Abstract
Reconfiguration of membrane protein channels for gated transport is highly regulated under physiological conditions. However, a mechanistic understanding of such channels remains challenging owing to the difficulty in probing subtle gating-associated structural changes. Herein, we show that charge neutralization can drive the shape reconfiguration of a biomimetic 6-helix bundle DNA nanotube (6HB). Specifically, 6HB adopts a compact state when its charge is neutralized by Mg2+; whereas Na+ switches it to the expanded state, as revealed by MD simulations, small-angle X-ray scattering (SAXS), and FRET characterization. Furthermore, partial neutralization of the DNA backbone charges by chemical modification renders 6HB compact and insensitive to ions, suggesting an interplay between electrostatic and hydrophobic forces in the channels. This system provides a platform for understanding the structure-function relationship of biological channels and designing rules for the shape control of DNA nanostructures in biomedical applications.
Keywords: DNA nanotechnology, molecular channels, molecular dynamics, SAXS, shape reconfiguration
The shape reconfiguration of membrane protein channels plays an important role in living systems. It is closely related to the gating of ions, water, and other entities that are vital for many cell functions. These shape changes are often stimulated by membrane tension and electric fields.[1] Although the ionic selectivity, rectification, and gating function of membrane protein channels have been well studied, elucidation of associated subtle structural changes remains challenging.
Artificial nanotubes (for example, carbon nanotubes) have emerged as promising models for biological channels owing to their nanoscale features and tailorable properties.[2] Because of the endogenous nature and high programmability of DNA, self-assembled DNA nanotubes have attracted intense interest.[3] These DNA nanotubes, when appropriately modified, can be readily inserted into membranes[4] to function as biomimetic channels. Molecular dynamics (MD) simulations have also revealed interesting properties,[5] including ion flow and gating-like behaviors in these DNA-based nanochannels.[6]
In this work, we designed a computational/experimental approach to study the charge neutralization-induced shape reconfiguration of a 6-helix bundle (6HB) DNA nanotube with three typical states, namely, expanded, compact, and partially compact states. Specifically, by combining MD simulation with structural analysis using small-angle X-ray scattering (SAXS) and Förster resonance energy transfer (FRET), we demonstrated that 6HB adopted a more compact and less expanded shape in a solution with Mg2+ as compared to that with Na+. At lower ion concentrations, 6HB underwent considerable shape expansion; whereas 6HB with an ethyl-phosphorothioate substitution of the DNA backbone remained compact within a volume with a smaller radius.
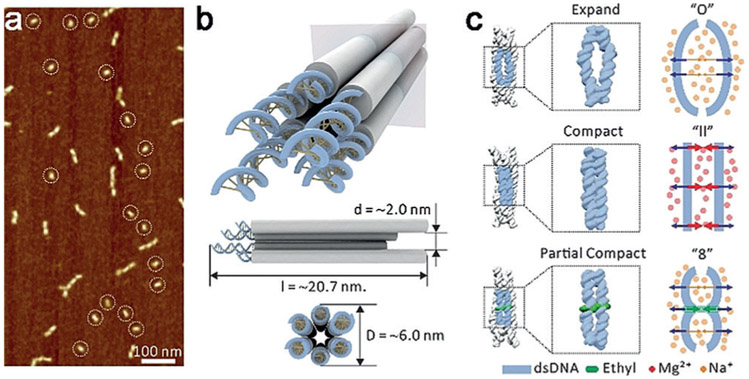
A typical membrane protein channel undergoes the transition from the closed state to the open state to function in physiological conditions,[7] probably via a partially or transiently closed state.[8] Similarly, our MD simulations of the biomimetic 6HB under three different physiological conditions (Na+ and Mg2+ conditions or with DNA backbone modification) show that the conformations of 6HB can adopt three states, an expanded state, a compact state, and a partially compact state (Figure 1). In the expanded state, the adjacent helixes within 6-HB were repelled by the electrostatic repulsion to form an “O” shape, whereas in the compact state, the adjacent helixes kept a “II” shape with balanced electrostatic repulsion, and in the partially compact state, owing to the elimination of the electrostatic repulsion by the ethyl-phosphorothioate, the adjacent helixes formed an “8” shape. The possibility of shape reconfiguration of the 6HB under different physiological conditions facilitates its potential applications as biomimetic channels.
