Significance
Retrograde transport from the cell surface to intracellular compartments is essential for homeostasis and cell physiology. We developed derivatized nanobodies to follow proteins from the cell surface to endosomes and the trans-Golgi network. Nanobodies are an emerging class of protein binders with many advantages over conventional antibodies: they are small and noncross-linking, and can be produced in bacteria. Using a nanobody against GFP, our tool is widely applicable to any GFP-tagged protein of interest. It allows the quantitative analysis of endocytic and retrograde transport biochemically, by fixed- and live-cell imaging and by electron microscopy. As proof-of-principle, we applied them to determine the contribution of adaptor protein-1/clathrin in retrograde transport of the mannose-6-phosphate receptors to the trans-Golgi.
Keywords: nanobodies, retrograde transport, tyrosine sulfation, AP-1, mannose-6-phosphate receptors
Abstract
Retrograde transport of membranes and proteins from the cell surface to the Golgi and beyond is essential to maintain homeostasis, compartment identity, and physiological functions. To study retrograde traffic biochemically, by live-cell imaging or by electron microscopy, we engineered functionalized anti-GFP nanobodies (camelid VHH antibody domains) to be bacterially expressed and purified. Tyrosine sulfation consensus sequences were fused to the nanobody for biochemical detection of trans-Golgi arrival, fluorophores for fluorescence microscopy and live imaging, and APEX2 (ascorbate peroxidase 2) for electron microscopy and compartment ablation. These functionalized nanobodies are specifically captured by GFP-modified reporter proteins at the cell surface and transported piggyback to the reporters’ homing compartments. As an application of this tool, we have used it to determine the contribution of adaptor protein-1/clathrin in retrograde transport kinetics of the mannose-6-phosphate receptors from endosomes back to the trans-Golgi network. Our experiments establish functionalized nanobodies as a powerful tool to demonstrate and quantify retrograde transport pathways.
Retrograde transport of proteins and lipids from the plasma membrane and endosomes to the Golgi and beyond is important for membrane homeostasis to counterbalance secretion and to recycle components of anterograde transport machineries (1, 2). Recycling from endosomes to the trans-Golgi network (TGN) is part of the functional cycle of several anterograde cargo receptors, such as the cation-dependent and -independent mannose-6-phosphate receptors (MPRs: CDMPR and CIMPR, respectively) sorting newly synthesized lysosomal proteins from the TGN toward late endosomes and lysosomes (3), sortilin and SorLA (4), and Wntless escorting Wnt through the secretory pathway (5). Other proteins retrieved back to the TGN include TGN46, amyloid precursor protein, the membrane proteases furin and carboxypeptidase D, several nutrient transporters, and SNAREs (soluble N-ethylmaleimide-sensitive fusion factor attachment receptors) involved in membrane fusion (1, 2). In addition, bacterial and plant toxins (e.g., Shiga and cholera toxins, ricin and abrin) exploit the cell’s retrograde machineries to reach the endoplasmic reticulum (ER) from the cell surface (6).
Several distinct machineries have been reported to mediate retrograde transport of different cargo from different endosomes. The most prominent ones are the retromer complexes, multimeric protein coats within the tubular early endosomal network (7). A core complex of Vps26–Vps29–Vps35 associates either with heterodimeric SNX-BAR (sorting nexin with a Bin/Amphiphysin/Rvs domain) tubulation subcomplexes or with SNX3 to generate transport carriers to the TGN. For example, it was shown that silencing of Vps26, Vps35, or multiple SNX-BAR proteins resulted in redistribution of CIMPR to peripheral endosomes (8–10). A pathway for both MPRs from late endosomes to the TGN was described to involve Rab9 (11) and controversially TIP47 (12, 13).
A further potential retrograde mechanism of MPRs involves clathrin-coated vesicles (CCVs) with the adaptor protein (AP)-1 complex that mainly localizes to the TGN and to early endosomes. They have a well-established role in anterograde transport of MPRs from the TGN to endosomes in cooperation with GGA (Golgi-localizing, γ-adaptin ear domain homology, ARF-binding) proteins (14, 15). However, in μ1 knockout cells lacking AP-1, the steady-state distribution of MPRs was unexpectedly shifted from the TGN to endosomes and CDMPR retrieval to the TGN was found to be defective in a resialylation assay (16). This suggested a defect in retrograde transport and led to the notion that AP-1 is involved in bidirectional transport of MPRs between the TGN and endosomes. A retrograde role of AP-1, clathrin, and ARF1 was also observed for the cholera toxin B subunit (17). EpsinR, an interactor of AP-1, likewise contributes to retrograde transport, since its depletion affected the distribution of the TGN/endosome SNARE vti1b and reduced its levels in CCVs (18), and produced deficits in delivering Shiga toxin (which, interestingly, was not sensitive to AP-1γ knockdown), CIMPR, as well as TGN46 from endosomes to the TGN (19).
