Abstract
Molecular characterization of the binding epitope of IL-23R and its cognate cytokine IL-23 is paramount to understand the role in autoimmune diseases and to support the discovery of new inhibitors of this protein–protein interaction. Our results revealed that HDX-MS was able to identify the binding epitope of IL-23R:IL-23, which opened the way to evaluate a peptide macrocycle described in the literature as disrupter of this autoimmune target. Thus, the characterization of the interactions of this chemotype by HDX-MS in combination with computational approaches was achieved. To our knowledge, this is the first reported structural evidence regarding the site where a small compound binds to IL-23R.
Keywords: IL-23R, HDX-MS, computational approaches, binding site, protein−protein interactions
IL-23 is a member of the IL-12 family of pro-inflammatory cytokines, which includes IL-12, IL-23, IL-27, and IL-35.1 IL-23 is mainly secreted by activated macrophages and dendritic cells located in peripheral tissues (skin, intestinal mucosa, and lung) and has been implicated in several autoimmune inflammatory disorders and cancer.2−4 IL-23 is a disulfide-linked composite cytokine that shares the p40 subunit with IL-12 but is distinguished from the latter by its cytokine unit p19.5 The receptor complex of IL-23 is composed of IL-12Rβ1 and IL-23R. The IL-12Rβ1 is also part of the IL-12 receptor complex, composed of IL-12Rβ1 and IL-12Rβ2. Recently, IL-23 binding to IL-23R has been observed crystallographically.6 However, the trimeric complex structure assembling IL-23, IL-23R, and IL-12Rβ1 is still elusive.
IL-23p19 is a nonglycosylated protein of 18.7 kDa and has the typical four-helix bundle structure characteristic of all IL-12 type cytokines.2 IL-23p40 is a glycosylated type 1 soluble protein of 34.7 kDa and consists of three domains.7 The binding of p40 to p19 is mediated by p40 domains D2 and D3 and p19 site I and strengthened by an intermolecular disulfide bridge, D1 is needed for binding of p40 to IL-12Rβ1.8
IL-12Rβ1 is a glycosylated type 1 membrane protein of 70.5 kDa with five extracellular fibronectin type III domains, of which D2 and D3 appear to comprise a cytokine binding module with the typical WSXWS motif in D2, a single trans-membrane domain and a cytoplasmic domain (Figure 1A).9 The IL-23R is a glycosylated type 1 membrane protein of 69.0 kDa, with three extracellular domains, a 37-amino acid long-stalk region, a single transmembrane domain, and a cytoplasmic domain. The extracellular domain of IL-23R contains a signal sequence, an N-terminal Ig-like domain, and two fibronectin type III domains. There are seven potential N-linked glycosylation sites in IL-23R. The transmembrane-proximal fibronectin type III domain contains a sequence WQPWS similar to the cytokine receptor signature WSXWS motif.10
Figure 1.
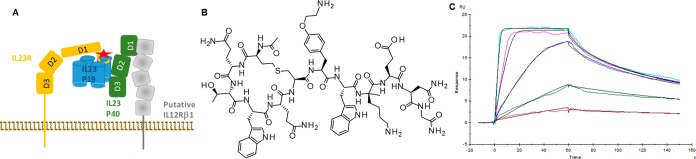
Schematic based on Bloch et al.6 showing the domains of IL-23R (orange), the p19 helical bundle of IL-23 (blue), the three domains of IL-23p40 (green, structurally identical to IL-12p40), and the binding site of compound C on D1 of IL-23R as identified in this paper (red star). The membrane (brown) and putative location of IL-12Rβ1 (light gray, required along with IL-23R to form the full IL-23 receptor complex) are also shown (A). Structure of the macrocyclic dodecapeptide Compound C inhibitor of IL-23R (B). SPR Sensorgrams (colors) of different concentrations of Compound C against IL-23R and their fit (black) to a 1:1 binding model (C). SPR revealed the following binding parameters: ka = 1.51 × 107 (1/Ms), kd = 2.74 × 10–2 (1/s), KD = 1.81 (nM). Because of the limited curvature observed from the sensorgrams, the SPR binding experiment appeared to be mass transport limited. Therefore, the binding parameters should be considered an estimate.