Figure 1.
a) Atomic force microscopy (AFM) image of mono dispersed 6HB. The circles highlight the 6HB monomers. b) Conformation of the designed 6HB. c) Schematic of expanded, compact, and partially compact states.
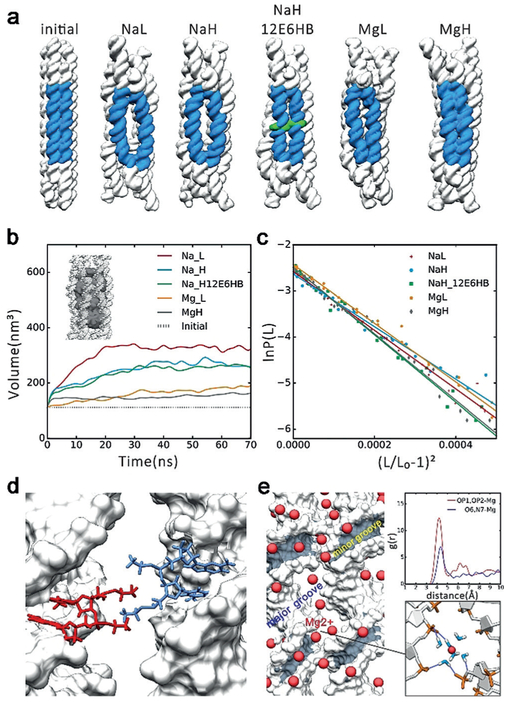
The 70-ns MD simulations were carried on the following four systems: 6HB solvated with 250 mM Na+ (NaL) and 500 mM Na+ (NaH) and 6HB solvated with 125 mM Mg2+ (MgL) and 250 mM Mg2+ (MgH). 6HB is composed of 936 bases, with a channel inner diameter of 2.0 nm, an outer diameter of approximately 6.0 nm, and a length of 20.7 nm (Supporting Information, Figures S1-S4 and Tables S4-S8). The simulation results (Figure 2a) show that the nanostructures of 6HB with both Na+ and Mg2+ counterions at low concentrations are in expanded state. With the increase of ion concentration, 6HB shows a more compact conformation. In the presence of Mg2+, 6HB nanostructures have much smaller inner volumes (Figure 2b and the Supporting Information, Figure S10), inter-helix distances, and cross-sectional area compared to the case with Na+ (Figure S5). Moreover, our calculations of the stretch modulus and persistence length of 6HB in Na+ and Mg2+ (Figure 2c and the Supporting Information, Figure S9 and Table S1) showed that 6HB with Mg2+ are more rigid along the length direction. Experimentally, the structural difference of 6HB in different solutions (12.5 mM Mg2+, 50.0 mM Mg2+, 125.0 mM Mg2+, and 100.0 mM Na+) was first examined by gel electrophoresis on native 6% polyacrylamide gel (PAGE). As shown in the Supporting Information, Figure S11 a, there were no obvious differences between the bands of the magnesium buffered samples (12.5, 50.0, 125.0 mM Mg2+), indicating well-folded 6HB-DNA nanostructures, while the 100.0 mM sodium buffered sample exhibited evident dispersion on the band, indicating a possible loosely folded 6HB structure under this condition.
Figure 2.