Most of these experiments relied on gene silencing by either knockdown or knockout. Concomitant gradual and long-term protein depletion has the risk of compensatory or indirect effects (e.g., cellular adaptation or an altered steady-state distribution of involved factors, like SNAREs). Robinson et al. (20, 21) therefore developed knocksideways, a method for the rapid depletion of a protein of interest within a few minutes. This method takes advantage of rapamycin-induced heterodimerization between the prolyl isomerase FKBP12 (FK506-binding protein of 12 kDa) and the FKBP–rapamycin-binding domain (FRB) of mammalian target of rapamycin (mTOR). FRB is anchored in the outer mitochondrial membrane as a trap (Mitotrap), while the FKBP domain is fused to the protein of interest. Upon silencing the endogenous protein (i.e., after siRNA-mediated silencing), rapamycin addition triggers rapid, essentially irreversible immobilization of the FKBP fusion to mitochondria within few minutes.
AP-1 inactivation by knocksideways produced more consistent results than knockdown or knockout: after 10 min of rapamycin addition, CIMPR and ARF1 levels in CCVs were substantially reduced and AP-2 unaffected, while in knockdown cells almost no reduction of CIMPR and ARF1 was found and AP-2 was increased (20, 22). However, longer rapamycin treatment reproduced CIMPR redistribution seen upon efficient AP-1 knockdown. Quantitative proteomics of CCV contents showed SNAREs and cargo receptors (MPRs and sortilin) to be more efficiently depleted after AP-1 than after GGA2 knocksideways, while lysosomal hydrolases were similarly reduced upon either inactivation (23). This result is consistent with bidirectional transport by AP-1 CCVs: anterograde together with GGAs for receptors and their cargo, and retrograde without for empty receptors.
To directly analyze retrograde traffic of a protein of interest, it has to be labeled at the cell surface for subsequent tracking to its intracellular destinations. Most frequently conventional antibodies are used. They are composed of two heavy and two light chains and are thus rather large (∼150 kDa). They contain intermolecular disulfide bonds, and can cross-link antigens with their two antigen-binding sites. Monovalent antigen-binding fragments (Fab) of ∼50 kDa can be generated by partial proteolysis. Fusion of the variable region of the heavy (VH) and light chains (VL) with a synthetic short linker yields even smaller (∼27 kDa) single-chain variable fragments (scFv) that can be functionally expressed in the cytosol and produced in bacteria. In recent years, many other protein binding scaffolds unrelated to antibodies have been developed (24) (e.g., the designed ankyrin repeat proteins) (25).
A new family of protein binders with rapidly growing applications is constituted by the nanobodies (26). They are derived from homodimeric heavy-chain antibodies (hcAbs) produced in camelids or cartilaginous fishes (27, 28). Nanobodies constitute the variable heavy-chain domain of heavy-chain–only antibodies (VHH). They are small (∼15 kDa), highly soluble, stable (heat-stable to ∼70 °C) and can be selected for high-affinity binding (29). They structurally consist of a β-sandwich of 4 + 5 β-strands with three hypervariable loops for target binding, frequently—but not necessarily—stabilized by a disulfide bond. Nanobodies without this disulfide bond can be functionally expressed in cytosol or produced in bacteria.
In this study, we developed a versatile nanobody-based approach to track GFP-tagged surface proteins from the cell surface to intracellular compartments either biochemically, by fixed- and live-cell imaging, or by electron microscopy (EM). The nanobodies were derivatized by fusing them to functional tags and produced in bacteria. We demonstrate their usefulness by assessing the involvement of AP-1/clathrin in retrograde endosome-to-TGN transport of the MPRs, demonstrating a reduction of their kinetics of TGN arrival upon rapid AP-1 depletion by knocksideways.
Results
Functionalized Nanobodies to Track GFP-Tagged Receptors.
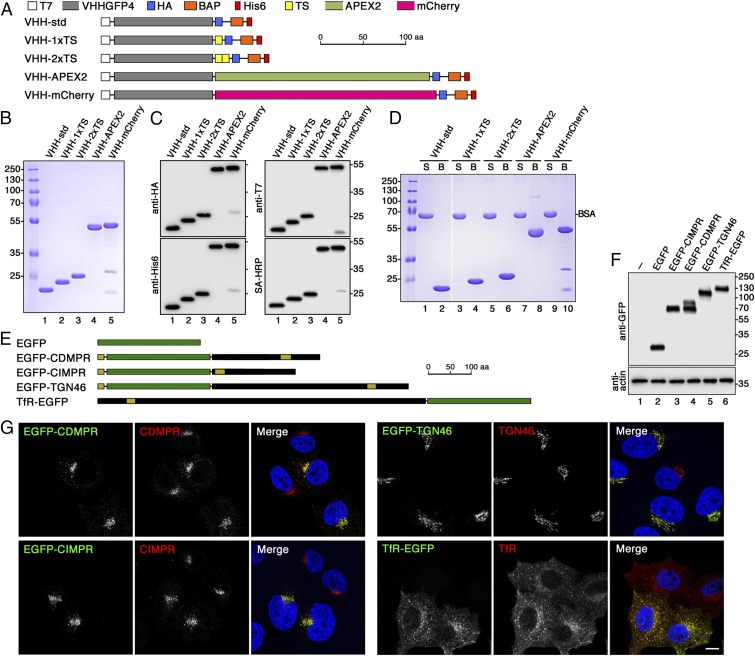
To investigate retrograde transport from the plasma membrane, we took advantage of the anti-GFP nanobody VHHGFP, a camelid single-chain antibody specifically recognizing folded GFP in a 1:1 stoichiometry with nanomolar affinity (30–32). This nanobody can bind any protein of interest with an extracellular GFP or related fluorescent protein tag (such as YFP and mCerulean, but not mCherry or RFP). As illustrated in Fig. 1A, the VHH domain was fused in its standard derivatized form (VHH-std) to a T7 and an HA epitope tag for subsequent detection by specific antibodies, to a carboxyterminal hexahistidine-tag for purification, and a biotin acceptor peptide (BAP) sequence for biotinylation and high-affinity avidin pull-down experiments. Additional variants were generated with one or two tyrosine-sulfation (TS) sequences of rat cholesystokinin precursor (VHH-1xTS and VHH-2xTS) to be potentially sulfated in the TGN, with mCherry (VHH-mCherry) to directly visualize the nanobody, or with a modified soybean ascorbate peroxidase APEX2 (VHH-APEX2) for ultrastructural visualization by cytochemical staining and compartment ablation.