IL-23p40 interacts directly with IL-12Rβ1 and inhibits IL-12 and IL-23 signaling.11 It appears that IL-23 binds IL-23R with an affinity of 44 nM, while binding IL-12Rβ1 with an affinity of 2 μM; nonetheless, the affinity of the IL-23:IL-23R complex for IL-12Rβ1 has been described as 25 nM, despite no apparent interaction of IL-23R with IL-12Rβ1, implying that there is a cooperative effect, likely to be due to a conformational change of IL-23 upon binding IL-23R, which is indeed observed crystallographically.6
The discovery and development of highly effective anti-IL-23 and anti-IL-23R monoclonal antibodies in the past few years have shown significant efficacy in human studies for psoriasis, psoriatic arthritis, etc.12,13 Cases in point are Ustekinumab (targeting the p40 subunit of IL-12 and IL-23), Tildrakizumab, and Mirikizumab (targeting the p19 subunit of IL-23) that have revolutionized the treatment of plaque psoriasis.14−16 Similarly, small protein therapeutics (i.e., Adnectins, Alphabodies, etc.) have been developed to neutralize human IL-23.17,18 In analogy to the development of these biologic drugs, researches have also focused their attention on disrupting the IL-23:IL-23R protein–protein interaction (PPI) by the use of small molecules specifically targeting the IL-23R. For example, an octapeptide as modulator of IL-23R that exhibited anti-inflammatory effects has been recently reported.19 In addition to this, Protagonist Therapeutics published a patent application describing orally stable macrocycle peptide inhibitors of the IL-23R for treating inflammatory bowel disease (IBD).20 Although biochemical/cell-based assays and biophysical technical assessments have probed the binding of this chemo-type to IL-23R, no structural information on the IL23-R in complex with a small molecule has been published yet.
Small molecules or peptides that modulate the interactions of IL-23R with IL-23 would potentially represent a more convenient and less costly treatment for autoimmune diseases. In addition, such molecules would complement the existing therapies if orally active. However, deeper structural insights are needed to identify the molecular sites of interactions between IL-23R and small molecules, particularly if SAR optimization of the small molecule is required. Hydrogen/deuterium exchange coupled to mass spectrometry (HDX-MS) has proved to be a well-suited biophysical tool to study the characterization of protein–protein dynamics in solution and to evaluate the interactions of new chemical diversity with PPI targets.21−27
Here, we report the application of HDX-MS and computational approaches to map and characterize the binding interactions between IL-23 and protagonist peptide (named Compound C, Figure 1B) with IL-23R. Our results demonstrate the sensitivity of HDX-MS to detect and localize the sites that contribute to the interactions of IL-23R with IL-23 and the binding mode for the protagonist peptide, which up to today has not been accessible by conventional structure elucidation techniques. Furthermore, additional biophysical techniques like surface plasmon resonance (SPR) and functional assays were used to corroborate the binding site proposal. To our knowledge, this is the first reported structural evidence regarding the site where a small compound binds to IL-23R.
To identify the dynamic of the IL-23R interface and to evaluate how IL-23 binding affects the conformation of this receptor, HDX-MS of the receptor alone and in complex with the cytokine was carried out. HDX-MS experiments were performed as described elsewhere and in the Supporting Information.24,27 A key step in the HDX-MS protocol is the digestion of the intact protein to yield proteolytic peptides for measurement by LC–MS. IL-23R is a heavily glycosylated protein that contains a single transmembrane domain. Finding conditions to yield high sequence coverage was challenging. However, upon optimization of the quench and digestion conditions, a total of 48 reproducible peptides resulted from the proteolysis step. These peptides covered 63% of the linear sequence of IL-23R (Figure S1 in Supporting Information). A model of the IL-23R based upon the recent crystal structure (PDB ID 5MZV)6 shows the incorporation of deuterium into IL-23R in the absence of IL-23 (see Figure 2). HDX data revealed that part of the N-terminal domain (residues 24–30 and 55–61) and the C-terminal domain (last 60 amino acids) exhibited the highest deuterium uptake even at the earliest time points and across the entire labeling course, consequently suggesting highly dynamic regions in the protein. Contrary to this effect, residues 154–160, 196–224, and 282–292 were identified as the less flexible regions (solvent protected). The HDX-MS on the apo-receptor therefore suggested that these domains exhibit different degrees of hydrogen-bonding and/or solvent accessibility.
