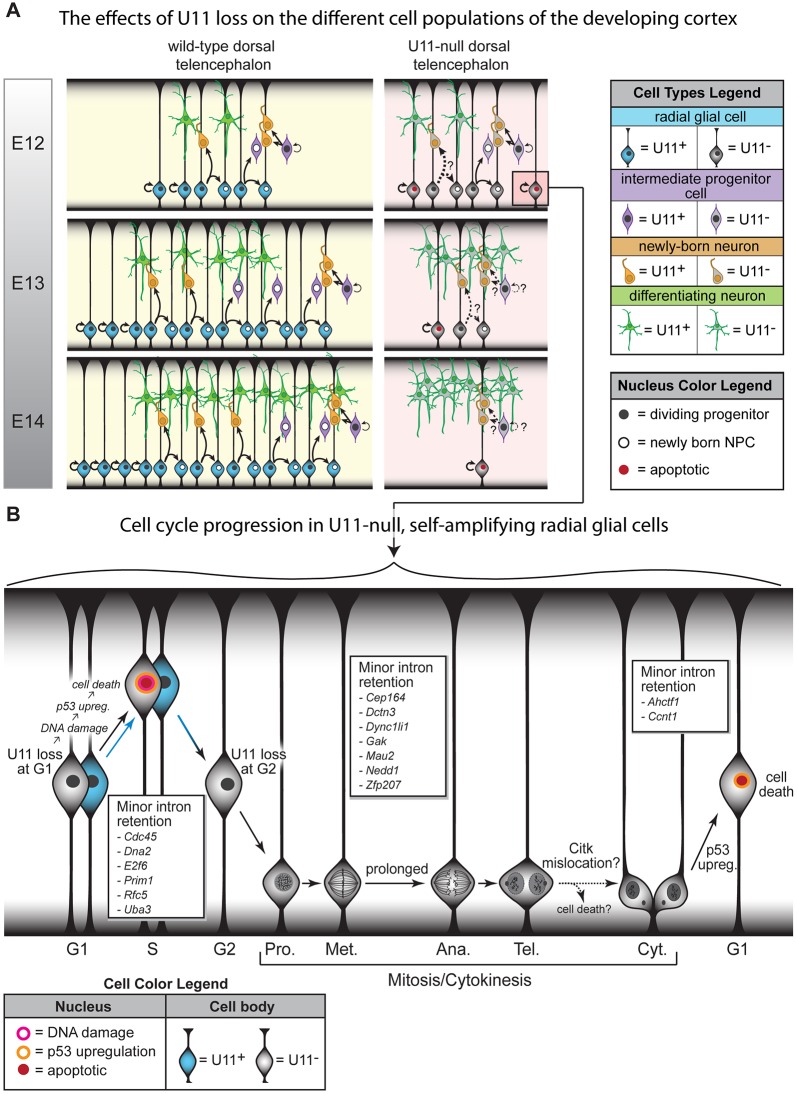
Here, we present the first mammalian model of microcephaly caused by minor spliceosome inactivation (Fig. 1D), allowing us to study the pathogenesis of microcephaly in disorders caused by minor spliceosome disruption. Unexpectedly, we found that even within the developing cortex, different cells showed different sensitivities to U11 loss. Specifically, self-amplifying RGCs were highly sensitive to minor spliceosome inactivation, while RGCs undergoing differentiative divisions, IPCs and neurons were less affected (Figs 3, 7A and S2C-F). We did not observe a significant difference between the percentage of nonpyknotic BrdU+ cells that were NeuN+ neurons in the E13 control and mutant pallium, suggesting similar rates of neurogenesis from the control and U11-null NPCs labeled at E12 (Fig. S2F,G). These findings are discordant with the similar numbers of neurons observed in the control and mutant at E13 (Fig. 3D), which could be explained by compensatory neuron production prior to the BrdU pulse. Together, our findings show that minor spliceosome inactivation in the developing mouse cortex predominantly affects the survival of self-amplifying RGCs and might shift RGCs to neurogenic divisions. Ultimately, this would deplete the RGC pool (Figs 3H and 7A), resulting in the observed microcephaly (Fig. 1D).
Fig. 7.
Model of cortical development in U11 cKO mice. (A) Schematic representing cortical development in wild-type and mutant embryos, from E12 to E14. Black arrows show type of cell division; arrow thickness denotes relative frequency of that division. Question marks and dashed lines indicate events that remain to be explored. (B) Schematic of proliferating RGCs progressing through the cell cycle in the late E11/early E12 pallium. Arrow color denotes the presence (blue) or absence (black) of U11. MIGs with significantly elevated minor intron retention, and with known functions at specific cell cycle phases, are listed in boxes. Ana., anaphase; Cyt., cytokinesis; Met., metaphase; Pro., prophase; Tel., telophase. See also Table S5.