Fig. 1.
Design and production of derivatized nanobodies to track EGFP-tagged surface proteins. (A) Schematic representation of the derivatized nanobodies. The standard nanobody (VHH-std) consists of the GFP-specific VHH domain, T7 and HA epitope tags, a BAP, and a hexahistidine (His6) purification tag. Other nanobodies in addition contain one or two TSs, APEX2, and/or mCherry. Scale bar is in amino acids (aa). (B) Bacterially expressed and purified nanobodies (30 µg) were analyzed by SDS-gel electrophoresis and Coomassie stained. Marker proteins with molecular mass in kiloDaltons are shown on the left. (C) Immunoblot analysis of nanobodies (10 µg) with antibodies against the T7, HA, or His6 epitopes, or with streptavidin-HRP (SA-HRP). (D) The extent of biotinylation was assessed by mixing the nanobodies 1:1 with BSA, incubating the mixture with streptavidin-agarose, pelleting and washing the beads, and analyzing equal fractions of supernatant (S) and material bound to the beads (B) by gel electrophoresis and Coomassie staining. Complete recovery in the bound fraction indicates complete biotinylation of the nanobodies. Recovery of both VHH-mCherry fragments with the beads suggests degradation during sample preparation for SDS-gel electrophoresis. The white line between lanes 2 and 3 indicates deletion of two unrelated lanes. (E) Schematic representation of EGFP (in green) and EGFP fusion proteins. Sequences derived from receptor proteins are shown in black with N-terminal signal peptides and internal transmembrane segments in yellow. EGFP was fused to full-length CDMPR, TGN46, and TfR, and to the transmembrane segment and cytoplasmic sequence of CIMPR. Scale bar in amino acids. (F) HeLa cells stably expressing EGFP-tagged reporter proteins were analyzed by SDS-gel electrophoresis and immunoblotting using anti-GFP antibodies. The positions of size markers with molecular mass in kiloDaltons are indicated. (G) HeLa cells stably expressing the EGFP-tagged reporter proteins were mixed with parental HeLa cells and stained with antibodies targeting the respective endogenous proteins. Except for CIMPR, the antibodies recognize both the endogenous and the EGFP-tagged proteins. The distribution patterns were not altered by expression of the EGFP reporters. Nuclei were stained with DAPI (blue). (Scale bar, 10 µm.)
These constructs were expressed in bacteria also producing the biotin ligase BirA in the presence of excess biotin and purified by Ni-NTA (Ni2+-nitrilotriacetate) chromatography. They were all isolated at high yield (20–30 mg/L) and high purity (Fig. 1B). Only VHH-mCherry showed minimal clipping between the VHH and the mCherry domains. They were all efficiently detected by immunoblot analysis with anti-HA, anti-His6, and anti-T7 antibodies, as well as with streptavidin-horseradish peroxidase (HRP) (Fig. 1C). Biotinylation was indeed complete, because the nanobodies from a 1:1 mixture with BSA were fully recovered by streptavidin-agarose (Fig. 1D).
To test our nanobody toolkit for retrograde transport studies, we prepared stable HeLa cell lines expressing either cytosolic EGFP as a negative control or EGFP-tagged surface proteins with different intracellular itineraries: the TfR, cycling between the plasma membrane and early (sorting and recycling) endosomes; TGN46, between the plasma membrane and the TGN via early endosomes; and the MPRs, between the TGN, plasma membrane, and early and late endosomes. EGFP was fused to the extracellular end of each protein (i.e., between the signal peptide and the receptor sequence of CDMPR, CIMPR, and TGN46) and to the carboxyterminus of the TfR, leaving the cytoplasmic domains containing all known sorting signals unobstructed (Fig. 1E). For CIMPR, the very large extracellular domain was truncated in the same way that was previously shown to retain normal trafficking behavior (33, 34).
Stable cell lines were generated by retroviral transduction followed by FACS of homogeneous cell pools of moderate and comparable expression levels (Fig. 1F). Only EGFP-CDMPR appeared as a double band on immunoblots (as is also the case for the endogenous receptor) (35), indicating heterogeneous glycosylation.