Figure 2.
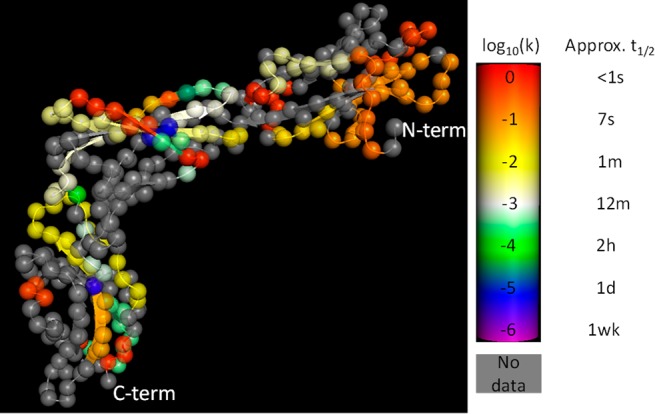
Apo heat map illustrated on a model of IL-23R built from 5MZV(6) by removal of the nanobody and glycosyl groups, addition of missing side chains, selection of one from alternate residue positions, appropriate capping of terminal atoms, and restrained minimization. The color (generated using the pymol spectrum command) of each α carbon represents the magnitude of the deuterium uptake in calculated log10 H–D exchange rate constant (purple ≤ −6.0; blue, green, white, yellow, orange, red ≥ 0.0). An additional 56 residues at the C-terminus showed a highly dynamic region with log10(k) in the range 0.0 to −0.5, consistent with residues beyond Pro316 (crystallographic numbering) not being located in the crystal structure of 5MZV. The label “C-term” refers to the last crystallographically visible C-terminal residue.
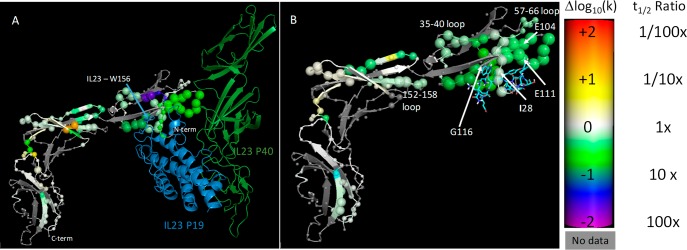
The differential HDX analysis of IL-23R in the presence and absence of IL-23 revealed two regions located within the N-terminal domain (residues 24–42 and 88–113) that exhibited strong protection from deuterium uptake (see Figure 3A). The region 24–42 covered by overlapping peptide fragments (24–30, 24–32, and 31–42) showed a large decrease in deuterium uptake at the shortest time point, but negligible difference at the longest time point (see uptake profile plots Figure S2A in Supporting Information). In contrast, peptide fragments located in region 88–113 display the largest difference in deuterium uptake at all the exposure time points. Upon IL-23 binding, peptide fragments 88–99, 92–99, and 100–113 exhibited strong protection from 10 s to 2 h. These results suggested two discontinuous epitopes that are important for binding of the cytokine and of the two regions in the receptor; 88–113 might be the primary site for IL-23 interaction. Our HDX analysis is thereby in excellent agreement with the X-ray data, demonstrating that IL-23 binds to the N-terminal immunoglobulin domain of IL-23R not only in the solid state but also under more physiologically relevant conditions.
Figure 3.