MD simulation results. a) The initial conformations and averaged conformations of five systems in the last 10 ns. The big “O” rings are shown in blue, the ethyl-phosphorothioate-substituted DNA in green. NaL represents 6HB with 250 mM Na+, NaH represents 6HB with 500 mM Na+, NaH_12E6HB represents 6HB with 12 ethyl-phosphorothioated nucleotides with 500 mM Na+, MgL represents 6HB with 125 mM Mg2+, MgH represents 6HB with 250 mM Mg2+. b)Plot of inner volume vs. time for the center 42 bp of 6HBs. c)Stretch modulus of center 42 bp of 6HBs. d) Interaction model of 12E6HB, the hydrophobic ethyl group interacts in finger crossed conformation. e) Binding model of Mg2+ߝ6H2O. Left: Distribution of Mg2+ around helix (Mg2+ in red). Top right: Radial pair distribution function g(r) of Mg2+ around OP atoms and O6, N7 atoms of guanine. Bottom right: Bridging of the phosphates of two adjacent helixes by Mg2+–6H2O, the hydrogen bonds are shown in dark blue.
FRET and SAXS were utilized to track the change in distance between two DNA helixes. The ideal initial distance between the donor Cy3 fluorophore and the acceptor Cy5 is 4.2 nm (Figure S11c). At a higher ionic strength, the fluorescent intensity of Cy5 (acceptor) was higher, suggesting a higher FRET efficiency and closer distance between Cy3–Cy5 FRET pairs (Figure S11 b,c and Table S2). This implies that 6HB becomes more rigid when the ionic environment changes from 100.0 mM Na+, 12.5 mM Mg2+, and 50 mM Mg2+ to 125 mM Mg2+. The global reconfiguration of 6HB was further validated using SAXS[9] (see detailed descriptions in the Supporting Information, Figures S11-S14).
The molecular model of 6HB at different expanded states are presented in Figure S11 f. In the presence of 125 mM MgCl2, 6HB showed a most compact structure, which is consistent with MD simulations. With the decrease of ion concentration, or when changing from a divalent to a monovalent cation (Mg2+ to Na+), 6HB expanded, which is also reflected in their 2D SAXS profiles. In comparison to the initial models, some regions in the refined 6HB models swelled and the helices at the ends of the bundle slightly bent outwards, which means a more severe electrostatic repulsion force observed in experiments than in the theoretical simulation. These phenomena are consistent with the observation of large DNA origami objects.[10] Taken together, these data suggest an important role for the electrostatic neutralization, which can stabilize the highly negatively charged DNA nanotube.
The combination of MD simulations with SAXS and FRET characterization shows that 6HB adopts a compact state when its charge is neutralized with divalent ions (Mg2+) at high concentrations; whereas monovalent ions (Na+) switch it to the expanded state. Counterions can form an ionic atmosphere around DNA, hence neutralizing the negatively charged phosphate and stabilizing the DNA system.[11] Mg2+ has stronger interactions than Na+ with water molecules, so Mg2+ can form a tighter and more stable Mg2+–6H2O complex, which can function as hydrogen bond donors and interact with DNA mainly through hydrogen bonds. In contrast, Na+ ions prefer a direct interaction with phosphate. Moreover, Mg2+–6H2O can bridge two hydrogen bond acceptors at a distance of 10 Å. Moreover, the hydrogen bonds between negatively charged atoms and hydrated Mg2+ ions are much stronger than those of hydrated Na+. Our MD trajectory shows that Mg2+ is mainly distributed in following zones: Hydrated Mg2+ bridging the phosphate outside the minor groove, hydrated Mg2+ binding to O6 and N7 of the CG base in the major groove (Figure 2e and the Supporting Information, Figure S6), which is consistent with previous reports.[11b,12] These two kinds of distribution make DNA more rigid in hydrated Mg2+ systems. Moreover, hydrated Mg2+ can also bridges phosphates of two adjacent helixes in 6HB (Figure 2e). The strong interactions between 6HB and hydrated Mg2+ kept 6HB in the compact state, which make the DNA helix more rigid with Mg2+ than with Na+.
