Abstract
Phospholipids are synthesized at the endoplasmic reticulum (ER), the largest membrane bound organelle that forms membrane contact sites (MCS) with almost every other organelle. MCS are locations at which the membrane
es of two organelles are closely positioned to provide a microenvironment where proteins in one membrane can interact with the opposite membrane. Thus, MCS provide an ideal location at which lipid transfer proteins (LTPs) can achieve the efficient transfer of individual classes of lipids from the ER to other organelles via non-vesicular transport. Here we provide an overview of emerging findings on the localization and biochemical activity of LTPs at MCS between the ER and other cellular membranes. The localization of LTPs at MCS offers an elegant cell biological solution to tune local lipid composition to ongoing cell physiology.
Current Opinion in Cell Biology 2018, 53:52–60
This review comes from a themed issue on Membrane trafficking
Edited by Anne Spang and Satyajit Mayor
For a complete overview see the Issue and the Editorial
Available online 30 May 2018
https://doi.org/10.1016/j.ceb.2018.04.011
0955-0674/© 2018 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
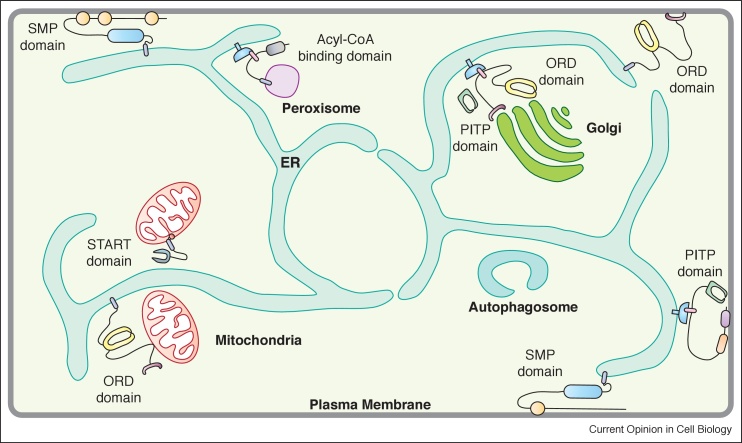
The endoplasmic reticulum (ER) is the main site of phospholipid synthesis and provides lipids to other membrane compartments by vesicular and non-vesicular transport. Non-vesicular transport relies on lipid transfer proteins (LTPs) that can move lipids between membranes through aqueous cytosol. The ER is an elaborate network of membranes making contact with nearly all organelles including mitochondria, plasma membranes (PM), endosomes, lysosomes, peroxisomes, Golgi apparatus, lipid droplets and autophagosomes (Figure 1). These areas of close contact, referred to as membrane contact sites (MCS), are formed by transient associations or can be stably present depending on cell type and context. The gap between two membranes at MCS is generally 10–30 nm spanned by tethering proteins. One of the many functions of MCS is the transfer of lipids by LTPs. LTPs are distinguished by the presence of domains such as the START (StaR related lipid-transfer), ORD (OSBP-related domain), Acyl-CoA, SMP (synaptotagmin-like mitochondrial-lipid binding protein) and PITP (phosphatidylinositol transfer protein) domains (Figure 1). Most LTP domains contain hydrophobic cavities that can accommodate a single lipid and are highly selective. LTPs fall into two categories: single domain proteins constituted of solely the lipid binding domain and multi-domain proteins where an LTP domain is associated with additional domains [1,2] (see Figure 3 for examples).
Figure 1.
Lipid transfer at membrane contact sites. The ER is the main site of lipid synthesis and makes contact with many organelles. At these membrane contact sites, lipid transfer proteins from different families defined by the presence of specific domains such as the ORD, START, Acyl-CoA, SMP and PITP domains mediate lipid exchange. Abbreviations: ORD, OSBP-related domain; START, StAR-related lipid-transfer; SMP, synaptotagmin-like mitochondrial; PITP, phosphatidylinositol transfer protein domain.
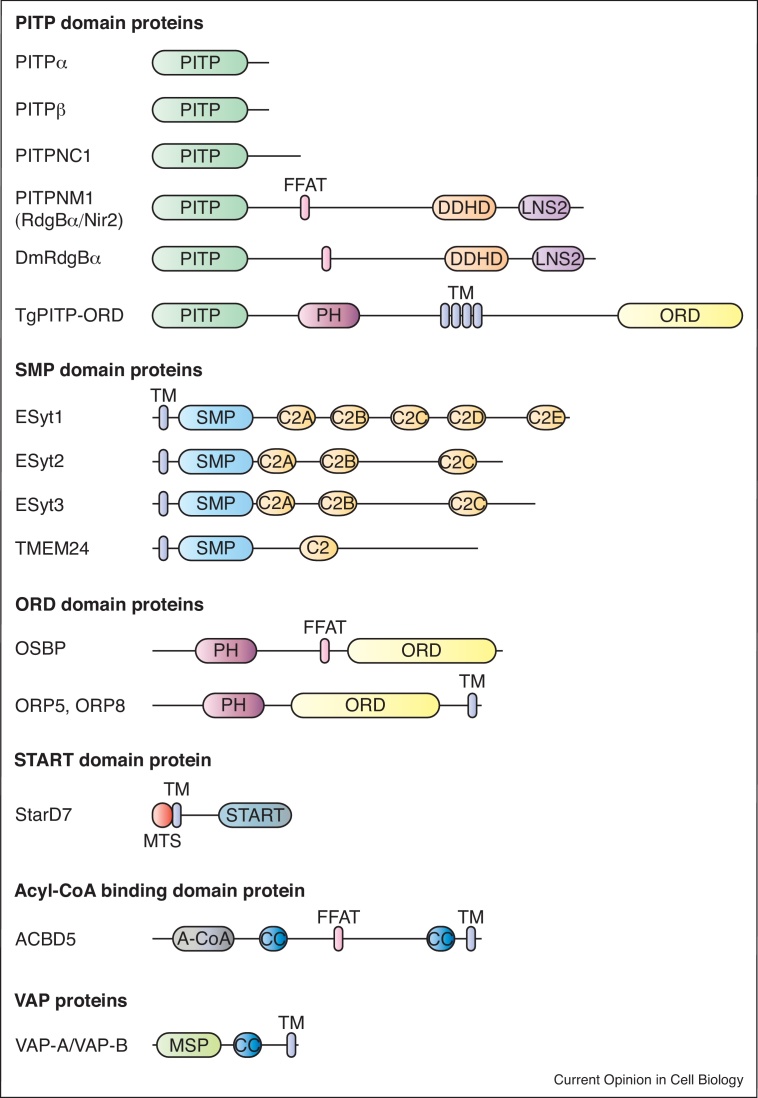
Figure 3.