To compare the steady-state distribution of the EGFP fusion proteins and expression levels with those of the endogenous counterparts, stably expressing cells of each kind were mixed with parental HeLa cells and analyzed by confocal fluorescence microscopy (Fig. 1G). The same distribution was observed for the EGFP signal as for the immunostaining of the endogenous proteins. As expected, CDMPR, CIMPR, and their EGFP fusions were predominantly found in the perinuclear region reflecting their TGN and late endosome localization and additionally in peripheral endosomes. EGFP-tagged and endogenous TGN46 localized almost exclusively to the perinuclear region of the TGN, whereas the TfR proteins showed the typical early endosome pattern of peripheral sorting and perinuclear recycling endosomes. Even though the antibodies recognized both forms of each protein except in the case of CIMPR, the staining was not visibly stronger in cells expressing both forms, indicating that the fusion proteins were not strongly overexpressed.
Kinetic Analysis of Internalization and Recycling of EGFP-Reporters Using Nanobodies.
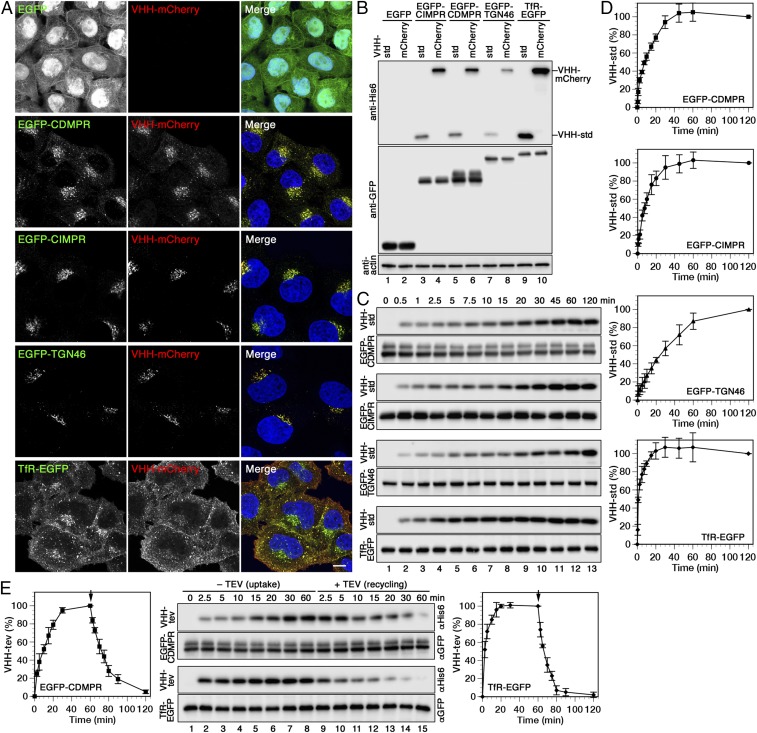
Upon incubation of standard or mCherry-containing nanobodies for 1 h at 37 °C with HeLa cells expressing EGFP fusion proteins, considerable amounts of nanobodies were found associated with the cells (Fig. 2 A and B). In contrast, no signal was observed with cells expressing cytosolic EGFP, indicating that there is negligible nonspecific binding or uptake. Because nanobodies were detected by fluorescence microscopy closely colocalizing with the respective reporter protein (Fig. 2A), they were indeed internalized by the EGFP-tagged surface proteins
Fig. 2.
Uptake, intracellular localization, and recycling of nanobodies by different EGFP-reporters. (A) Cells expressing cytosolic EGFP or the indicated EGFP fusion proteins were incubated for 1 h at 37 °C with 5 µg/mL VHH-mCherry, fixed, and imaged by fluorescence microscopy. (Scale bar, 10 µm.) (B) The same cell lines were incubated for 1 h at 37 °C with 2 µg/mL VHH-std or 5 µg/mL VHH-mCherry (∼0.1 µM), washed, lysed, and analyzed by immunoblotting for the nanobodies and the EGFP reporters. (The intensity difference between VHH-std and VHH-mCherry is most likely due to better transfer or retention of the larger protein to the blotting membrane.) (C) Kinetics of nanobody uptake were analyzed by incubating the cells at 37 °C for up to 2 h with 2 µg/mL VHH-std. At different times, the cells were lysed, and immunoblotted for cell-associated nanobody (anti-His6) and the EGFP fusion proteins (anti-GFP). (D) Quantitations of nanobody uptake (mean and SD of three independent experiments) are shown in percent of the 120-min values. (E) To determine nanobody uptake and recycling kinetics, cells expressing EGFP-CDMPR or TfR-EGFP were incubated at 37 °C for up to 60 min with 2 µg/mL VHH-tev. Then, cells were washed and further incubated for up to 60 min with 100 µg/mL (2 µM) MBP-TEV protease to remove the hexahistidine tag of nanobody reappearing at the cell surface. Uptake of intact VHH-tev and subsequent TEV cleavage was monitored by immunoblot analysis using anti-His6 antibody, and quantified in percent of the value after 60 min (mean and SD of three independent experiments).