Although counterions (such as Mg2+ at high concentration) can reduce the electrostatic repulsion of adjacent bundles and hence stabilize 6HB, the repulsion between two negative helical bundles will still deform the 6HB. Removing the negative charges on the DNA backbone would be an alternative to stabilize 6HB. To demonstrate this concept, we replaced 12 phosphate groups of 6HB with ethyl-phosphor-othioate. A 70-ns MD simulation with 500 mM Na+ (NaH_12E6HB) (see Table S5 for the position of ethyl-phosphorothioate 6HB) indicated that ethyl-phosphoro- thioate nucleotides can reduce the expansion of 12E6HB with a finger crossed conformation in the middle of O-ring, which can keep two adjacent helixes tight (Figure 2a and Figure 3a). The inner volume of last 20 ns (of MD) of NaH_12E6HB is 259 nm3 and cross-sectional area is 31.3 nm2, while for NaH system, these two values are 275 nm3 and 33.4 nm2, respectively (Figure 2b and Figure S5). Especially, the “O” ring in unmodified 6HB in the presence of Na+ is converted to an “8” shape (Figure 1 and the Supporting Information, Figure S8). Both of the distances between helixes 1 and 2 and between helixes 4 and 5 decrease by about 1 nm. Moreover, the stretch modulus and persistence length analysis of 6HB (Table S1, Figure 2c, and Figure S9) show that with ethyl-phosphorothioate substitution, 12E6HB was more rigid in the length direction and bending angle than the NaH system.
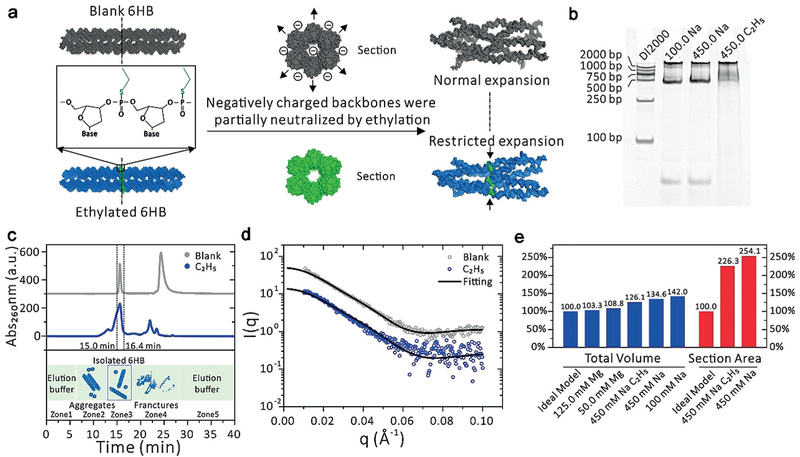
Figure 3.
Experimental verification of the MD results for 6HB with and without ethylation {noted as C2H5 and blank). a) Schematic of ethylation experiment and the restricted expansion caused by a two-base-wide “ethyl ring”. b) Non-denaturing gel electrophoresis of samples on native 6% PAGE gel synthesized in 450 mM NaCl buffer. c) Elution profile of C2H5 and blank. The five zones correspond to the elution buffer (1), aggregates (2), isolated 6HB-DNA nanostructures (3), fractures and excess ssDNA chain (4), and elution buffer (5). d) Experimental data and theoretical fitting of SAXS intensities for C2H5 and blank sample group. The PDB models used for fitting are shown in Figure 3a, on the right. e) Total volume change for all the samples, and section area change for C2H5 and blank sample group, calculated from the PDB files. We defined the section area as the plane crossing through the center of the “ethyl ring”.
Experimentally, we synthesized partially ethylated 6HB (Figure 3b) and diluted it in 450.0 mM Na+ solution for SAXS analysis (Figure 3d). The slight partial reconfiguration resulted in two very similar scattering curves of 6HB with and without ethylation. To describe the changes of 6HB-DNA brought about by the “ethyl ring”, the changes of the total volume and section area were calculated (Figure 3e). Both the total volume and section area of 6HB-DNA decrease by introducing the “ethyl ring” because the electrostatic repulsion force is reduced by the uncharged two-base-wide ethyl group. The radius of gyration (Rg) for a hollow cylinder[13] can be calculated from Equation (1):
| (1) |
where ri andro are the inner and outer shell radii, respectively, and h is the length of cylinder. Judging from the geometrical parameters of the PDB models, since h did not change much, the Rg decrease of C2H5 sample (Table S3) was caused by the decrease of the radius near the ethyl ring area.