Domain structures of lipid transfer proteins discussed. The PITP domain-containing proteins comprise of five genes encoding the single domain proteins, PITPα and β, which bind and transfer either phosphatidylinositol or phosphatidylcholine. In contrast, PITPNC1 (also known as RdgBβ), also a single domain protein with a disordered C-terminal extension, binds and transfers either PI or PA. This lipid binding and transfer property is shared with the multi-domain PITPNM1 and the Drosophila RdgBα. The FFAT motif of PITPNM1/RdgBα binds to the integral ER-localized VAP proteins. Extended synaptotagmins (E-Syts) comprise of a transmembrane domain that localizes the protein to the ER followed by the SMP lipid transfer domain, and multiple C2 domains. OSBP binds and transfers either cholesterol or PI4P facilitating their counter-exchange between the ER and the Golgi. The FFAT motif of OSBP localizes the protein to the ER via binding to VAP. ORP5/ORP8 are integral ER membrane proteins that can associate with the mitochondria by binding to the outer mitochondrial protein, PTP1P5. The ORD domain of ORP5/8 binds PS allowing its transfer to the mitochondria from the ER. Acyl-CoA binding domain containing protein 5 (ACBD5) is a peroxisomal membrane protein with a cytosolic acyl-CoA binding domain. It binds to VAP at the ER due to its FFAT motif. The acyl-CoA binding domain allows for the transfer of very long chain fatty acids from the ER to the peroxisomes. Abbreviations: PITP, phosphatidylinositol transfer protein domain; PH, pleckstrin homology domain; FFAT motif, two phenylalanines in an acidic tract; DDHD domain, domain named after these four conserved residues and may form a metal binding site; LNS2 (Lipin/Ned1/Smp2) domain, found in lipins and lipin homologues from S. cerevisiae (Smp2) and S. pombe (Ned1); TM, Transmembrane; SMP, synaptotagmin-like mitochondrial lipid binding domain; C2 domain, a structural domain that can bind Ca2+ and phospholipids; ORD domain, OSBP-related domain; OSBP, oxysterol binding proteins; ORP, OSBP-related proteins; MTS, mitochondrial targeting sequence; START domain, stAR-related lipid transfer domain; stAR; Steroidogenic acute regulatory protein; A-CoA domain, acyl-CoA binding domain; CC, coiled coil; MSP, Major sperm protein domain; VAP-A/VAP-B, VAMP-associated proteins, A and B.
The purpose of this review is to discuss emerging concepts of how lipid transfer between membrane compartments is facilitated at these MCS. In depth reviews on LTPs and MCS can be consulted for background information [2, 3, 4, 5].
Lipid exchange at ER–PM contact sites
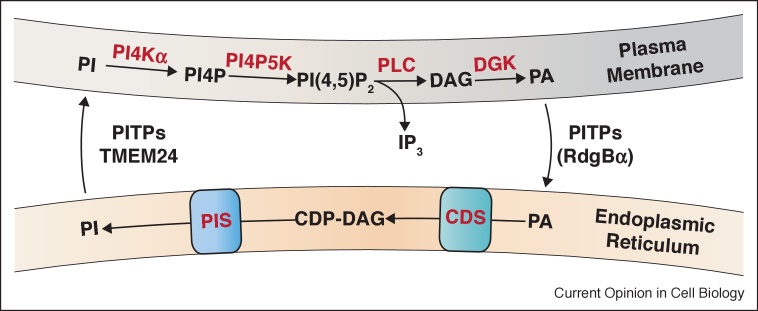
The PM of cells has a unique lipid composition being enriched in phosphoinositides and phosphatidylserine. Phosphoinositides are low abundance lipids generated by the phosphorylation of the precursor lipid phosphatidylinositol (PI) which is synthesized in the ER (Figure 2). The two most abundant phosphoinositides, phosphatidylinositol(4,5)bisphosphate (PI(4,5)P2) and its precursor phosphatidylinositol 4-phosphate (PI4P) are enriched in the inner leaflet of the PM where they serve many functions including regulation of the actin cytoskeleton, ion channel activity and exo-endocytosis. In addition, receptor-regulated phospholipase C (PLC) hydrolyses PI(4,5)P2 to generate the second messengers, inositol(1,4,5)trisphosphate (IP3) and diacylglycerol (DAG). During PLC signaling, PI(4,5)P2 levels can drop rapidly at the PM requiring compensatory resynthesis to ensure stable levels of this key lipid. The biochemical pathway triggered by PI(4,5)P2 hydrolysis and leading to its resynthesis includes five lipid intermediates that are distributed between the ER and the PM (PI(4,5)P2 cycle); this leads to a topological constraint requiring transfer of lipid intermediates between the ER and PM [4] (Figure 2).
Figure 2.
Transfer of PI and PA during the PI(4,5)P2 cycle triggered by PLC activation. The enzymes are distributed between two membrane compartments, the ER and PM. Lipid transfer between these compartments is required at two points in the cycle; transfer of PI from the ER to the plasma membrane and transfer of phosphatidic acid (PA) from the plasma membrane to the ER. Abbreviations: PIS, PI synthase; CDS, CDP-diacylglycerol synthase; DGK, diacylglycerol kinase; PI4K, PI 4-kinase; PIP5K, PI4P 5-kinase; PLC, phospholipase C; PA, phosphatidic acid; DAG, diacylglycerol.