Differential HDX of IL-23R in complex with IL-23 (A: IL-23p19 is shown in blue and IL-23p40 in green) and Compound C (B: ligand shown as sticks) vs the apo state, illustrated on a model of IL-23-IL-23R built from 5MZV. The color (generated using the pymol spectrum command) of each α carbon represents the magnitude of the change in calculated H–D exchange rate constant upon binding of the corresponding ligand, with purple ≤ −2.0; blue, green, white = 0.0; yellow, orange, red ≥ +2.0 log units. The size of each sphere indicates confidence that the apparent signal is truly nonzero, based upon the mean and standard deviation of the acceptable models from the Bayesian analysis, with 0.25 Å radius indicating zero confidence and 2.0 Å indicating a confidence of 1.0.28
To further study the interaction between IL-23R and protagonist peptide observed with HDX-MS, binding activity was probed through a SPR assay. Internal SPR data suggested that it is a fast binding peptide with low single digit nanomolar affinity. A Kd value of 2 nM was found for the interaction between this peptide and IL-23R (see Figure 1C and Supporting Information). In addition, the functional activity of this molecule to disrupt the protein–protein interaction between IL-23 and IL-23R was assayed with two different detection technologies, trFRET and alphaLISA. The IC50 values for Compound C to disrupt the interaction between these two proteins was also in the single digit nanomolar range, being 2.7 ± 0.1 and 5.1 ± 1.0 nM for alphaLISA and TR-FRET assays, respectively (n = 3 for both assays; mean ± SD).
Figure 3B shows the differential HDX analysis of the IL-23R in complex with Compound C. Two discontinuous regions in the receptor, spanning residues 67–75 and segments 88–99 and 100–113 located within the N-terminal in domain 1, experienced significant decreases in deuterium uptake when bound to Compound C (see uptake profile plots Figure S2B in Supporting Information). A constant degree in reduced deuterium uptake is observed across the exposure time points with the exception of the longest time point, which exhibits a minor difference. It is worthy of note that domain 2 and domain 3 were hardly affected upon ligand binding. Thus, the HDX profiles suggest that the N-terminal immunoglobulin-like domain provides the entire binding interface for this peptide ligand. Our HDX-MS data allow locating the binding epitope of a macrocyclic small molecule against IL-23R for the first time.
It is immediately apparent from comparison of Figure 3A,B that IL-23 and Compound C broadly affect the same regions of the IL-23R protein, with most of the effects occurring in the N-terminal Ig-like domain, and perhaps some indirect effects being observed in the proximal β sheet and turn regions of domain 2 of IL-23R. The N-terminal “hook” of IL-23R may be slightly more protected by IL-23 than by the much smaller Compound C, while the binding site of IL-23 W-156, indicated in Figure 3A and known to be a critical residue for binding of the cytokine to its receptor,6,8 is close to the region of maximal protection of IL-23R by IL-23. The similar if slightly reduced protection seen in this region by Compound C suggests that this ligand also occupies this site.