To biochemically measure the kinetics of nanobody binding and uptake, 0.1 µM VHH-std was added to the HeLa cell lines in full medium at 37 °C for different times before washing the cells with cold buffer, lysis, and immunoblot analysis (Fig. 2 C and D). Nanobody uptake showed reporter-specific kinetics to steady state with TfR-EGFP displaying the fastest, with an apparent half-life of 1–3 min and reaching saturation at ∼20 min, consistent with previous measurements using iodinated or fluorescently labeled Tf (e.g., refs. 36 and 37). EGFP-CDMPR and -CIMPR showed slower uptake with apparent t1/2 of 8–10 min and saturation after ∼40–45 min, in agreement with their distribution to more distant compartments in the cell. The slowest uptake with apparent t1/2 of 25–30 min and saturation after more than 60 min was measured for EGFP-TGN46, which is most strongly localized to the TGN and least to the plasma membrane.
To biochemically distinguish between intracellular and surface-bound nanobodies, we produced VHH-tev, a derivative of the standard nanobody with an additional cleavage site for the tobacco etch virus (TEV) protease inserted between the VHH domain and the HA-BAP-His6 tags (SI Appendix, Fig. S1 A–C). After 1 h of VHH-tev uptake by TfR-EGFP, ∼15% of cell-associated nanobody was sensitive to TEV protease (purified as a maltose-binding protein fusion protein) at 4 °C on the surface (SI Appendix, Fig. S1D), in agreement with the established steady-state distribution to the TfR. Similarly, nanobody binding at 4 °C to cells expressing TfR-EGFP at 4 °C amounted to ∼15% of the steady-state value, all of which was sensitive to subsequent protease incubation (SI Appendix, Fig. S1D).
VHH-tev can also be used to monitor the kinetics of nanobody-bound reporter proteins recycling back to the cell surface. After VHH-tev uptake by EGFP-CDMPR or TfR-EGFP for 1 h, cells were quickly washed and incubated further at 37 °C with TEV protease. Probing for cell-associated nanobodies by anti-HA immunoblot analysis showed their uptake kinetics as before, followed by the inverse loss of the signal due to protease cleavage upon their reappearance at the cell surface (Fig. 2E). The apparent half-lives of 12 min and 5 min for recycling of nanobodies by EGFP-CDMPR and TfR-EGFP, respectively, are slightly higher than the corresponding half-lives of uptake of ∼10 min and ∼3 min, most likely because TEV cleavage is slower than nanobody binding at the chosen protease concentration.
Live-Cell Imaging of VHH-mCherry Uptake.
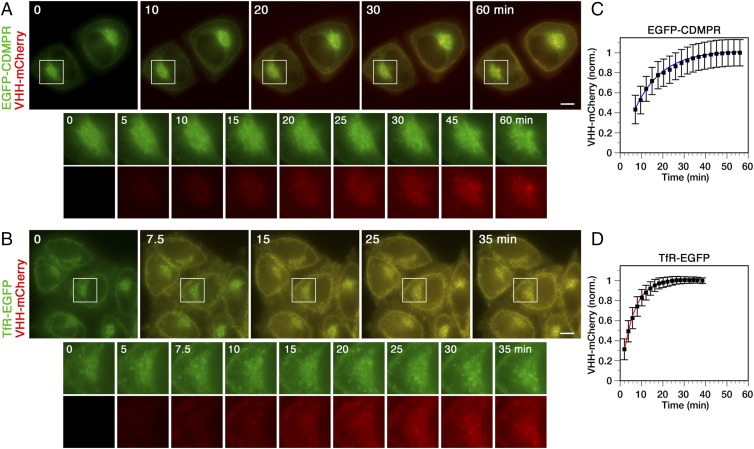
Using mCherry-tagged nanobodies, retrograde transport of EGFP-tagged reporter proteins can also be monitored by live-cell imaging. For this, we used as examples cells expressing EGFP-CDMPR and TfR-EGFP. The cells were imaged over time with an inverted widefield fluorescent microscope upon addition of VHH-mCherry to the medium (Movies S1 and S2, respectively). Still images at various time points are shown in Fig. 3. Uptake was quantified by measuring the signal in the mCherry channel, subtracting the autofluorescence background, and normalization to the EGFP signal to eliminate fluctuations due to the movement of labeled compartments or small shifts in the focal plane. The fluorescence of VHH-mCherry in the medium at 25 nM was negligible and did not interfere with the measurements. Analysis of transport of the reporters from the cell surface to their intracellular compartments, to the steady-state distribution, yielded the same kinetic results as the biochemical experiments shown in Fig. 2, with apparent half-lives of uptake of ∼9 min for EGFP-CDMPR and ∼4 min for TfR-EGFP, and saturation after ∼43 min and ∼20 min, respectively.
Fig. 3.
Live-cell imaging of nanobody uptake kinetics by EGFP-CDMPR and TfR-EGFP. Upon addition of 25 nM VHH-mCherry to cells expressing EGFP-CDMPR (A) or TfR-EGFP (B) in phenol red-free complete medium at 37 °C. Cells were imaged in the GFP and mCherry channels with a semiautomated widefield fluorescence microscope every 36 s. Merged still images are shown with enlarged perinuclear regions (magnification: 2.2×) in separated channels below. (Scale bars, 10 µm.) See also Movies S1 and S2. Quantitation of uptake kinetics by EGFP-CDMPR (C) or TfR-EGFP (D) of three independent experiments, each recording ∼40 cells. The mCherry signal was normalized (norm) to the GFP signal to correct for fluctuations of organelles or focal plane and plotted as the mean and SD of all cells from all three experiments. The average saturation value was set to 1. The uptake curves of all cells were individually fitted to first order kinetics. The lines are based on the average of the resulting rate constants.