Phosphatidylinositol transfer proteins
Phosphatidylinositol transfer proteins (PITPs) were originally identified as soluble factors supporting PLC signaling in mammalian cells [6,7]. The most compelling in vivo evidence of their requirement for PI transfer from the ER to the PM comes from studies in Drosophila photoreceptors. In fly photoreceptors, the microvillar PM is arranged in close contact with the ER-derived sub-microvillar cisternae (SMC); this is reminiscent of an ER–PM MCS [8]. PLCβ is activated at the microvillar PM, whereas RdgBα, a multi-domain protein with an N-terminal PITP domain (Figure 3), is localized to the SMC. RdgBα mutants show depletion of PM PI(4,5)P2 in the resting state and reduced rates of PI(4,5)P2 resynthesis during PLC activation [9,10]. These biochemical defects affect photoreceptor function resulting in reduced responses to light and retinal degeneration. The PITP domain of RdgBα can bind and transfer PI in vitro, and in vivo analyses have shown that RdgBα mutants can be rescued with wild type protein but not with a version that is unable to bind and transfer PI [9]. Conceptually similar results have been shown for the mammalian orthologs of RdgBα, PITPNM1/2 (alt. names: Nir2/Nir3); these studies have been performed in cultured cell-lines with both PITPNM1 (Nir2) and receptors over-expressed [11, 12, 13]. A role for endogenous PITPNM1/2 in supporting endogenous PLC signaling remains to be established; this underscores the need to find mammalian cell types in which the function of endogenous PITPNM proteins and receptors can be studied. Another PITP that supports PLC signaling is PITPα [14•,15]. Platelets from mouse knockouts in this single domain PITP show reduced PI4P and PI(4,5)P2 basal levels; upon stimulation with thrombin, IP3 production is minimal and the rise in cytosol Ca2+ is reduced [14•].
Central to current thinking on the cell biology of lipid transfer at MCS is the idea that LTPs localize to and mediate lipid transfer at these sites. The localization of PITPα to an MCS remains to be determined. However for RdgBα, the protein is detected at the MCS between the SMC and the microvillar PM [16,17]. A recent study demonstrated the importance of this localization. When the PITP domain of RdgBα is delocalized from this interface, Drosophila photoreceptors are unable to support PI(4,5)P2 resynthesis during high rates of PLC activation [18••]. The localization of RdgBα at this MCS is dependent on protein-protein interaction between its FFAT motif and the protein dVAP-A that is enriched at the SMC [18••].
In contrast to fly photoreceptors, the MCS in mammalian cells seems dynamically formed during PLC activation and PITPNM1 (Nir2) translocates to these sites to support PM PI(4,5)P2 synthesis [12,13,19]. The size of the membrane contact area at MCS is reported to be oblong with the dimensions of ∼120 nm × ∼80 nm in HeLa cells [19]. Multiple signals have been proposed to mediate this PM recruitment including the binding to PA by the LNS2 domain of PITPNM1 [11,20], DAG binding to the DGBL domain of PITPNM1 [13], Ca2+ binding to the C2 domain of Extended-Synaptotagmin 1 (E-Syt1) [12] and most recently cortical actin [19]. However, these studies in mammalian cells have only been done where the receptor has been over-expressed suggesting that these PITPNM proteins may only be required during intense stimulation as seen with the Drosophila photoreceptors.
SMP domain proteins
Recent studies have highlighted a new class of LTPs that contain an SMP domain. The extended synaptotagmins (E-Syts) are ER-localized integral membrane proteins comprising an SMP (Synaptotagmin-like mitochondrial lipid binding) domain with multiple C2 domains. The SMP domains dimerize to form a 90 Å cylinder housing two lipids [21,22••,23]; the SMP domains of E-Syts appear not to have selectivity for any particular glycerolipid. The mammalian E-Syt family has three members: E-Syt1 with 5 C2 domains and E-Syt2/E-Syt3, each with three C2 domains (Figure 3). The C2C domain of E-Syt2 and E-Syt3 form membrane contacts with the PM by binding to PI(4,5)P2. In contrast, E-Syt1 translocates to ER–PM junctions after an increase in intracellular Ca2+ mediated by its C2A and C2C domain [22••,24]; Ca2+ binding to C2C promotes membrane tethering by C2E binding to PI(4,5)P2 at the PM [25••]. The lipid transfer activity of E-Syt1 is strictly dependent on the binding of Ca2+ to both the C2A and C2C domains. Since E-Syts are only active at elevated cytosol Ca2+, lipid transfer can only occur during Ca2+ signaling subsequent to PLC activation. Moreover, entry of Ca2+ also triggers activation of PLC, and consequently PI(4,5)P2 hydrolysis, conditions that would cause E-Syt1 to dissociate from the PM. This raises the question of the relevance of E-Syt1 in replenishing PI(4,5)P2 levels at the PM during intense PLC activation. It is notable that mice devoid of all three E-Syts develop normally and are viable and fertile. These animals show upregulation of genes encoding Orp5/8, Orai1, STIM1 and TMEM110, ER–PM MCS proteins that could compensate for loss of E-Syts [26•,27•].
TMEM24 (C2CD2L) is a protein containing an SMP domain followed by a C2 domain (Figure 3). This SMP domain binds a single PI molecule unlike that of E-Syt2, which can bind two phospholipids. TMEM24 is an ER anchored transmembrane protein that concentrates at ER–PM MCS under resting conditions. TMEM24 binding to the PM is regulated by a phosphorylation cycle mediated by protein kinase C (PKC) and the phosphatase, PP2B both of which are Ca2+ dependent enzymes. When phosphorylated by PKC, TMEM24 dissociates from the PM and therefore ceasing transfer; it can only re-associate after dephosphorylation for transfer to resume. Thus TMEM24 maintains basal PI4P and PI(4,5)P2 levels; over-expression of TMEM24 in cells leads to increased levels of PI4P and PI(4,5)P2 [28••]. The TMEM24 protein is highly enriched in pancreatic β-cells and plays a key role in regulating glucose-sensitive insulin release [29].
PI4P and PI(4,5)P2 transfer
Recent studies have identified proteins that can mediate PI(4,5)P2 and PI4P removal from the PM. ORP5 and ORP8, localized to ER–PM MCS, can transfer phosphatidylserine (PS) to the PM while removing PI(4,5)P2 [30]. RASSF4 was also identified as a regulator of PI(4,5)P2 homeostasis by mediating ER–PM junction formation through tethering via E-Syts [31]. ORP5 and ORP8 have previously been implicated in a PI4P/PS exchange cycle that can facilitates PS transfer to the PM coupled to PI4P transfer to the ER [32,33]. Several studies have noted the localization of the PI4P phosphatase Sac1 at ER–PM contact sites depleting PI4P in this microdomain of the ER thus generating a PI4P gradient for this exchange [32, 33, 34, 35].