Our computational/experimental method shows that partial ethylation of the 6HB backbone can switch the 6HB from expanded state to partially compact state, mainly owing to the elimination of the negative charge on the DNA backbone and addition of Van der Waals (vdW) interactions between helixes (Figures 2d and 3a). Finally, we simulated a neutral 6HB, the fully ethyl-phosphorothioated 6HB (Full- E6HB). After a 70-ns MD simulation, FullE6HB holds a more compact conformation than unmodified 6HB in the presence of Mg2+ of high concentration (simulation data are shown in Figure S7 in the Supporting Information). We thus expect that the design of DNA nanostructures with reduced electrostatic repulsive forces might be promising drug delivery tools.
In summary, we have demonstrated that a biomimetic 6HB can be switched from an expanded state to a (partially) compact state in aqueous solution by charge neutralization through changing ion type and concentration or through chemical modification of the DNA backbone. 6HB is a user-defined model channel based on its programmable structural design and controllable modification features. As DNA is a biocompatible material present in the organisms themselves, DNA-based 6HB nanostructures could have potential biomedical and biological applications. For example, drug molecules can be incorporated into the DNA nanotube and released through shape reconfiguration. The charge neutralization-driven shape reconfiguration of 6HB is highly relevant to functional membrane protein channels. The changes in ion type and ionic strength are very common in living organisms, and are closely related to vital biological functions, such as for voltage-gated potassium channels[14] and calcium-activated potassium channels.[15] Thus, our finding has important implications in uncovering the physiological function of natural ion channels. The mechanism presented herein helps explain previous observations on interhelical spacing of DNA nanostructures.[16] Furthermore, the concept of charge neutralization-driven shape reconfiguration of 6HB, either by ion-atmosphere variation or chemical modification, provides an innovative, simple, and convenient route for controlled release in drug delivery.
Supplementary Material
Acknowledgements
J.L. thanks the National Science Foundation of China (No. 31070640) for supporting this study. C.F. thanks the National Science Foundation of China (No. 21329501, 21390414, 21373260, 31371015 and 21603262), National Key R&D Program of China (2016YFA0201200, 2016YFA0400900), Key Research Program of Frontier Sciences, CAS (QYZDJSSW-SLH031), Natural Science Foundation of Shanghai (No. 17ZR1442200) for supporting this study. S.Y. thanks the NIH (grant no. R01GM114056). The authors thank the staff from BL19U2 beamline of National Facility for Protein Science Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility (SSRF), for assistance during data collection.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Pi Liu, State Key Laboratory of Medicinal Chemical Biology, College of Pharmacy and Tianjin Key Laboratory of Molecular Drug Research, Nankai University Tianjin 300353 (China); Biodesign Center, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences Tianjin 300308 (China).
Yan Zhao, Division of Physical Biology & Bioimaging Center, Shanghai Synchrotron Radiation Facility, CAS Key Laboratory of Interfacial Physics and Technology, Shanghai Institute of Applied Physics, Chinese Academy of Sciences Shanghai 201800 (China).
Xiaoguo Liu, Division of Physical Biology & Bioimaging Center, Shanghai Synchrotron Radiation Facility, CAS Key Laboratory of Interfacial Physics and Technology, Shanghai Institute of Applied Physics, Chinese Academy of Sciences Shanghai 201800 (China).
Jixue Sun, State Key Laboratory of Medicinal Chemical Biology, College of Pharmacy and Tianjin Key Laboratory of Molecular Drug Research, Nankai University Tianjin 300353 (China).