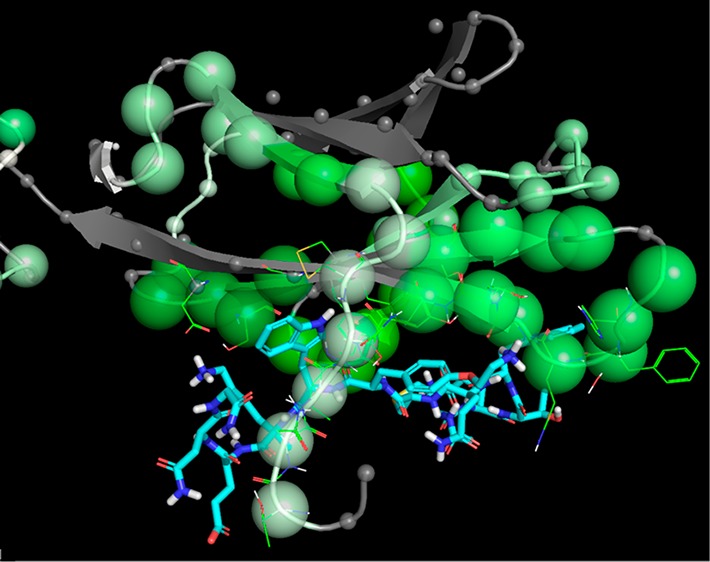
In order to provide some support for the hypothesis that a binding mode could exist for Compound C that would be consistent with the observed effects on HDX upon ligand binding, we carried out a docking study of compound C using the protein structure of IL-23R derived from the crystal structure with PDB ID 5MZV. Compound C was prepared for docking using the standard ligprep procedure, and docking was carried out using the SP_peptide method of GLIDE, with van der Waals scaling of 0.6, using macrocycle sampling with PRIME, enhanced sampling (by 4 times default values), and the GLIDE screening setting set to keep 1,000,000 poses per ligand with a scoring window of 200 and the best 20,000 poses kept for energy minimization, with the ligand pose filter set to 20 kcal/mol rather than the default 0.29 The best 1000 poses were subjected to postdocking minimization, and the best 100 of those retained. The poses thus obtained were viewed with the HDX information, and it was immediately apparent that one of the 100 poses (the second-best by docking score with a value of −8.061 vs −8.072 for the “best” pose) satisfied two key criteria that the IL-23-W156 binding site should be occupied by an aromatic amino acid side chain from Compound C (see above) and that the ligand should be in contact with many of the regions that were highlighted by the HDX experiment (see below). This pose is illustrated in Figure 4, and while it is clearly not guaranteed to be the true binding pose, it does show that at least it is possible to find hypothetical binding poses that are consistent with the HDX data and other known features of importance (Figure S3 in Supporting Information). In this pose, the ligand is in direct hydrogen-bonding contact with I28, which is in a region somewhat protected from exchange by this ligand (the apparently weak signal is likely due to very fast exchange in the apo state). Five additional backbone N–H groups are within 4 Å of at least one ligand atom in this pose, four of which (K72, E111–L113) are also strongly protected to exchange, while the remaining G116 is not in the covered region. Equally, only one of the regions where significant protection was detected had no residues in contact with this ligand pose; the exception is the loop from K57–F66, which might undergo some conformational change upon ligand binding since it is stacked above and disulfide-linked to the E104–E111 loop that is directly in contact with the ligand in this putative pose. There were also smaller trends toward protective effects seen in some other regions not in contact with the ligand, possibly due to modulation of the stability of the secondary structure (e.g., 35–40, 90–93) and/or tertiary structure (e.g., 152–158) of the protein in response to ligand binding.
Figure 4.
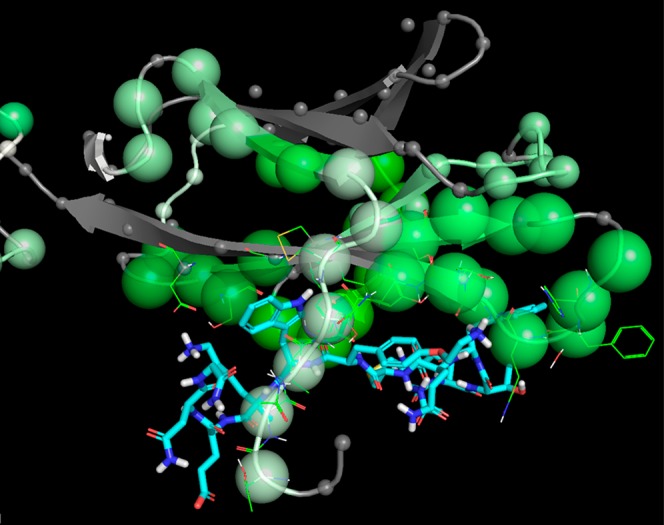
Closeup of the hypothetical binding mode of Compound C to IL-23R using docking study.