In the developing mouse cortex, the U11-null RGCs experience multiple cell cycle defects. For example, fewer U11-null RGCs were in S-phase (Fig. 5G), suggesting faster S-phase progression or death in late G1/early S-phase. U11-null RGCs also displayed prolonged prometaphase/metaphase (Fig. 5C-E), indicative of spindle assembly defects. Because prolongation of mitosis – and particularly of prometaphase/metaphase – has also been linked to increased neurogenic drive in cortical NPCs (Pilaz et al., 2016), this mitotic defect might drive U11-null RGCs to produce neurons. In addition, we observed fewer Citk+ cytokinetic midbodies along the ventricular edge where RGCs divide, without a change in the fraction of mitotic RGCs in telophase (Fig. 5E,F). This could be caused by RGC death prior to cytokinesis or improper Citk localization to the midbody. Because proper Citk localization is required for cytokinesis, mislocalization would cause aberrant cytokinesis, resulting in RGC death postdivision (Di Cunto et al., 2000). In addition to these cell cycle defects, we observed accumulation of DNA damage at E11, followed by further DNA damage accumulation and p53 upregulation in RGCs at E12 (Fig. 4E-G). The majority of RGCs with DNA damage also upregulated p53 (Fig. 4E). Thus, activation of the DNA damage response pathway in these cells likely results in the stabilization of p53 protein, as has been reported (Williams and Schumacher, 2016). However, the vast majority of p53-upregulating RGCs did not have DNA damage, suggesting that DNA damage accumulation is not the sole cause of p53 upregulation and cell death in the mutant pallium (Fig. 4E). For these RGCs, we expect that p53 upregulation is triggered by prometaphase/metaphase prolongation and cytokinesis defects, as has been proposed (Fig. 5C-E) (Uetake and Sluder, 2010; Ganem et al., 2014). This is consistent with the observation that most p53-upregulating cells were not in S-, G2- or M-phases, suggesting that the majority of p53-upregulating cells were either NPCs in G1 phase or neurons (Fig. 6B,C). Because the vast majority of p53-upregulating cells were Pax6+ (Fig. 4G), most of these p53-upregulating cells were likely RGCs in G1 phase (Fig. 6B,C). RNAseq of the pallium revealed that upregulated genes enriched for ‘intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator’, which is consistent with the observed DNA damage and p53 upregulation in RGCs (Fig. 4F,G, Table S2). Two of the genes enriching for this functional category were the well-known p53-mediated apoptosis effectors Puma (Bbc3) and Noxa (Pmaip1) (Table S2). Upregulation of these p53-mediated apoptosis effectors is likely specific to RGCs, particularly those in G1 phase, because p53 upregulation was observed in RGCs (Figs 4G and 6B-D) and the majority of cells in the E12 pallium are RGCs (Fig. 3E).
In essence, all of these RGC-specific cellular defects are caused by minor spliceosome inactivation, i.e. mis-splicing of MIGs. In the U11-null pallium, 62% of minor introns were significantly retained compared with the control (Table S3), 21 of which are involved in cell cycle regulation (Fig. S3B, Table S5). Notably, four of these cell cycle-regulating MIGs – Cdc45, Dna2, Prim1 and Rfc5 – regulate DNA replication and S-phase progression (Table S5); therefore, disruption of their function likely results in DNA damage and cell death in S-phase, which is consistent with the observed cellular defects (Figs 4D-E, 5G and 7B). Ineffective DNA damage repair, owing to minor intron retention in the 13 MIGs regulating this process (Table S6), would further contribute to DNA damage accumulation and the subsequent p53 upregulation. This pathway might underlie the DNA damage observed in the E11 mutant pallium, prior to p53 upregulation (Figs 4E and 7B). Disrupted function of many of the remaining cell cycle-regulating MIGs, such as Ahctf1, Dctn3, Dync1li1, Gak, Mau2 and Zfp207, is reported to cause spindle assembly defects, chromosome mis-segregation, and aberrant cytokinesis (Table S5). Thus, minor intron retention in these genes likely contributes to the prolonged prometaphase/metaphase and cytokinesis defects observed in U11-null RGCs at E12, which might in turn trigger the observed p53 upregulation in the absence of DNA damage (Figs 4E, 5E,F and 7B). The identification of these subsets of MIGs, which show significantly elevated minor intron retention and the functions of which track with the observed cellular defects in the U11-null pallium, opens new avenues of research for scientists investigating the pathogenesis of the developmental disorders linked to minor spliceosome inactivation.
Of the 186 minor introns that showed significantly elevated retention in the U11-null pallium, we predicted that the majority would result in NMD (Fig. 4B, Table S4). However, none of the 147 MIGs containing these retained minor introns were downregulated in the E12 U11-null pallium (Tables S1 and S4). This is in agreement with the finding that mutation in an essential minor spliceosome protein (Rnpc3) in zebrafish did not affect expression levels of 90% of MIGs, while still resulting in elevated minor intron retention (Markmiller et al., 2014). These findings suggest that inactivation of the minor spliceosome, while disrupting minor intron splicing, does not necessarily result in downstream changes in overall gene expression. This could be a result of a transcriptional feedback loop that drives compensatory upregulation of MIG transcription in response to impaired function of these MIGs (Merico et al., 2015). Translation of these transcripts could result in aberrant protein production, with loss- or gain-of-function effects, contributing to the RGC-specific cellular defects and death.