PA transfer activity
It is well established that DAG generated by PLC activity is rapidly converted to PA by DAG kinases at the PM [36]. A precursor–product relationship between PA disappearance and new PI synthesis has been demonstrated. Recent studies have shown that the PITP domain of Drosophila and mammalian RdgBα/PITPNM1 and PITPNC1 are capable of transporting PA [37,38]. Thus in contrast to PITPα and PITPβ that are PI/PC LTPs, the PITP domains of the RdgB family are PI/PA transfer proteins [38]. Loss of RdgBα in Drosophila [9] or PITPNM1 in mammalian cells [13] results in altered PA dynamics during PLC activation. This PI/PA transfer function of RdgBα offers an efficient mechanism for coupling the removal of PA with the supply of PI for PI(4,5)P2 resynthesis (Figure 2).
Diacylglycerol transfer activity
A recent study in mammalian cells depleted of all E-Syts demonstrated sustained accumulation of PM DAG following stimulation by histamine [39,40]. These studies assumed DAG to be derived from PI(4,5)P2 hydrolysis. However, DAG at the PM can be derived from both PLC (directly) or phospholipase D (indirectly via PA) activation and both these phospholipases are activated by histamine receptor activation. The accumulation of DAG was rescued by expression of E-Syt1, but not by mutant E-Syt1 lacking the SMP domain. As the SMP domain was found to transfer DAG, this is a potential mechanism for E-Syt1 to regulate the PI(4,5)P2 cycle [25••].
Lipid transfer from the ER to other organelles
ER–Golgi
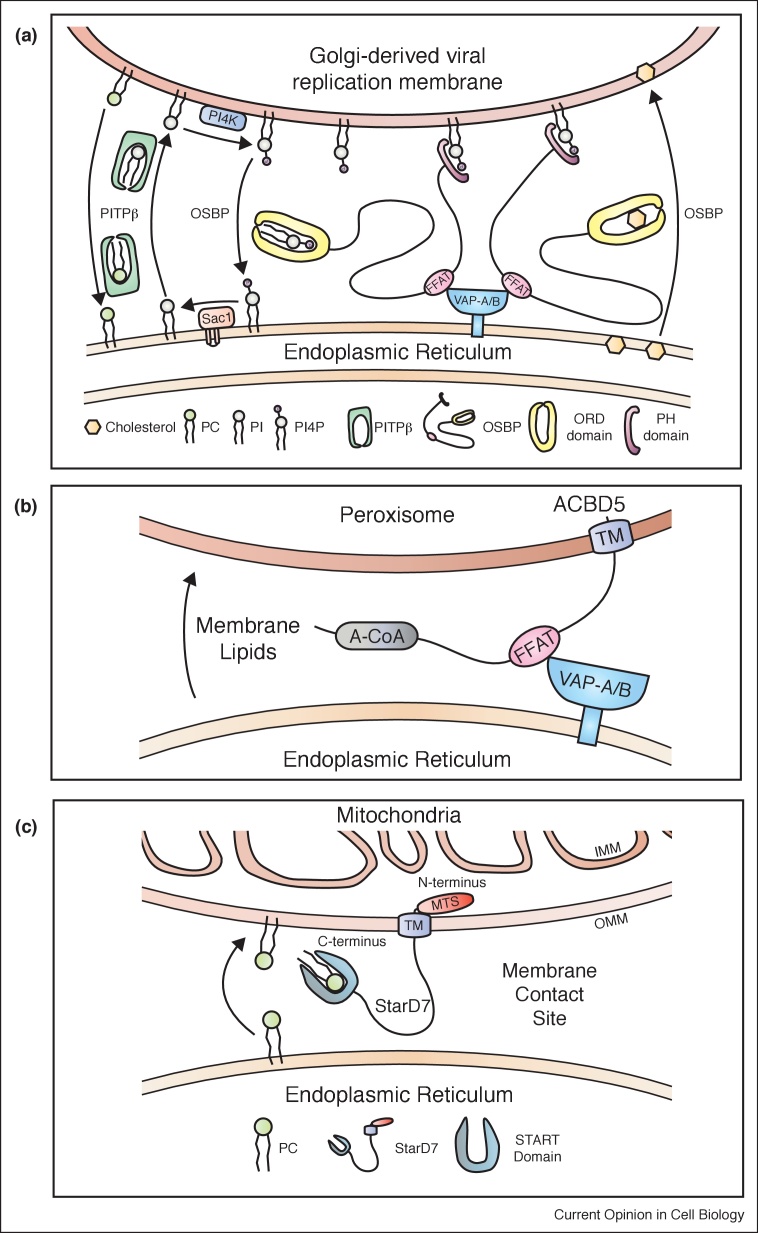
PI synthesized at the ER (Figure 2) is also used for non-PLC dependent processes at other organelle membranes. In this case, PI transfer takes place at MCS using the single domain PITPβ. Oxysterol binding protein (OSBP) is a modular LTP (Figure 3) localized to ER–Golgi contact sites; the FFAT motif permits OSBP to bind VAP at the ER while the PH domain binds to PI4P at the Golgi membrane (Figure 4a). Unlike the ER, the Golgi is enriched in PI4P due to the presence of the PI 4-kinase, IIIβ. The ORD domain drives cholesterol export from the ER to the Golgi through the reciprocal transfer of PI4P from the Golgi to the ER [32,41]. At the ER, Sac1 converts PI4P into PI. Since PI synthesis is restricted to the ER, a supply of PI to the Golgi for PI4P synthesis is necessary. PITPβ is localized to the Golgi and a previous study reported that its depletion led to decreased PI4P levels and disrupted COP1 mediated retrograde transport from the Golgi to the ER [42]. Thus PITPβ is ideally localized for maintaining Golgi PI4P levels that is consumed during cholesterol transfer. Evidence to support this comes from recent studies which has identified PITPβ as a host factor required for positive-strand RNA viral replication [43,44]. For replication, the virus builds a membrane-associated replication complex which is Golgi-derived that is tightly associated with the ER. Interestingly OSBP, PI4KIIIβ and Sac1 all of which are also localized at this MCS, are required for both PI4P homeostasis and viral replication [43,44] (Figure 4a). What recruits PITPβ to these MCS is unclear. In Toxoplasma gondii, a multi-domain protein of 1912 a.a. incorporates a PITP domain with a PH domain, 4 transmembrane domains and an oxysterol binding domain (PITP-PH-4TM-OSBP) (Figure 3). Thus the concept that a PITP domain works in concert with an OSBP domain protein appears to be amalgamated into a single protein in some organisms.
Figure 4.