In conclusion, HDX-MS and computational analyses revealed valuable information about the interactions of IL-23R and its cognate cytokine IL-23 as well as locating the binding epitope of a macrocyclic small molecule against IL-23R. These studies provide new insights into the quaternary structure of this protein–protein interaction and protein–small molecule interaction in solution. Our HDX analysis is in excellent agreement with the X-ray data, demonstrating that IL-23 binds to the N-terminal immunoglobulin domain of IL-23R not only in the solid state but also under more physiologically relevant conditions. Importantly, the HDX data allowed validation of the docking hypothesis about the existence of a binding mode for Compound C in IL-23R. Indeed, this new chemo-type might be used as a foundation for discovery and characterization of small molecules targeting this cytokine receptor.
Acknowledgments
We thank Leticia Cano and Ruben Haro for technical assistance.
Glossary
ABBREVIATIONS
- IL-23
interleukin 23
- IL-23R
interleukin 23 receptor
- IL-23Rβ1
interleukin 23 receptor beta1
- IBD
inflammatory bowel disease
- PPI
protein–protein interaction
- HDX-MS
hydrogen–deuterium exchange mass spectrometry
- SPR
surface plasmon resonance.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00255.
Protein expression and purification, synthesis of Compound C, SPR methodology, biochemical assays, HDX-MS and computational experimental details, HDX peptide map of IL-23R, HDX kinetic plots, and 2D representation of the interactions between Compound C and IL-23R (PDF)
Author Present Address
# Proteomics Unit, Spanish National Cancer Research Centre. Madrid, Spain.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by Eli Lilly and Company (salaries for all authors, reagents and supplies).
The authors declare no competing financial interest.
Supplementary Material
References
- Vignali D. A.; Kuchroo V. IL-12 Family Cytokines: Immunological Playmakers. Nat. Immunol. 2012, 13, 722–728. 10.1038/ni.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie B. S.; Kastelein R. A.; Cua D. J. Understanding the IL-23-IL-17 immune pathway. Trends Immunol. 2006, 27, 17–23. 10.1016/j.it.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Tang C.; Chen S.; Qian H.; Huang W. Interleukin-23: as a drug target for autoimmune inflammatory diseases. Immunology 2012, 135, 112–124. 10.1111/j.1365-2567.2011.03522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxford A. L.; Mair F.; Becher B. IL-23: one cytokine in control of autoimmunity. Eur. J. Immunol. 2012, 42, 2263–2273. 10.1002/eji.201242598. [DOI] [PubMed] [Google Scholar]
- Oppmann B.; Lesley R.; Blom B.; Timans J. C.; Xu Y.; Hunte B.; Vega F.; Yu N.; Wang J.; Singh K.; Zonin F.; Vaisberg E.; Churakova T.; Liu M.; Gorman D.; Wagner J.; Zurawski S.; Liu Y.; Abrams J. S.; Moore K. W.; Rennick D.; de Waal-Malefyt R.; Hannum C.; Bazan J. F.; Kastelein R. A. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 2000, 13, 715–725. 10.1016/S1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- Bloch Y.; Bouchareychas L.; Merceron R.; Składanowska K.; Van des Bossche L.; Detry S.; Govindarajan S.; Elewaut D.; Haerynck F.; Dullaers M.; Adamapoulos I. E.; Savvides S. N. Structural activation of pro-inflammatory human cytokine IL-23 cognate IL-23 receptor enables recruitment of the shared receptor IL-23Rβ1. Immunity 2018, 48, 1–14. 10.1016/j.immuni.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C.; Johnston S. C.; Tang J.; Stahl M.; Tobin J. F.; Somers W. S. Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12. EMBO J. 2000, 19, 3530–3541. 10.1093/emboj/19.14.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder J.; Moll J. M.; Baran P.; Grötzinger J.; Scheller J.; Floss D. M. Non-Canonical Interleukin 23 Receptor Complex Assembly. J. Biol. Chem. 2015, 290, 359–370. 10.1074/jbc.M114.617597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua A. O.; Chizzonite R.; Desai B. B.; Truitt T. P.; Nunes P.; Minetti L. J.; Warrier R. R.; Presky D. H.; Levine J. F.; Gately M. K.; Gubler U. Expression Cloning of a Human IL-12 Receptor Component. A New Member of the Cytokine Receptor Superfamily with Strong Homology to gp130. J. Immunol 1994, 153, 128–136. [PubMed] [Google Scholar]