In both the U11-null pallium and in MOPD1 patients, the observed differential sensitivity to minor spliceosome inactivation might be informed by differences in MIG expression across cell types, where higher MIG expression is correlated with increased sensitivity to minor spliceosome inactivation. Furthermore, if minor introns themselves display differential sensitivity to minor spliceosome inactivation, as our data suggest (Table S3), then the identity of MIGs expressed in specific cell types could inform that cell type's susceptibility to minor spliceosome inactivation. In all, our findings establish how minor spliceosome inactivation in the developing mammalian cortex leads to microcephaly. Moreover, we are the first to identify cell type-specific requirements for minor spliceosome activity within the developing cortex, raising new questions about the mechanisms underlying cell type-specific sensitivity to minor spliceosome activity in mammalian development and disease.
MATERIALS AND METHODS
Animal procedures and generation of the Rnu11 cKO mouse
All mouse procedures were performed according to the protocols approved by the University of Connecticut Institutional Animal Care and Use Committee, which ensures adherence to the U.S. Public Health Service Policy on the treatment of laboratory animals. The Rnu11 cKO mouse was generated by the University of Connecticut Health Center. A single targeting construct was utilized to introduce both loxP sites into the Rnu11 locus in mouse ES cells (Fig. S1A). This construct contained a 5′ loxP site 1090 bp from the Rnu11 gene. Immediately upstream of the 3′ loxP site was a phosphoglycerine kinase (PGK)-neomycin (Neo) cassette flanked by Frt sites. Additionally, this construct contained a PGK-diphtheria toxin A (dTA)-negative selection cassette downstream of the 3′ arm of homology. This construct was electroporated into 129X1/SvJ mouse ES cells. Successful targeting was verified by G418-mediated positive selection. Subsequently, nested long-range PCR was employed to confirm successful homologous recombination at both the 5′ and 3′ loxP sites (Fig. S1C). Two ES cell clones, F4 and H8, were then used for morula aggregation using C57BL/6 embryos to generate a chimera. Germline transmission and ablation of the Neo cassette was verified by introducing germline Flp recombinase. Proper loxP site placement in the resulting mouse line was confirmed by PCR (Fig. 1A, Table S7) to identify loss of the WT allele in the homozygous floxed line and by breeding in the germline EIIa-Cre (Lakso et al., 1996) (Fig. 1A). Emx1-Cre was bred into the Rnu11 cKO line to target Rnu11 for removal in the developing forebrain (Gorski et al., 2002). The primers used for Rnu11, Cre and Emx1-Cre zygosity genotyping PCRs are listed in Table S7. All experiments were performed using male and female Rnu11WT/Flx and Rnu11Flx/Flx mice that were Emx1-Cre+/−. For matings intended for embryonic harvests, the plug date was considered E0; on the harvest date, the embryonic timepoint was further verified by Theiler staging of the embryos.
qPCR
qPCR using genomic DNA
To determine Rnu11 heterozygosity and loss of the WT allele, 25 ng genomic DNA (gDNA) from Rnu11WT/KO and Rnu11WT/WT mice was used for qPCR with Rnu11 cKO, 5′ loxP primers (Table S7). Equal gDNA input was controlled with amplification for Hist1h1a, an intronless gene, which was then used to normalize amplification for Rnu11.
qRT-PCR
Loss of U11 expression was confirmed by qRT-PCR with U11 primers (Table S7) on cDNA prepared from total RNA extracted from the pallium of control (n=3) and mutant (n=4) E12 embryos. In this case, Neat1, a noncoding RNA that did not show change in expression between the control and mutant samples by RNAseq, was used to normalize expression of U11 by qRT-PCR (Hutchinson et al., 2007) (Table S1). The same cDNA was used to determine expression of Spc24, using primers designed in the 3′ UTR of the gene (Table S7). The Cq values obtained for Sfrs10 (Tra2b) (Table S7) were used to normalize expression of Spc24 (Banday et al., 2014). For minor intron retention in Coa3, Parp1 and Pten, expression values of exon-intron (unspliced) product were normalized using Cq values generated using intergenic primers (Table S7) to offset any gDNA contamination (Banday et al., 2014). Expression values of exon-exon (spliced) product were normalized using Sfrs10 Cq values (Banday et al., 2014). Minor intron retention for each of these MIGs was calculated by dividing the normalized fold expression values of the unspliced product by the normalized fold expression values of the spliced product. Statistical significance was determined by two-tailed Student's t-tests (Table S8).
Hematoxylin and Eosin staining
Frozen 16 µm coronal sections of P0 mouse brains were cured at room temperature for 15 min, followed by incubation in PBS for 15 min. The tissue was then dehydrated using an ethanol gradient (25%, 50% and 75%). Slides were washed twice in 100% ethanol for 30 s each, with agitation, then washed twice in 95% ethanol for 30 s each, with agitation. Following a rinse in running tap water, the sections were incubated in Harris Hematoxylin (Thermo Fisher Scientific, 6765001) for 7 min. Excess Hematoxylin was removed by washing in running water for 30 s. The slides were briefly agitated in 1% acid alcohol (1% HCl in ethanol), then briefly rinsed in running water. Sections were then incubated for 2-3 min in saturated lithium carbonate (1.54% in dH2O), followed by a 5 min incubation in tap water. After a 30 s wash in 80% ethanol with agitation, slides were incubated in an Eosin Y-Phloxine B solution (0.1% Eosin Y, 0.01% Phloxine B, 0.45% glacial acetic acid, 82.9% ethanol) for 1 min. The sections were then washed in 95% ethanol for 30 s with agitation, followed by two 30 s washes in 100% ethanol with agitation and incubation in 100% ethanol for 1 min. After washing in xylene for 30 s with agitation, the slides were washed in xylene for 1 min, then mounted using Permount mounting medium (Fisher Scientific, SP15-100).