Dede Xu, State Key Laboratory of Medicinal Chemical Biology, College of Pharmacy and Tianjin Key Laboratory of Molecular Drug Research, Nankai University Tianjin 300353 (China).
Yang Li, State Key Laboratory of Medicinal Chemical Biology, College of Pharmacy and Tianjin Key Laboratory of Molecular Drug Research, Nankai University Tianjin 300353 (China).
Qian Li, Division of Physical Biology & Bioimaging Center, Shanghai Synchrotron Radiation Facility, CAS Key Laboratory of Interfacial Physics and Technology, Shanghai Institute of Applied Physics, Chinese Academy of Sciences Shanghai 201800 (China).
Lihua Wang, Division of Physical Biology & Bioimaging Center, Shanghai Synchrotron Radiation Facility, CAS Key Laboratory of Interfacial Physics and Technology, Shanghai Institute of Applied Physics, Chinese Academy of Sciences Shanghai 201800 (China).
Sichun Yang, Center for Proteomics and Department of Nutrition Case Western Reserve University 10900 Euclid Ave, Cleveland, OH 44106-4988 (USA).
Chunhai Fan, Division of Physical Biology & Bioimaging Center, Shanghai Synchrotron Radiation Facility, CAS Key Laboratory of Interfacial Physics and Technology, Shanghai Institute of Applied Physics, Chinese Academy of Sciences Shanghai 201800 (China).
Jianping Lin, State Key Laboratory of Medicinal Chemical Biology, College of Pharmacy and Tianjin Key Laboratory of Molecular Drug Research, Nankai University Tianjin 300353 (China); Biodesign Center, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences Tianjin 300308 (China).
References
- [1].a) Liu B, Cao YY, Huang ZH, Duan YY, Che SN, Adv. Mater 2015, 27, 479–497; [DOI] [PubMed] [Google Scholar]; b) Konijnenberg A, Yilmaz D, Ingólfsson HI, Dimitrova A, Marrink SJ, Li Z, Vénien-Bryan C, Sobott F, Kocer A, Proc. Natl. Acad. Sci. USA 2014, 111, 17170–17175; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Reeves D, Ursell T, Sens P, Kondev J, Phillips R, Phys. Rev. E 2008, 78, 041901; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Ollila OHS Louhivuori M Marrink SJ Vattulainen I Biophys. J 2011, 100, 1651–1659; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Wiggins P, Phillips R, Biophys. J 2005, 88, 880–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Hummer G, Rasaiah JC, Noworyta JP, Nature 2001, 414, 188–190; [DOI] [PubMed] [Google Scholar]; b) Holt JK, Park HG, Wang Y, Stadermann M, Artyukhin AB, Grigoropoulos CP, Noy A, Bakajin O, Science 2006, 312, 1034. [DOI] [PubMed] [Google Scholar]
- [3].a) Kallenbach NR, Ma R-I, Seeman NC, Nature 1983, 305, 829–831; [Google Scholar]; b) Mathieu F, Liao S, Kopatsch J, Wang T, Mao C, Seeman NC, Nano Lett. 2005, 5, 661–665; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang T, Schiffels D, Cuesta SM, Fygenson DK, Seeman NC, J. Am. Chem. Soc 2013, 135, 1606–1616; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Pei H, Liang L, Yao G, Li J, Huang Q, Fan C, Angew. Chem. Int. Ed 2012, 51, 9020–9024; [DOI] [PubMed] [Google Scholar]; Angew. Chem 2012, 124, 9154–9158; [Google Scholar]; e) Ye D Zuo X, Fan C, Prog. Chem 2017, 29, 36–46. [Google Scholar]