- Parham C.; Chirica M.; Timans J.; Vaisberg E.; Travis M.; Cheung J.; Pflanz S.; Zhang R.; Singh K. P.; Vega F.; To W.; Wagner J.; O’Farrell A.-M.; McClanahan T.; Zurawski S.; Hannum C.; Gorman D.; Rennick D. M.; Kastelein R. A.; de Waal Malefyt R.; Moore K. W. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rβ1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 2002, 168, 5699–5708. 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- Mattner F.; Fischer S.; Guckes S.; Jin S.; Kaulen H.; Schmitt E. The interleukin-12 subunit p40 specifically inhibits effects of the interleukin-12 heterodimer. Eur. J. Immunol. 1993, 23, 2202–2208. 10.1002/eji.1830230923. [DOI] [PubMed] [Google Scholar]
- Campa M.; Mansouri B.; Warren R.; Menter A. A review of biologic therapies targeting IL-23 and IL-17 for use in moderate-to-severe plaque psoriasis. Dermatol. Ther (Heidelb) 2016, 6, 1–12. 10.1007/s13555-015-0092-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K. F.; Isaacs J. D. Novel therapies for immune-mediated inflammatory diseases: What can we learn from their use in rheuma-toid arthritis, spondyloarthritis, systemic lupus erythematosus, pso-riasis, Crohn’s disease and ulcerative colitis?. Ann. Rheum. Dis. 2018, 77, 175–187. 10.1136/annrheumdis-2017-211555. [DOI] [PubMed] [Google Scholar]
- Papp K.; Gottlieb A. B.; Naldi L.; Pariser D.; Ho V.; Goyal K.; Fakharzadeh S.; Chevrier M.; Calabro S.; Langholff W.; Krueger G. Safety surveillance for Ustekinumab and other psoriasis treatments from the psoriasis longitudinal assessment and registry (PSOLAR). J. Drugs Dermatol 2015, 14, 706–714. [PubMed] [Google Scholar]
- Papp K.; Thaci D.; Reich K.; Rield E.; Langley R. G.; Krueger J. G.; Gottlieb A. B.; Nakagawa H.; Bowman E. P.; Mehta A.; Li Q.; Zhou Y.; Shames R. Tildrakizumab (MK-3222), an anti-interleukin-23p19 monoclonal antibody, improves psoriasis in a phase IIb randomized placebo-controlled trial. Br. J. Dermatol. 2015, 173, 930–939. 10.1111/bjd.13932. [DOI] [PubMed] [Google Scholar]
- Beidler C. B.; Bright S. W.; Girard D. S.; Kikly K. K.. Antibodies that bind to IL-23. Patent Appl. US20140255422 A1, filed Mar 04, 2014.
- Simeon R.; Chen Z. In vitro-engineered non-antibody protein therapeutics. Protein Cell 2018, 9, 3–14. 10.1007/s13238-017-0386-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet J.; Verstraete K.; Bloch Y.; Lorent E.; Wen Y.; Devreese B.; Vandenbroucke K.; Loverix S.; Hettmann T.; Deroo S.; Somers K.; Henderikx P.; Lasters I.; Savvides S. N. Structural basis of IL-23 antagonism by an Alphabody protein scaffold. Nat. Commun. 2014, 5, 5237–5249. 10.1038/ncomms6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiniou C.; Domínguez-Punaro M.; Cloutier F.; Erfani A.; Ennaciri J.; Sivanesan D.; Sanchez M.; Chognard G.; Hou X.; Rivera J. C.; Beauchamp C.; Charron G.; Vilquin M.; Kuchroo V.; Michnick S.; Rioux J. D.; Lesage S.; Chemtob S. Specific targeting of the IL-23 receptor, using a novel small peptide noncompetitive antagonist, decreases the inflammatory response. Am. J. Physiol. Regul. Integr. Comp. Physiol 2014, 307, 1216–1230. 10.1152/ajpregu.00540.2013. [DOI] [PubMed] [Google Scholar]
- Bhandari A.; Bourne G.; Cheng X.; Frederick B. T.; Zhang J.; Patel D. V.; Liu D.. Oral peptide inhibitors of interleukin-23 receptor and their use to treat inflammatory bowel diseases. Patent Appl. US 2018/0079782 A1, published Mar 22, 2018.