Emerging map of lipid transfer reactions at diverse MCS. (A) Lipid transfer coordinated by OSBP and PITPβ moving cholesterol to Golgi-derived viral replication membranes. PITPβ transfers PI from the ER to the Golgi-derived viral replication membranes where PI4KIIIβ converts it to PI4P. OSBP utilizes the PI4P to co-ordinate the reciprocal transfer of cholesterol to the replication membranes in exchange for PI4P. At the ER, Sac1 dephosphorylates PI4P to PI to maintain the PI4P gradient. (B) Transfer of long chain fatty acids by ACBD5 and ER-peroxisome MCS. (C) Transfer of phosphatidylcholine by STARD7 at ER-mitochondria MCS. Additional tethering complexes will be required to connect the two membranes.
ER–peroxisomes
Peroxisomes depend on the ER for their lipid composition and the ER receives lipid precursors for plasmalogen biosynthesis (ether phospholipids) from peroxisomes. The tether that links these two organelles is the ER protein VAP-B interacting via its MSP domain with the FFAT-like motif of acyl-CoA binding domain-containing 5 (ACBD5), a peroxisomal tail-anchored membrane protein [45••,46••,47] (Figure 4b). An intact VAP-ACBD5 tether is required for peroxisome growth, plasmalogen synthesis and maintenance of cellular cholesterol levels [45••]. The ACB domain of ACBD5 preferentially binds very long chain fatty acyl-CoAs and transfers them to peroxisomes. Mutations in ACBD5 show elevated levels of very long chain fatty acids and a defect in peroxisomal β-oxidation of very long chain fatty acids [48]; patients with ACBD5 deficiency manifest with retinal dystrophy [49].
ER–autophagosome
The transfer of PI from the ER is also required for autophagosome biogenesis [50]. PI is synthesized in the ER from CDP-DAG by PI synthase (PIS) (Figure 2). Over-expressed PIS localizes to a highly dynamic compartment of the ER and at leading edges of tubules [51,52]. Early autophagic structures are formed in close apposition to the ER and recent studies reveal that autophagosome formation requires a subdomain of the ER, which is highly enriched in PIS. The ULK complex first localizes to the PIS-enriched ER subdomain and then translocates to the ATG9A-positive autophagosome precursors in a PI3P-dependent manner. PI in the PIS-enriched membrane is required for autophagosome formation. LTPs that could transfer PI from the PIS-enriched subdomain to the ATG9A vesicles [50,53] remain to be identified.
ER–mitochondria
Mitochondria can synthesize PA, PG, cardiolipin and PE. However, PC, PI and PS have to be imported from the ER. PS imported into mitochondria is used by PS decarboxylase to produce PE at the inner mitochondrial membrane. Close contacts between the ER and mitochondria are important for lipid transfer; ER to mitochondria PS transfer slows down significantly in yeast cells missing both the ER-shaping reticulon proteins and the ERMES complex. This defect in PS transfer could be corrected by expression of a protein that artificially tethers the ER and mitochondria [54]; these findings have now been extended to mammalian cells [55]. The ER that associates with mitochondria is enriched in PS synthase [56,57]. Recent studies have identified ORP5/ORP8 (Figure 3) to localize to ER–mitochondria contacts, interact with the outer mitochondrial protein, PTP1P5 and transfer PS from the ER to mitochondria [58••]. Depletion of ORP5/ORP8 leads to altered mitochondrial morphology and function. Together, these findings indicate that PS production and transport at the ER–mitochondrial MCS is required to support mitochondrial function.
PC is the dominant lipid of mitochondria and recent studies emphasize StarD7, a member of the START family facilitates PC transfer from the ER to the outer mitochondrial membrane (OMM). StarD7-I, the longer isoform, contains a mitochondrial targeting sequence followed by a transmembrane domain anchoring the protein to the OMM [59] (Figure 3). The C-terminal START domain would then extend into the cytoplasm and shuttle PC from the ER to OMM at the ER–mitochondria contact sites [60••] (Figure 4c). Loss of StarD7 results in embyronic lethality and compromised mitochondrial function [61••]. Interestingly, loss of StarD7 results in only a partial loss of PC in mitochondria and suggests that other PC transfer proteins such as PCTP (StarD2) and StarD10 may have a role in PC transfer.
General conclusions
LTPs were initially identified as soluble single domain proteins that could transfer lipids between membrane compartments in vitro. However, in recent years, it has emerged that such lipid transfer domains occur in a diverse range of proteins in conjunction with other protein domains that in themselves appear to have no LTP activity. One emerging function of these additional domains is their ability to act as protein targeting signals thus ensuring the positioning of lipid transfer activity at specific and in some cases unique locations with cells. One such location that has emerged are MCS between the ER and multiple cellular organelles where LTPs seem to localize suggesting their ability to transfer lipid locally at these subcellular locations.
Numerous cell biological studies have informed on the localization and biochemical activity of LTPs in cultured cells. However, most are performed with the LTP overexpressed; while this has provided initial insights, it will be essential that going forward the localization of the endogenous LTPs in cells be established. Such studies will help to identify in vivo cell types where a given LTP is enriched leading to the development of model cell types where the function and regulation of endogenous LTPs can be studied.
In contrast to cell biological studies, there have been limited analyses of the role of LTPs in physiological processes in vivo. Where they have been done, in many cases, phenotypes have been surprisingly limited. This observation may reflect functional redundancy between multiple LTPs that can perform the same biochemical activity; this possibility is reflected in the finding that multiple genes encoding an LTP are found in mammalian genomes. Studies to address this redundancy will be important to understand the overall contribution of this class of molecules in regulating lipid homeostasis in cell physiology.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Work in the Cockcroft Laboratory is supported by grants from British Heart Foundation, Grant numbers FS/15/73/31672 and FS/12/49/29729 and from Biotechnology and Biological Sciences Research Council, grant number BB/J005606/1. We thank Nicholas Blunsom for making the illustrations and for commenting on the manuscript. P. Raghu Laboratory is supported by the National Centre for Biological Sciences-TIFR and a Senior Fellowship from the Wellcome-DBT India Alliance (IA/S/14/2/501540).