- [4].a) Kerstin G, Li CY, Maria R, Prathyusha BS, Jejoong Y, Bertalan G, Alexander O, Mathias W, Aleksei A, Keyser UF, ACS Nano 2016, 10, 8207–8214;27504755 [Google Scholar]; b) Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC, Science 2012, 338, 932–936; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Burns JR, Stulz E, Howorka S, Nano Lett. 2013, 13, 2351–2356; [DOI] [PubMed] [Google Scholar]; d) Burns JR, Göpfrich K, Wood JW, Thacker VV, Stulz E, Keyser UF, Howorka S, Angew. Chem. Int. Ed 2013, 52, 12069–12072; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem 2013, 125, 12291–12294. [Google Scholar]
- [5].a) Qian P, Seo S, Kim J, Kim S, Lim BS, Liu WK, Kim BJ, Labean TH, Park SH, Kim MK, Nanotechnology 2012, 23, 105704; [DOI] [PubMed] [Google Scholar]; b) Joshi H, Dwaraknath A, Maiti PK, Phys. Chem. Chem. Phys 2015, 17, 1424–1434; [DOI] [PubMed] [Google Scholar]; c) Joshi H, Kaushik A, Seeman NC, Maiti PK, ACS Nano 2016, 10, 7780–7791; [DOI] [PubMed] [Google Scholar]; d) Burns JR, Seifert A, Fertig N, Howorka S, Nat. Nanotechnol. 2016, 11, 152–156. [DOI] [PubMed] [Google Scholar]
- [6].a) Yoo J, Aksimentiev A, J. Phys. Chem. Lett 2015, 6, 4680–4687; [DOI] [PubMed] [Google Scholar]; b) Vishal M, Mickaël L, Stefan H, Sansom MSP, ACS Nano 2015, 9, 11209–11217.26506011 [Google Scholar]
- [7].a) Derler I, Jardin I, Romanin C, Am. J. Physiol. Cell Physiol 2016, 310, C643–C662; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Boscardin E, Alijevic O, Hummler E, Frateschi S, Kellenberger S, Br. J. Pharmacol 2016, 173, 2671–2701; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Craven KB, Zagotta WN, Annu. Rev. Physiol 2006, 68, 375–401. [DOI] [PubMed] [Google Scholar]
- [8].Cao E, Liao M, Cheng Y, Julius D, Nature 2013, 504, 113 –118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ravikumar KM, Huang W, Yang S, J. Chem. Phys 2013, 138, 024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bruetzel LK, Gerling T, Sedlak SM, Walker PU, Zheng W, Dietz H, Lipfert J, Nano Lett. 2016, 16, 4871–4879. [DOI] [PubMed] [Google Scholar]
- [11].a) Jayaram AB, Beveridge DL, Annu. Rev. Biophys. 1996, 25, 367–394; [DOI] [PubMed] [Google Scholar]; b) Maffeo C, Yoo J, Comer J, Wells DB, Luan B, Aksimentiev A, J. Phys. Condens. Matter 2014, 26, 413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Li W, Nordenskiöld L, Mu Y, J. Phys. Chem. B 2011, 115, 14713–14720; [DOI] [PubMed] [Google Scholar]; b) Baumann CG, Smith SB, Bloomfield VA, Bustamante C, Proc. Natl. Acad. Sci. USA 1997, 94, 6185–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mittelbach P, Acta Phys. Austriaca 1964, 19, 53–102. [Google Scholar]
- [14].Long SB, Campbell EB, Mackinnon R, Science 2005, 309, 897–903. [DOI] [PubMed] [Google Scholar]
- [15].Jiang Y, Lee A, Chen J, Cadene M, Chait BT, Mackinnon R, Nature 2002, 417, 515–522. [DOI] [PubMed] [Google Scholar]
- [16].a) Wu N, Czajkowsky DM, Zhang J, Qu J, Ye M, Zeng D, Zhou X, Hu J, Shao Z, Li B, Fan C, J. Am. Chem. Soc 2013, 135, 12172–12175; [DOI] [PubMed] [Google Scholar]; b) Fischer S, Hartl C, Frank K, Rädler JO, Liedl T, Nickel B, Nano Lett. 2016, 16, 4282–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.