In principle, kinetics of transport into subcellular regions of interest, such as the perinuclear region of highest concentration of MPRs, can also be analyzed. However, the perinuclear region contains not only Golgi/TGN, but is also enriched in late endosomes and recycling endosomes. Kinetics of nanobody uptake into the perinuclear region is not sufficiently specific to analyze retrograde transport to a defined organelle. Ultrastructural or functional analysis would be required for this.
Analysis of Retrograde Transport by Peroxidase Labeling Using APEX2-Nanobodies.
Frequently, EM will be required to morphologically identify the compartments to which nanobodies are transported from the cell surface by a protein of interest. Because there is an anti-myc antibody we successfully used before for immunogold EM (38), we constructed and produced VHH-myc, a version of VHH-std with an additional myc epitope (SI Appendix, Fig. S2 A–D). HeLa cells expressing TfR-EGFP loaded to steady state with VHH-myc were prepared for immunogold EM using anti-GFP and anti-myc antibodies with secondary antibodies coupled to 20- and 10-nm colloidal gold, respectively (SI Appendix, Fig. S2E). Endosomal compartments positive for both markers could be detected in this manner.
An alternative to immunogold staining with potentially higher sensitivity is peroxidase-mediated compartment labeling. HRP was frequently used for this in the past coupled to antibodies or receptor ligands (e.g., Tf-HRP) (39) and internalized by their target protein. The peroxidase catalyzes H2O2-dependent polymerization of diaminobenzidine (DAB) into local precipitates that recruit osmium tetroxide for EM contrast. For our purpose, we took advantage of the improved soybean ascorbate peroxidase APEX2 (40), which, unlike HRP, is relatively small and monomeric and lacks disulfide bonds and calcium binding sites, allowing the protein to be expressed in the reducing and low-calcium environment of the bacterial cytosol.
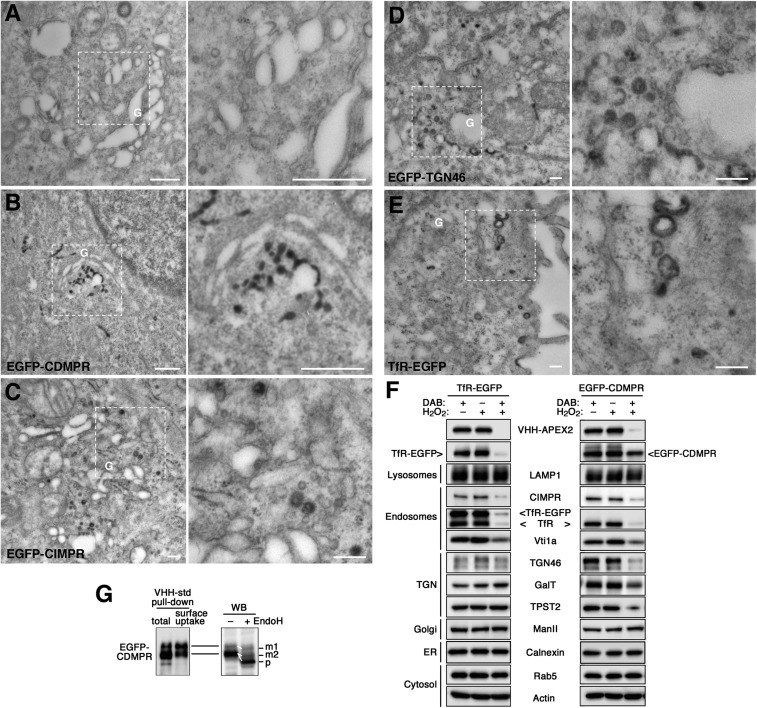
VHH-APEX2 nanobodies were produced in bacteria and purified (Fig. 1 A–D). Upon incubation with the EGFP-reporter cell lines for 1 h at 37 °C, they were internalized and colocalized with EGFP fusion proteins (SI Appendix, Fig. S3) just like the other nanobodies (Fig. 2 A and B). VHH-APEX2 incubated with control HeLa cells did not produce staining upon DAB/H2O2 incubation (Fig. 4A). Upon internalization by TfR-EGFP, however, DAB precipitations were found in peripheral endosomal compartments, vesicles, and plasma membrane invaginations (Fig. 4E), similar to what had been observed before with Tf-HRP (e.g., ref. 41). VHH-APEX2 imported by EGFP-CDMPR, -CIMPR, and -TGN46 in addition stained the TGN (i.e., tubulo-vesicular structures next to the Golgi) (Fig. 4 B–D). The APEX2 nanobody thus can be used to morphologically detect the retrograde target compartments of EGFP-tagged proteins of interest.
Fig. 4.