References
- 1.Chiapparisno A., Maeda K., Turei D., Saez-Rodriguez J., Gavin A.C. The orchestra of lipid-transfer proteins at the crossroads between metabolism and signaling. Prog Lipid Res. 2016;61:30–39. doi: 10.1016/j.plipres.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Wong L.H., Copic A., Levine T.P. Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem Sci. 2017;42:516–530. doi: 10.1016/j.tibs.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gatta A.T., Levine T.P. Piecing together the patchwork of contact sites. Trends Cell Biol. 2017;27:214–229. doi: 10.1016/j.tcb.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Cockcroft S., Raghu P. Topological organisation of the phosphatidylinositol 4,5-bisphosphate-phospholipase C resynthesis cycle: PITPs bridge the ER–PM gap. Biochem J. 2016;473:4289–4310. doi: 10.1042/BCJ20160514C. [DOI] [PubMed] [Google Scholar]
- 5.Saheki Y., De Camilli P. Endoplasmic reticulum–plasma membrane contact sites. Annu Rev Biochem. 2017;86:659–684. doi: 10.1146/annurev-biochem-061516-044932. [DOI] [PubMed] [Google Scholar]
- 6.Thomas G.M.H., Cunningham E., Fensome A., Ball A., Totty N.F., Troung O., Hsuan J.J., Cockcroft S. An essential role for phosphatidylinositol transfer protein in phospholipase C-mediated inositol lipid signalling. Cell. 1993;74:919–928. doi: 10.1016/0092-8674(93)90471-2. [DOI] [PubMed] [Google Scholar]
- 7.Kauffmann-Zeh A., Thomas G.M.H., Ball A., Prosser S., Cunningham E., Cockcroft S., Hsuan J.J. Requirement for phosphatidylinositol transfer protein in epidermal growth factor signalling. Science. 1995;268:1188–1190. doi: 10.1126/science.7761838. [DOI] [PubMed] [Google Scholar]
- 8.Yadav S., Cockcroft S., Raghu P. The Drosophila photoreceptor as a model system for studying signalling at membrane contact sites. Biochem Soc Trans. 2016;44:447–451. doi: 10.1042/BST20150256. [DOI] [PubMed] [Google Scholar]
- 9.Yadav S., Garner K., Georgiev P., Li M., Espinosa E.G., Panda A., Mathre S., Okkenhaug H., Cockcroft S., Raghu P. RDGBα, A PI-PA transfer protein regulates G-protein coupled PtdIns(4,5)P2 signalling during Drosophila phototransduction. J Cell Sci. 2015;128:3330–3344. doi: 10.1242/jcs.173476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardie R.C., Liu C.H., Randall A.S., Sengupta S. In vivo tracking of phosphoinositides in Drosophila photoreceptors. J Cell Sci. 2015;128:4328–4340. doi: 10.1242/jcs.180364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C.L., Liou J. Phosphatidylinositol 4,5-bisphosphate homeostasis regulated by Nir2 and Nir3 at endoplasmic reticulum–plasma membrane junctions. J Biol Chem. 2015;290:14289–14301. doi: 10.1074/jbc.M114.621375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C.L., Hsieh T.S., Yang T.T., Rothberg K.G., Azizoglu D.B., Volk E., Liao J.C., Liou J. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 2013;5:813–825. doi: 10.1016/j.celrep.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y.J., Guzman-Hernandez M.L., Wisniewski E., Balla T. Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER–PM contact sites maintains phosphoinositide signaling competence. Dev Cell. 2015;33:549–561. doi: 10.1016/j.devcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Zhao L., Thorsheim C.L., Suzuki A., Stalker T.J., Min S.H., Lian L., Fairn G.D., Cockcroft S., Durham A., Krishnaswamy S. Phosphatidylinositol transfer protein-α in platelets is inconsequential for thrombosis yet is utilized for tumor metastasis. Nat Commun. 2017;8:1216. doi: 10.1038/s41467-017-01181-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that PITPα, a single domain protein, can regulate PI(4,5)P2 homeostasis at the plasma membrane in platelets. Loss of PITPα leads to a decrease in PI4P and PI(4,5)P2 levels under basal conditions and when stimulated with thrombin, IP3 production is disrupted.
- 15.Xie Y., Hong Y., Ma X.Y., Ren X.R., Ackerman S., Mei L., Xiong W.C. DCC-dependent phospholipase C signaling in netrin-1-induced neurite elongation. J Biol Chem. 2006;281:2605–2611. doi: 10.1074/jbc.M512767200. [DOI] [PubMed] [Google Scholar]
- 16.Vihtelic T.S., Goebl M., Milligan S., O’Tousa S.E., Hyde D.R. Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J Cell Biol. 1993;122:1013–1022. doi: 10.1083/jcb.122.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki E., Hirosawa K. Immunolocalization of a Drosophila phosphatidylinositol transfer protein (rdgB) in normal and rdgA mutant photoreceptor cells with special reference to the subrhabdomeric cisternae. J Electron Microsc. 1994;43:183–189. [PubMed] [Google Scholar]
- 18••.Yadav S., Thakur R., Georgiev P., Deivasigamani S., Krishnan H., Ratnaparkhi G., Raghu P. RDGBalpha localization and function at membrane contact sites is regulated by FFAT–VAP interactions. J Cell Sci. 2018;131 doi: 10.1242/jcs.207985. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies that the main function of the FFAT/VAP interaction is the ensure a high concentration of RdgBα at the MCS. Expression of the PITP domain alone can recapitulate the RdgBα phenotype as long as the domain is expressed at much higher concentrations compared to RdgBα.
- 19.Hsieh T.S., Chen Y.J., Chang C.L., Lee W.R., Liou J. Cortical actin contributes to spatial organization of ER–PM junctions. Mol Biol Cell. 2017;28:3171–3180. doi: 10.1091/mbc.E17-06-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S., Kedan A., Marom M., Gavert N., Keinan O., Selitrennik M., Laufman O., Lev S. The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep. 2013;14:891–899. doi: 10.1038/embor.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schauder C.M., Wu X., Saheki Y., Narayanaswamy P., Torta F., Wenk M.R., De Camilli P., Reinisch K.M. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510:552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Yu H.J., Liu Y.H., Gulbranson D.R., Paine A., Rathore S.S., Shen J.S. Extended synaptotagmins are Ca2+-dependent lipid transfer proteins at membrane contact sites. Proc Natl Acad Sci U S A. 2016;113:4362–4367. doi: 10.1073/pnas.1517259113. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper together with [25••] show that the SMP domain of E-Syts transfer glycerophospholipids between membrane bilayers only in the presence of Ca2+. The SMP domains themselves cannot transport lipids unless the two membranes are tightly tethered by Ca2+-bound C2 domains. The Ca2+ -regulated lipid transfer activity of E-Syts is fully recapitulated when the SMP domain is fused to the cytosolic domain of synaptotagmin-1, the Ca2+ sensor in synaptic vesicle fusion, indicating that a common mechanism of membrane tethering governs the Ca2+ regulation of lipid transfer and vesicle fusion.