ISH
Cryosections (16 µm) of mouse heads (for E10-E12 and P0), telencephalons (E13-E14) and whole brains (for P21) (n=3 for each timepoint and genotype) were used for section ISH. Section ISH was performed using an antisense, digoxigenin-labeled U11 RNA probe, which was detected using either alkaline phosphatase or fluorescent labeling (FISH), as described in Baumgartner et al. (2014). The U11 probe was generated using the U11 expression primers in Table S7.
TUNEL assay
Cryosections (16 µm) of either embryonic mouse heads (for E10-E12) or telencephalons (E13-E14) were used for TUNEL analysis. The in situ cell death detection kit, TMR red (Roche, 12156792910), was used, according to the manufacturer's instructions.
IF
IF was performed using one of three protocols. Staining for Tbr2 was performed as described by Arnò et al. (2014). For staining with anti-NeuN antibody, heat-mediated antigen retrieval was performed using citrate buffer, according to the manufacturer's instructions (Vector Laboratories, H-3300), followed by 10 min PBS washes (3×). After three 10 min washes in 0.5% Triton X-100 in PBS (PBT), sections were incubated in 10% fetal bovine serum (FBS) in PBS for 1 h at room temperature. Sections were then incubated in a PBS solution containing 1% FBS, 0.1% Triton X-100, and the primary antibody overnight at 4°C. Following 5 min washes in PBT (10×), incubation with the secondary antibody (1:500) was performed similarly to the primary antibody incubation, except for 2 h at room temperature. The slides were then washed five times with PBT, followed by three PBS washes and mounting with ProLong Gold Antifade Mountant (Thermo Fisher Scientific, P36930). For all other antibodies, staining was performed as described in (Karunakaran et al., 2015). Primary antibodies were diluted to 1:50 (mouse anti-CRIK/CITK, BD Biosciences, 611376; rabbit anti-cyclin B1, Cell Signaling Technology, 4138; MoBu-1 clone mouse anti-BrdU, Santa Cruz Biotechnology, sc-51514), 1:100 (mouse anti-Aurora-B/AIM1, BD Biosciences, 611082; mouse anti-Pax6, Developmental Studies Hybridoma Bank, PAX6-b), 1:200 (mouse anti-γH2AX, MilliporeSigma, 05-636; rabbit anti-NeuN, Abcam, ab17787), 1:300 (rabbit anti-cleaved caspase 3, Cell Signaling Technology, 9665S; mouse anti-KI67, BD Biosciences, 556003; rabbit anti-p53, Leica Microsystems, p53-CM5p; rabbit anti-Pax6, MilliporeSigma, AB2237), or 1:500 (rabbit anti-PH3, Bethyl Laboratories, IHC-00061; rabbit anti-TBR2, Abcam, ab23345).
BrdU and EdU pulse experiments
Timed-pregnant Rnu11WT/Flx::Emx1-Cre+/+ females were first weighed and peritoneally injected with 0.2 µl 25 mM BrdU or 5-ethynyl-2′deoxyuridine (EdU) in PBS per 100 mg body weight, 1 h prior to embryonic harvest. BrdU incorporation was detected using the BrdU-specific MoBu-1 clone anti-BrdU antibody (Santa Cruz Biotechnology, sc-51514) (Liboska et al., 2012). EdU detection was then performed on sections of the pallium, using the Click-iT EdU Alexa Fluor 647 imaging kit (Thermo Fisher Scientific, C10340) in accordance with the manufacturer's instructions.
Image acquisition and quantification
Processed slides were imaged with a Leica SP2 confocal microscope, where settings for laser intensity, excitation-emission windows, gain and offset conditions were identical between control and mutant sections on a given slide. For every slide that was processed for ISH, FISH or IF, the slide contained at least one control and one mutant section. Sections serial to those that revealed loss of U11 by ISH analysis were used for IF staining, and images were collected in regions of the pallium that showed U11 loss in the mutant by ISH (Fig. 1E). For each channel for every experimental condition, confocal imaging settings were optimized for fluorescence in the control section on a given slide, and maintained for all other sections on that slide. For TUNEL, CC3, γH2AX and p53 imaging, collection settings were adjusted using the control section. These settings were maintained when imaging the mutant section(s) on the same slide. Further processing was performed on IMARIS v8.3.1 (Bitplane) and Adobe Photoshop CS4 (Adobe Systems). All image processing in IMARIS and Photoshop was identical between images of control and mutant sections from the same slide. Manual quantification was performed using the spot tool in the IMARIS software, on 240 µm-wide columns of the pallium; statistical significance was calculated using two-tailed Student's t-tests (Table S8). For quantification of mitotic phases, as identified by Aurora B staining (Fig. S4A), prometaphase and metaphase cells were summed together into a single category (prometaphase/metaphase). For analysis of Pax6+ and Tbr2+ cell quantification across mutant cortical development, statistical significance was determined by one-way ANOVA, followed by the post hoc Tukey test (Table S8). For comparisons between the γH2AX+, p53+ or CC3+ subpopulation and all 4′,6-diamidino-2-phenylindole-positive (DAPI+) cells in the E12 mutant pallium, statistical significance was determined by Fisher exact test (Table S8). Specifically, for each n, a separate Fisher exact test was performed on quantification data from a 240 µm-wide column of the mutant pallium. The P-values from these Fisher exact tests were then combined using Fisher's method (Table S8).