- Wei H.; Mo J.; Tao L.; Russell R. J.; Tymiak A. A.; Chen G.; Iacob R. E.; Engen J. R. Hydrogen/deuterium exchange mass spectrometry for probing higher order structure of protein therapeutics: methodology and applications. Drug Discovery Today 2014, 19, 95–102. 10.1016/j.drudis.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R. Y.; Chen G. Higher order structure characterization of protein therapeutics by hydrogen/deuterium exchange mass spectrometry. Anal. Bioanal. Chem. 2014, 406, 6541–6558. 10.1007/s00216-014-7924-3. [DOI] [PubMed] [Google Scholar]
- Iacob R. E.; Krystek S. R.; Huang R. C-H.; Wei H.; Tao L.; Lin Z.; Morin P. E.; Doyle M. L.; Tymiak A. A.; Engen J. R.; Chen G. Hydrogen deuterium exchange mass spectrometry applied to IL-23 interaction characteristics: potential impact for therapeutics. Expert Rev. Proteomics 2015, 12, 159–169. 10.1586/14789450.2015.1018897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada A.; Broughton H.; Jones S.; Chalmers M. J.; Dodge J. A. A binding site on IL-17A for Inhibitory macrocycles revealed by hydrogen/deuterium exchange mass spectrometry. J. Med. Chem. 2016, 59, 2255–2260. 10.1021/acs.jmedchem.5b01693. [DOI] [PubMed] [Google Scholar]
- Li J.; Wei H.; Krystek S. R. Jr.; Bond D.; Brender T. M.; Cohen D.; Feiner J.; Hamacher N.; Harshman J.; Huang R. Y-C.; Julien S. H.; Lin Z.; Moore K.; Mueller L.; Noriega C.; Sejwal P.; Sheppard P.; Stevens B.; Chen G.; Tymiak A. A.; Gross M. L.; Schneeweis L. A. Mapping the energetic epitope of an antibody/interleukin-23 interaction with hydrogen/deuterium exchange, fast photochemical oxidation of proteins mass spectrometry, and alanine shave mutagenesis. Anal. Chem. 2017, 89, 2250–2258. 10.1021/acs.analchem.6b03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R. Y.; Krystek S. R. Jr; Felix N.; Graziano R. F.; Srinivasan M.; Pashine A.; Chen G. Hydrogen/deuterium exchange mass spectrometry and computational modeling reveal a discontinuous epitope of an antibody/TL1A Interaction. MAbs 2018, 10, 95–103. 10.1080/19420862.2017.1393595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting J. P.; Tung F.; Antonysamy S.; Wasserman S.; Jones S. B.; Zhang F. F.; Espada A.; Broughton H.; Chalmers M. J.; Woodman M. E.; Bina H. A.; Dodge J. A.; Benach J.; Afshar S. Utilization of peptide phage display to investigate hotspots on IL-17A and what it means for drug discovery. PLoS One 2018, 13, 1–18. 10.1371/journal.pone.0190850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzberg D. J.; Broughton H. B.; Pellarin R.; Chalmers M. J.; Espada A.; Dodge J. A.; Pascal B. D.; Griffin P. R.; Humblet C.; Sali A. A residue-resolved Bayesian approach to quantitative interpretation of hydrogen–deuterium exchange from mass spectrometry: Application to characterizing protein–ligand interactions. J. Phys. Chem. B 2017, 121, 3493–3501. 10.1021/acs.jpcb.6b09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubert-Brohman I.; Sherman W.; Repasky M.; Beuming T. Improved docking of polypeptides with Glide. J. Chem. Inf. Model. 2013, 53, 1689–1699. 10.1021/ci400128m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.