- 23.Fernandez-Busnadiego R., Saheki Y., De Camilli P. Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum–plasma membrane contact sites. Proc Natl Acad Sci U S A. 2015;112:E2004–E2013. doi: 10.1073/pnas.1503191112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giordano F., Saheki Y., Idevall-Hagren O., Colombo S.F., Pirruccello M., Milosevic I., Gracheva E.O., Bagriantsev S.N., Borgese N., De Camilli P. PI(4,5)P2-Dependent and Ca(2+)-regulated ER–PM interactions mediated by the extended synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Bian X., Saheki Y., De Camilli P. Ca(2+) releases E-Syt1 autoinhibition to couple ER–plasma membrane tethering with lipid transport. EMBO J. 2018;37:219–234. doi: 10.15252/embj.201797359. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifies how Ca2+ binding to the C2A and C2C domains regulates E-Syt1 function. Ca2+ binding to C2C promotes E-Syt1 mediated membrane tethering by releasing an inhibition that prevents C2E from interacting with plasma membrane PI(4,5)P2. Ca2+ binding to C2A enables lipid transport by releasing a charge-based autoinhibitory interaction between this domain and the SMP domain.
- 26•.Tremblay M.G., Moss T. Loss of all 3 extended synaptotagmins does not affect normal mouse development, viability or fertility. Cell Cycle. 2016;15:2360–2366. doi: 10.1080/15384101.2016.1203494. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper can be seen as a companion paper to Sclipet al. [27•] that show that all three ESyt proteins can be deleted from mice with no consequences suggesting that multiple redundant mechanisms exist to maintain membrane contact sites and phosphoinositide levels at the plasma membrane
- 27•.Sclip A., Bacaj T., Giam L.R., Sudhof T.C. Extended synaptotagmin (ESyt) triple knock-out mice are viable and fertile without obvious endoplasmic reticulum dysfunction. PLOS ONE. 2016;11:e0158295. doi: 10.1371/journal.pone.0158295. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation to Ref [26•]
- 28••.Lees J.A., s M., Sun E.W., Wheeler H., Torta F., Wenk M.R., De Camilli P., Reinisch K.M. Lipid transport by TMEM24 at ER–plasma membrane contacts regulates pulsatile insulin secretion. Science. 2017;355 doi: 10.1126/science.aah6171. [DOI] [PMC free article] [PubMed] [Google Scholar]; Release of insulin occurs in two phases; in the second phase granules are docked and primed and then released in a series of bursts, each triggered by a spike of cytosolic Ca2+. TMEM24, an ER-localized protein is associated with PM by binding to PI(4,5)P2 under resting conditions. The SMP domain can transfer PI from the ER to the PM to maintain PI4P and PI(4,5)P2 concentrations. When cytosol Ca2+ levels increase, TMEM24 is phosphorylated by PKC and dissociates from the PM. Only after dephosphorylation by the Ca2+-dependent phosphatase, PP2B, can TMEM24 re-associate with the PM and permit PI transfer to occur.
- 29.Pottekat A., Becker S., Spencer K.R., Yates J.R., 3rd, Manning G., Itkin-Ansari P., Balch W.E. Insulin biosynthetic interaction network component, TMEM24, facilitates insulin reserve pool release. Cell Rep. 2013;4:921–930. doi: 10.1016/j.celrep.2013.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghai R., Du X., Wang H., Dong J., Ferguson C., Brown A.J., Parton R.G., Wu J.W., Yang H. ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane. Nat Commun. 2017;8:757. doi: 10.1038/s41467-017-00861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y.J., Chang C.L., Lee W.R., Liou J. RASSF4 controls SOCE and ER–PM junctions through regulation of PI(4,5)P2. J Cell Biol. 2017;216:2011–2025. doi: 10.1083/jcb.201606047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moser von Filseck J., Vanni S., Mesmin B., Antonny B., Drin G. A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat Commun. 2015;6:6671. doi: 10.1038/ncomms7671. [DOI] [PubMed] [Google Scholar]
- 33.Chung J., Torta F., Masai K., Lucast L., Czapla H., Tanner L.B., Narayanaswamy P., Wenk M.R., Nakatsu F., De Camilli P. Intracellular transport. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER–plasma membrane contacts. Science. 2015;349:428–432. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dickson E.J., Jensen J.B., Vivas O., Kruse M., Traynor-Kaplan A.E., Hille B. Dynamic formation of ER–PM junctions presents a lipid phosphatase to regulate phosphoinositides. J Cell Biol. 2016;213:33–48. doi: 10.1083/jcb.201508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moser von Filseck J., Copic A., Delfosse V., Vanni S., Jackson C.L., Bourguet W., Drin G. Intracellular transport. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015;349:432–436. doi: 10.1126/science.aab1346. [DOI] [PubMed] [Google Scholar]
- 36.Michell R.H. Inositol phospholipids in cell surface receptor function. Biochim Biophys Acta. 1975;415:81–147. doi: 10.1016/0304-4157(75)90017-9. [DOI] [PubMed] [Google Scholar]
- 37.Garner K., Hunt A.N., Koster G., Somerharju P., Grover E., Li M., Raghu P., Holic R., Cockcroft S. Phosphatidylinositol transfer protein, Cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. J Biol Chem. 2012;287:32263–32276. doi: 10.1074/jbc.M112.375840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cockcroft S., Garner K., Yadav S., Gomez-Espinoza E., Raghu P. RdgBalpha reciprocally transfers PA and PI at ER–PM contact sites to maintain PI(4,5)P2 homoeostasis during phospholipase C signalling in Drosophila photoreceptors. Biochem Soc Trans. 2016;44:286–292. doi: 10.1042/BST20150228. [DOI] [PubMed] [Google Scholar]
- 39.Saheki Y., Bian X., Schauder C.M., Sawaki Y., Surma M.A., Klose C., Pincet F., Reinisch K.M., De Camilli P. Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat Cell Biol. 2016;18:504–515. doi: 10.1038/ncb3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bian X., Saheki Y., De Camilli P. Ca2+ releases E‐Syt1 autoinhibition to couple ER–plasma membrane tethering with lipid transport. EMBO J. 2018;37:219–234. doi: 10.15252/embj.201797359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mesmin B., Bigay J., Polidori J., Jamecna D., Lacas‐Gervais S., Antonny B. Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J. 2017;36:3156–3174. doi: 10.15252/embj.201796687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carvou N., Holic R., Li M., Futter C., Skippen A., Cockcroft S. Phosphatidylinositol- and phosphatidylcholine-transfer activity of PITPβ is essential for COP1-mediated retrograde transport from the Golgi to the endoplasmic reticulum. J Cell Sci. 2010;123:1262–1273. doi: 10.1242/jcs.061986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roulin P.S., Lotzerich M., Torta F., Tanner L.B., van Kuppeveld F.J., Wenk M.R., Greber U.F. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER–Golgi interface. Cell Host Microbe. 2014;16:677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Ishikawa-Sasaki K., Nagashima S., Taniguchi K., Sasaki J. Model of OSBP-mediated cholesterol supply to Aichi virus RNA replication sites involving protein–protein interactions among viral proteins, ACBD3, OSBP, VAP-A/B, and SAC1. J Virol. 2018;92 doi: 10.1128/JVI.01952-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Hua R., Cheng D., Coyaud E., Freeman S., Di Pietro E., Wang Y., Vissa A., Yip C.M., Fairn G.D., Braverman N. VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J Cell Biol. 2017;216:367–377. doi: 10.1083/jcb.201608128. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper can be seen as a companion paper to Costelloet al. [46•] that show that peroxisomes make membrane contact by interactions between ER-localized VAP and peroxisome-localized ACBD5 due to the presence of a FFAT-like motif in ACBD5.