Retrograde transport analyzed by APEX2-nanobodies. (A–E) Cytochemical staining of VHH-APEX2–containing compartments for EM. Control and EGFP reporter cell lines were incubated with 5 μg/mL VHH-APEX2 for 1 h (2 h for EGFP-TGN46) at 37 °C to reach steady state. After fixation with 2% glutaraldehyde, cells were treated with DAB/H2O2 to allow DAB polymerization in APEX2-containing compartments, stained with osmium tetroxide and then uranyl acetate, and further processed for EM and sectioned. The panel on the Right is an enlargement of the selected area in the image on the left. (Scale bars, 500 nm.) G, Golgi. (F) Biochemical ablation of VHH-APEX2–containing compartments. To inactivate compartments reached by the TfR-EGFP or EGFP-CDMPR from the cell surface, cells were loaded with VHH-APEX2 for 1 h at 37 °C and washed before incubation with 1 mg/mL DAB, 0.03% H2O2, or both for 90 min at 4 °C. Cells were lysed and subjected to immunoblot analysis to detect the nanobody, the EGFP-target proteins as well as compartment-specific endogenous markers. ManII, α-mannosidase II. Experiments have been repeated three times with consistent results. (G, Left) VHH-std was added either to EGFP-CDMPR cell lysate to bind total receptor protein (total) or to intact cells for 1 h at 37 °C to capture only protein cycling to the surface (surface uptake). VHH-std was then isolated with Ni-NTA beads before immunoblotting for GFP. All of the upper band but only a fraction of the lower one was captured by internalized nanobodies. (Right) Lysate of EGFP-CDMPR expressing cells was treated with or without endoglycosidase H (EndoH) before Western blotting (WB). The upper band and part of the lower one were partially EndoH-resistant (reduced in apparent size by ∼3 kDa), as expected for the mature proteins (m1 and m2). The rest of the lower band showed a larger shift (∼10 kDa), indicating complete Endo-H sensitivity of the precursor (p) still in the ER.
Biochemical Compartment Ablation by APEX2 Nanobodies.
On a biochemical level, DAB/H2O2 cytochemistry can be used for the ablation of compartments containing peroxidase activity (41–43). DAB polymerization traps the proteins within the organelle to make them unavailable for immunoprecipitation and even gel electrophoresis and immunoblot analysis. Sensitivity of marker proteins to DAB polymerization thus provides evidence for colocalization with the peroxidase.
Cells expressing TfR-EGFP or EGFP-CDMPR were loaded for 1 h at 37 °C with VHH-APEX2 nanobodies to reach steady state, washed, and incubated with DAB. As expected, hardly any VHH-APEX2 could be recovered after DAB polymerization with H2O2 (Fig. 4F). Similarly, TfR-EGFP was almost entirely lost. Of EGFP-CDMPR, the larger glycoform was completely eliminated, while the smaller one was only partially affected. The resistant molecules of both reporters most likely represent the newly synthesized population present in the early secretory pathway (mostly in the ER). To test this possibility directly for EGFP-CDMPR, standard nanobody VHH-std was either mixed with total cell lysate for binding to all cellular EGFP-CDMPR or incubated with intact cells to only capture receptor appearing on the cell surface during 1 h at 37 °C. Nanobodies were then purified on Ni-NTA-beads (Fig. 4G, Left). All of the larger form of EGFP-CDMPR, but only a fraction of the lower band, were recovered from nanobodies taken up by intact cells and thus correspond to mature protein in post-ER compartments, suggesting a significant amount of biosynthetic precursors. This was further confirmed by endoglycosidase H (EndoH) digestion (Fig. 4G, Right): the upper band of EGFP-CDMPR and a fraction of the lower one were partially sensitive, as previously shown for purified CDMPR (35), while the rest of the lower band was fully deglycosylated by EndoH. This indicated a pool of precursors in the ER that is not conspicuous in fluorescence microscopy and not affected by ablation. Indeed, calnexin and mannosidase II, markers for ER and medial-Golgi, respectively, were also resistant to VHH-APEX2–mediated ablation, as were the cytosolic proteins actin and Rab5 and the lysosome marker LAMP1 (Fig. 4F).
Both EGFP-tagged proteins mediated ablation of endosomes, because endogenous TfR and CIMPR were both strongly reduced. The endosomal SNARE protein Vti1a was also affected, even though it presents only approximately five residues into the endosomal lumen. However, TfR-EGFP did not notably impact the TGN markers TGN46, galactosyltransferase (GalT), or the tyrosylprotein sulfotransferase 2 (TPST2), indicating that it does not return to the TGN from its plasma membrane/endosome pool. This is in contrast to EGFP-CDMPR, which mediated an obvious loss of these markers. The extent of elimination was highest for TGN46 and TPST2, and lower for GalT, suggesting differences in the localization of these proteins with respect to that of CDMPR within the trans-Golgi/TGN compartments. The results show that VHH-APEX2 nanobodies with DAB/H2O2 cytochemistry can be used to identify the compartments reached by a protein of interest in retrograde transport from the plasma membrane.
Nanobody Sulfation to Determine Kinetics of Retrograde Transport to the TGN.