Bioinformatics analysis
RNAseq
The pallium was dissected from E12 (n=5 Rnu11WT/Flx::Emx1-Cre+/−, n=5 Rnu11Flx/Flx::Emx1-Cre+/−) embryos, followed by RNA extraction using TRIzol (Thermo Fisher Scientific, 15596018), according to the manufacturer's instructions. Sample preparation and sequencing were performed by the University of Connecticut Center for Genome Innovation. The total RNA library was prepared by Illumina TruSeq Stranded Total RNA Library Sample Prep Kit (RS-122-2201) with Ribozero to remove ribosomal RNA, and then sequenced using Illumina NextSeq 500. Each sample was barcoded so that the sequencing could be multiplexed. In total, we obtained 189,989,434 paired-end 151 bp reads for the control (n=5) and 219,849,711 paired-end 151 bp reads for the mutant (n=5) samples.
Gene expression
First, reads of individual replicates were merged and mapped to the mm10 genome (UCSC genome browser) using Hisat2 (options –no-discordant and –no-mixed), with an overall alignment rate of 85% (Kim et al., 2015). Reads that mapped to multiple locations in the genome were removed. Because the samples were obtained from C57BL/6-129X1/SvJ mice, SNVQ was run to call single nucleotide variations (SNVs) (Duitama et al., 2012), which were used to create a new reference genome. Reads from all replicates were then mapped separately to the newly generated reference genome using Hisat2. Isoform expression was determined by IsoEM2, an expectation-maximization algorithm that bootstraps samples in order to determine an expression value in FPKM (Mandric et al., 2017). Gene expression was then calculated by taking the sum of the FPKM values of a given gene's constituent isoforms. Finally, differential gene expression (≥2-FC, P≤0.01) was determined by IsoDE2 (Mandric et al., 2017). Briefly, this tool utilizes the 200 bootstrapping iterations to compute a confident log2FC, which is the maximum log2FC that can be computed within the specified P-value. A log2FC of 100 is reported when the FPKM value for one of the conditions was 0 in one of the bootstrapping iterations.
Splicing efficiency
To assess minor intron splicing, sequences of the 555 minor introns were curated from the U12DB (Alioto, 2007) and blasted using Ensembl83 to obtain the minor intron coordinates in the canonical transcript. Sequences of eight minor introns (found in Arpc5l, Cacna1c, Exosc2, Golga7, Ift80, Mlst8, Tmem161b and Morc4) that did not have 100% identity with any annotated intron in the Ensembl database were removed from our analysis. All gene and exon coordinates from the mm10 genome were obtained from a Biomart file (Ensembl v84). Major intron coordinates were downloaded from the UCSC genome browser (Ensembl v84) and randomized after removal of introns smaller than 4 nt.
Reads from each of the biological replicates were separately mapped with Hisat2 to the previously described post-SNVQ generated reference genome. Reads that mapped to multiple locations were then filtered out. To determine splicing efficiency of minor introns, we employed the strategy described by (Madan et al., 2015). Briefly, all reads mapping to an exon-intron boundary were extracted and quantified using BEDTools (Quinlan and Hall, 2010). The MSI was then calculated for each intron by dividing the number of exon-intron junction reads by the number of reads mapping to the canonical exon-exon splice-junctions. Only introns with >4 exon-intron boundary reads, ≥1 read aligning to either the 5′ or 3′ splice site, and >95% intron coverage in at least one sample were used for this analysis (Madan et al., 2015). As a control for the 545 minor introns subjected to these criteria, we also generated 100 lists of 545 randomized major introns (54,500 randomized major introns in total), to which we applied the same criteria. For each intron that passed the filtering criteria, we calculated the difference between the mutant and the control (mutant MSI – control MSI; ΔMSI). To determine which minor introns were retained significantly more in the mutant, we compared the average MSI of each minor intron between the control and mutant samples using two-tailed Student's t-tests (Table S3).