- 46•.Costello J.L., Castro I.G., Hacker C., Schrader T.A., Metz J., Zeuschner D., Azadi A.S., Godinho L.F., Costina V., Findeisen P. ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J Cell Biol. 2017;216:331–342. doi: 10.1083/jcb.201607055. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotations to Ref [45••]
- 47.Murphy S.E., Levine T.P. VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta. 2016;1861:952–961. doi: 10.1016/j.bbalip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Ferdinandusse S., Falkenberg K.D., Koster J., Mooyer P.A., Jones R., van Roermund C.W.T., Pizzino A., Schrader M., Wanders R.J.A., Vanderver A. ACBD5 deficiency causes a defect in peroxisomal very long-chain fatty acid metabolism. J Med Genet. 2017;54:330–337. doi: 10.1136/jmedgenet-2016-104132. [DOI] [PubMed] [Google Scholar]
- 49.Yagita Y., Shinohara K., Abe Y., Nakagawa K., Al-Owain M., Alkuraya F.S., Fujiki Y. Deficiency of a retinal dystrophy protein, acyl-CoA binding domain-containing 5 (ACBD5), impairs peroxisomal beta-oxidation of very-long-chain fatty acids. J Biol Chem. 2017;292:691–705. doi: 10.1074/jbc.M116.760090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishimura T., Tamura N., Kono N., Shimanaka Y., Arai H., Yamamoto H., Mizushima N. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 2017;36:1719–1735. doi: 10.15252/embj.201695189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim Y.J., Guzman-Hernandez M.L., Balla T. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev Cell. 2011;21:813–824. doi: 10.1016/j.devcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.English A.R., Voeltz G.K. Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol. 2013;15:169–178. doi: 10.1038/ncb2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishimura T., Mizushima N. The ULK complex initiates autophagosome formation at phosphatidylinositol synthase-enriched ER subdomains. Autophagy. 2017;13:1795–1796. doi: 10.1080/15548627.2017.1358344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voss C., Lahiri S., Young B.P., Loewen C.J., Prinz W.A. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisisae. J Cell Sci. 2012;125:4791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho I.T., Adelmant G., Lim Y., Marto J.A., Cho G., Golden J.A. Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum–mitochondrial contacts. J Biol Chem. 2017;292:16382–16392. doi: 10.1074/jbc.M117.795286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stone S.J., Vance J.E. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J Biol Chem. 2000;275:34534–34540. doi: 10.1074/jbc.M002865200. [DOI] [PubMed] [Google Scholar]
- 57.Kannan M., Lahiri S., Liu L.K., Choudhary V., Prinz W.A. Phosphatidylserine synthesis at membrane contact sites promotes its transport out of the ER. J Lipid Res. 2017;58:553–562. doi: 10.1194/jlr.M072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58••.Galmes R., Houcine A., van Vliet A.R., Agostinis P., Jackson C.L., Giordano F. ORP5/ORP8 localize to endoplasmic reticulum–mitochondria contacts and are involved in mitochondrial function. EMBO Rep. 2016;17:800–810. doi: 10.15252/embr.201541108. [DOI] [PMC free article] [PubMed] [Google Scholar]; ORP5/ORP8 have been previously shown to transport PS from the ER to the PM at ER–PM MCS. In this study, ORP5 and ORP8 are also localized to ER–mitochondria contacts and interact with the outer mitochondrial membrane protein, PTPIP51. At this membrane contact, PS is transferred from the ER to mitochondria.
- 59.Horibata Y., Sugimoto H. StarD7 mediates the intracellular trafficking of phosphatidylcholine to mitochondria. J Biol Chem. 2010;285:7358–7365. doi: 10.1074/jbc.M109.056960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Horibata Y., Ando H., Satou M., Shimizu H., Mitsuhashi S., Shimizu Y., Itoh M., Sugimoto H. Identification of the N-terminal transmembrane domain of StarD7 and its importance for mitochondrial outer membrane localization and phosphatidylcholine transfer. Sci Rep. 2017;(7):8793. doi: 10.1038/s41598-017-09205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper can be seen as a companion paper to Yanget al. [61••] that demonstrate that PC transfer to mitochondria requires a PC transfer protein that is localized to the outer membrane of the mitochondria.
- 61••.Yang L., Na C.L., Luo S., Wu D., Hogan S., Huang T., Weaver T.E. The phosphatidylcholine transfer protein Stard7 is required for mitochondrial and epithelial cell homeostasis. Sci Rep. 2017;7:46416. doi: 10.1038/srep46416. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotations to Ref [60••]