ORF analysis
To analyze the effect of minor intron retention on the ORF, we curated all annotated isoforms of the 178 MIGs containing minor introns with significantly elevated retention in the mutant from Ensembl (v84). From this list of isoforms, we extracted only isoforms that would require minor intron splicing for their production, i.e. the blasted minor intron coordinates match one of the annotated intron coordinates 100%, resulting in 330 isoforms. Comparison of the annotated ORF sequence, with the sequence of the isoform containing a retained minor intron, was performed to determine whether minor intron retention would result in introduction of a premature stop codon in these isoforms. Isoforms were then binned into one of four categories: (1) truncation of the ORF, owing to a premature stop codon; (2) elongation of the ORF, with usage of a premature stop codon; (3) elongation of the ORF, with usage of the isoform's annotated stop codon; and (4) no change in ORF size, with usage of a premature stop codon (Table S4). For the 273 isoforms binned in category 1, we identified the location of the stop codon relative to the downstream exon-exon junction. Based on this information, we further subdivided isoforms from category 1 into isoforms with premature stop codons >50 nt upstream of an exon-exon junction, which would be predicted to activate NMD of the transcript (Nagy and Maquat, 1998); and isoforms with premature stop codons >50 nt upstream of an exon-exon junction, which might escape NMD and be used for protein production (Table S4).
RNA extraction and cDNA preparation
Palliums were dissected from E12 Rnu11 control (n=3) and mutant (n=4) mice, and the tissue collected from each embryo was individually used for total RNA isolation. Tissue from each embryo was separately triturated in 100 µl TRIzol (Invitrogen, 15596026). RNA was extracted using the DirectZOL RNA MiniPrep kit (Zymo Research, R2050), according to the manufacturer's instructions. For total RNA samples, 500 ng RNA was used for cDNA synthesis, which was performed as previously described (Baumgartner et al., 2014).
Supplementary Material
Acknowledgements
We thank Drs Anastasios Tzingounis and Heun Soh for sharing the Emx1-Cre line; Dr Akiko Nishiyama for sharing the EIIa-Cre line; Dr Siu-Pok Yee from the Gene Targeting and Transgenic Facility at the UConn Health Center for generating the Rnu11 conditional knockout mouse; and Dr Bo Reese from the Institute for Systems Genomics Center for Genome Innovation for performing RNAseq. Finally, we acknowledge Dr Ion Mandoiu for his assistance with bioinformatics analyses and Ethan Cope for help with microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.B., A.M.O., R.N.K.; Methodology: M.B., A.M.O., K.C.H., C.L., S.a.S., R.N.K.; Software: A.M.O., S.a.S.; Validation: M.B., A.M.O., G.S.A., K.C.H., C.L., K.D., N.S., N.N., R.N.K.; Formal analysis: M.B., A.M.O.; Investigation: M.B., A.M.O., G.S.A., K.C.H., C.L., K.D., N.S., N.N., R.N.K.; Resources: R.N.K.; Data curation: A.M.O., K.C.H.; Writing - original draft: M.B., A.M.O., R.N.K.; Writing - review & editing: M.B., A.M.O., R.N.K.; Visualization: M.B., A.M.O., K.C.H., R.N.K.; Supervision: R.N.K.; Project administration: R.N.K.; Funding acquisition: R.N.K.
Funding
This work was supported by grants (to R.N.K) from the National Institute of Neurological Disorders and Stroke [1R21NS096684-01A1] and the University of Connecticut. Deposited in PMC for release after 12 months.
Data availability
RNAseq data are available at the NCBI Gene Expression Omnibus (GEO), under accession number GSE96616.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.166322.supplemental
References
- Alioto T. S. (2007). U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 35, D110-D115. 10.1093/nar/gkl796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnò B., Grassivaro F., Rossi C., Bergamaschi A., Castiglioni V., Furlan R., Greter M., Favaro R., Comi G., Becher B. et al. (2014). Neural progenitor cells orchestrate microglia migration and positioning into the developing cortex. Nat. Commun. 5, 5611 10.1038/ncomms6611 [DOI] [PubMed] [Google Scholar]
- Banday A. R., Baumgartner M., Al Seesi S., Karunakaran D. K. P., Venkatesh A., Congdon S., Lemoine C., Kilcollins A. M., Mandoiu I., Punzo C. et al. (2014). Replication-dependent histone genes are actively transcribed in differentiating and aging retinal neurons. Cell Cycle 13, 2526-2541. 10.4161/15384101.2015.941757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini G., Ciceri G. and Marín O. (2013). Integration of GABAergic interneurons into cortical cell assemblies: lessons from embryos and adults. Neuron 79, 849-864. 10.1016/j.neuron.2013.08.014 [DOI] [PubMed] [Google Scholar]
- Baumgartner M., Lemoine C., Al Seesi S., Karunakaran D. K., Sturrock N., Banday A. R., Kilcollins A. M., Mandoiu I. and Kanadia R. N. (2014). Minor splicing snRNAs are enriched in the developing mouse CNS and are crucial for survival of differentiating retinal neurons. Dev. Neurobiol . 75, 895-907. 10.1002/dneu.22257 [DOI] [PubMed] [Google Scholar]
- Calegari F. and Huttner W. B. (2003). An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J. Cell Sci. 116, 4947-4955. 10.1242/jcs.00825 [DOI] [PubMed] [Google Scholar]
- Chehrehasa F., Meedeniya A. C. B., Dwyer P., Abrahamsen G. and Mackay-Sim A. (2009). EdU, a new thymidine analogue for labelling proliferating cells in the nervous system. J. Neurosci. Methods 177, 122-130. 10.1016/j.jneumeth.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Cheng Q. and Chen J. (2010). Mechanism of p53 stabilization by ATM after DNA damage. Cell Cycle 9, 472-478. 10.4161/cc.9.3.10556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P. and Pines J. (1999). Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1, 82-87. 10.1038/10049 [DOI] [PubMed] [Google Scholar]
- Crosio C., Fimia G. M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C. D. and Sassone-Corsi P. (2002). Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22, 874-885. 10.1128/MCB.22.3.874-885.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cunto F., Imarisio S., Hirsch E., Broccoli V., Bulfone A., Migheli A., Atzori C., Turco E., Triolo R., Dotto G. P. et al. (2000). Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron 28, 115-127. 10.1016/S0896-6273(00)00090-8 [DOI] [PubMed] [Google Scholar]
- Duitama J., Srivastava P. K. and Măndoiu I. I. (2012). Towards accurate detection and genotyping of expressed variants from whole transcriptome sequencing data. BMC Genomics 13 Suppl. 2, S6 10.1186/1471-2164-13-S2-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C., Fink A., Lau C., Pham D., Daza R. A., Bulfone A., Kowalczyk T. and Hevner R. F. (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25, 247-251. 10.1523/JNEUROSCI.2899-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faleiro L., Kobayashi R., Fearnhead H. and Lazebnik Y. (1997). Multiple species of CPP32 and Mch2 are the major active caspases present in apoptotic cells. EMBO J. 16, 2271-2281. 10.1093/emboj/16.9.2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farach L. S., Little M. E., Duker A. L., Logan C. V., Jackson A., Hecht J. T. and Bober M. (2018). The expanding phenotype of RNU4ATAC pathogenic variants to Lowry Wood syndrome. Am. J. Med. Genet. A 176, 465-469. 10.1002/ajmg.a.38581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Cornils H., Chiu S.-Y., O'Rourke K. P., Arnaud J., Yimlamai D., Théry M., Camargo F. D. and Pellman D. (2014). Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell 158, 833-848. 10.1016/j.cell.2014.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J. A., Talley T., Qiu M., Puelles L., Rubenstein J. L. R. and Jones K. R. (2002). Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309-6314. 10.1523/JNEUROSCI.22-15-06309.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M. and Huttner W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777-788. 10.1038/nrm1739 [DOI] [PubMed] [Google Scholar]
- Götz M., Stoykova A. and Gruss P. (1998). Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21, 1031-1044. 10.1016/S0896-6273(00)80621-2 [DOI] [PubMed] [Google Scholar]
- Grant E., Hoerder-Suabedissen A. and Molnár Z. (2012). Development of the corticothalamic projections. Front. Neurosci. 6, 53 10.3389/fnins.2012.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley L. R., D'Anna J. A., Barham S. S., Deaven L. L. and Tobey R. A. (1978). Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. 84, 1-15. 10.1111/j.1432-1033.1978.tb12135.x [DOI] [PubMed] [Google Scholar]
- He H., Liyanarachchi S., Akagi K., Nagy R., Li J., Dietrich R. C., Li W., Sebastian N., Wen B., Xin B. et al. (2011). Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 332, 238-240. 10.1126/science.1200587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem C. C. F., Repic M. and Knoblich J. A. (2015). Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci. 16, 647-659. 10.1038/nrn4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hutchinson J. N., Ensminger A. W., Clemson C. M., Lynch C. R., Lawrence J. B. and Chess A. (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran D. K. P., Chhaya N., Lemoine C., Congdon S., Black A. and Kanadia R. (2015). Loss of citron kinase affects a subset of progenitor cells that alters late but not early neurogenesis in the developing rat retina. Invest. Ophthalmol. Vis. Sci. 56, 787-798. 10.1167/iovs.14-15272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B. and Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357-360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolossova I. and Padgett R. A. (1997). U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA 3, 227-233. [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk T., Pontious A., Englund C., Daza R. A. M., Bedogni F., Hodge R., Attardo A., Bell C., Huttner W. B. and Hevner R. F. (2009). Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb. Cortex 19, 2439-2450. 10.1093/cercor/bhn260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrylkova K., Kyryachenko S., Leid M. and Kioussi C. (2012). Detection of apoptosis by TUNEL assay. Methods Mol. Biol. 887, 41-47. 10.1007/978-1-61779-860-3_5 [DOI] [PubMed] [Google Scholar]
- Lakso M., Pichel J. G., Gorman J. R., Sauer B., Okamoto Y., Lee E., Alt F. W. and Westphal H. (1996). Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93, 5860-5865. 10.1073/pnas.93.12.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Huttner W. B. and Calegari F. (2009). Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 5, 320-331. 10.1016/j.stem.2009.05.026 [DOI] [PubMed] [Google Scholar]
- Liboska R., Ligasová A., Strunin D., Rosenberg I. and Koberna K. (2012). Most anti-BrdU antibodies react with 2′-deoxy-5-ethynyluridine -- the method for the effective suppression of this cross-reactivity. PLoS ONE 7, e51679 10.1371/journal.pone.0051679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., Kanojia D., Li J., Okamoto R., Sato-Otsubo A., Kohlmann A., Sanada M., Grossmann V., Sundaresan J., Shiraishi Y. et al. (2015). Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 6, 6042 10.1038/ncomms7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandric I., Temate-Tiagueu Y., Shcheglova T., Al Seesi S., Zelikovsky A. and Măndoiu I. I. (2017). Fast bootstrapping-based estimation of confidence intervals of expression levels and differential expression from RNA-Seq data. Bioinformatics 33, 3302-3304. 10.1093/bioinformatics/btx365 [DOI] [PubMed] [Google Scholar]
- Markmiller S., Cloonan N., Lardelli R. M., Doggett K., Keightley M.-C., Boglev Y., Trotter A. J., Ng A. Y., Wilkins S. J., Verkade H. et al. (2014). Minor class splicing shapes the zebrafish transcriptome during development. Proc. Natl. Acad. Sci. USA 111, 3062-3067. 10.1073/pnas.1305536111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M. L., Kallio M. J., Barrett-Wilt G. A., Kestner C. A., Shabanowitz J., Hunt D. F., Gorbsky G. J. and Stukenberg P. T. (2004). The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr. Biol. 14, 131-137. 10.1016/j.cub.2003.12.058 [DOI] [PubMed] [Google Scholar]
- Merico D., Roifman M., Braunschweig U., Yuen R. K. C., Alexandrova R., Bates A., Reid B., Nalpathamkalam T., Wang Z., Thiruvahindrapuram B. et al. (2015). Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman Syndrome by disrupting minor intron splicing. Nat. Commun. 6, 8718 10.1038/ncomms9718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E. and Maquat L. E. (1998). A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23, 198-199. 10.1016/S0968-0004(98)01208-0 [DOI] [PubMed] [Google Scholar]
- Noctor S. C., Martínez-Cerdeño V., Ivic L. and Kriegstein A. R. (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136-144. 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- Paramasivam M., Chang Y. J. and LoTurco J. J. (2007). ASPM and citron kinase co-localize to the midbody ring during cytokinesis. Cell Cycle 6, 1605-1612. 10.4161/cc.6.13.4356 [DOI] [PubMed] [Google Scholar]
- Patel A. A., McCarthy M. and Steitz J. A. (2002). The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 21, 3804-3815. 10.1093/emboj/cdf297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. A. and Steitz J. A. (2003). Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 4, 960-970. 10.1038/nrm1259 [DOI] [PubMed] [Google Scholar]
- Pilaz L.-J., Patti D., Marcy G., Ollier E., Pfister S., Douglas R. J., Betizeau M., Gautier E., Cortay V., Doerflinger N. et al. (2009). Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc. Natl. Acad. Sci. USA 106, 21924-21929. 10.1073/pnas.0909894106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz L.-J., McMahon J. J., Miller E. E., Lennox A. L., Suzuki A., Salmon E. and Silver D. L. (2016). Prolonged Mitosis of Neural Progenitors Alters Cell Fate in the Developing Brain. Neuron 89, 83-99. 10.1016/j.neuron.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R. and Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen T. and Gerdes J. (2000). The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 182, 311-322. [DOI] [PubMed] [Google Scholar]
- Sharma A., Singh K. and Almasan A. (2012). Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol. 920, 613-626. 10.1007/978-1-61779-998-3_40 [DOI] [PubMed] [Google Scholar]
- Siegenthaler J. A. and Pleasure S. J. (2011). We have got you ‘covered’: how the meninges control brain development. Curr. Opin. Genet. Dev. 21, 249-255. 10.1016/j.gde.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Gao J., Dong X., Liu M., Li D., Shi X., Dong J.-T., Lu X., Liu C. and Zhou J. (2008). EB1 promotes Aurora-B kinase activity through blocking its inactivation by protein phosphatase 2A. Proc. Natl. Acad. Sci. USA 105, 7153-7158. 10.1073/pnas.0710018105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Nowakowski R. S. and Caviness V. S. Jr. (1995). The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J. Neurosci. 15, 6046-6057. 10.1523/JNEUROSCI.15-09-06046.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W.-Y. and Steitz J. A. (1996a). Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science 273, 1824-1832. 10.1126/science.273.5283.1824 [DOI] [PubMed] [Google Scholar]
- Tarn W.-Y. and Steitz J. A. (1996b). A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84, 801-811. 10.1016/S0092-8674(00)81057-0 [DOI] [PubMed] [Google Scholar]
- Uetake Y. and Sluder G. (2010). Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 20, 1666-1671. 10.1016/j.cub.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. B. and Schumacher B. (2016). p53 in the DNA-damage-repair process. Cold Spring Harb. Perspect. Med. 6, a026070 10.1101/cshperspect.a026070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H. K., Buslei R., Schmidt-Kastner R., Schmidt-Kastner P. K., Pietsch T., Wiestler O. D. and Blümcke I. (1996). NeuN: a useful neuronal marker for diagnostic histopathology. J. Histochem. Cytochem. 44, 1167-1171. 10.1177/44.10.8813082 [DOI] [PubMed] [Google Scholar]
- Zhang J. and Jiao J. (2015). Molecular biomarkers for embryonic and adult neural stem cell and neurogenesis. Biomed. Res. Int. 2015, 727542 10.1155/2015/727542 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.