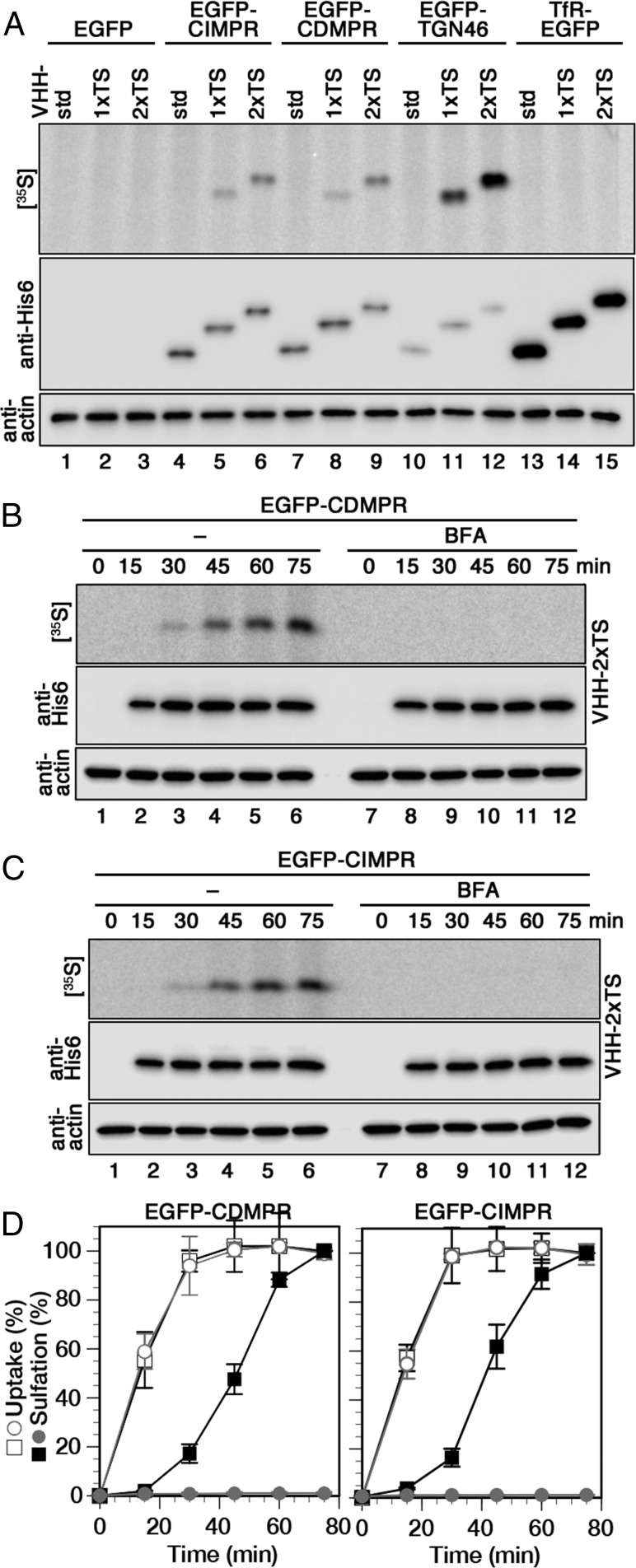
To be able to also determine kinetics of retrograde transport to the TGN, we took advantage of tyrosine sulfation, a posttranslational modification restricted to the trans-Golgi/TGN (44, 45). For this, VHH-1xTS and -2xTS were produced containing one or two tyrosine sulfation sequences (Fig. 1 A–D). These nanobodies (just like VHH-std, -mCherry, and -APEX2 shown above) were taken up specifically by the EGFP-tagged surface receptors to colocalize with their steady-state distribution (shown for VHH-2xTS in SI Appendix, Fig. S4). VHH-1xTS and -2xTS were radioactively labeled, when uptake was performed in the presence of [35S]sulfate for 60 min and was mediated by EGFP-CDMPR, -CIMPR, or -TGN46, but not by TfR-EGFP, even though the amount of imported nanobodies was the highest for TfR-EGFP (Fig. 5A). The experiment confirms again that the MPRs and TGN46, but not the TfR, are retrogradely transported from the cell surface to a compartment of sulfation (i.e., the TGN). The sulfation signal was proportional to the number of TS sequences within each cell line, but differed between uptake receptors: nanobodies taken up by EGFP-TGN46 showed considerably higher specific sulfation, then those captured by the EGFP-MPRs, most likely reflecting the residence time of these membrane proteins in the sulfation compartment.
Fig. 5.
Kinetics of retrograde transport to the TGN analyzed using sulfatable nanobodies. (A) Cells expressing cytosolic EGFP or the indicated EGFP-tagged membrane proteins were labeled with [35S]sulfate for 60 min with 2 μg/mL VHH-std, VHH-1xTS, or VHH-2xTS. The nanobodies were isolated and subjected to immunoblot analysis (anti-His6) and autoradiography ([35S]). Aliquots of cell lysates were immunoblotted for actin. (B and C) Cells expressing EGFP-CDMPR or -CIMPR were labeled with [35S]sulfate for up to 75 min in the presence of 2 μg/mL VHH-2xTS with and without 2 μg/mL BFA before analysis as for A. (D) Quantitation of VHH-2xTS uptake and sulfation from three independent experiments is shown in percent of the value without BFA after 75 min (mean and SD). Black squares, without BFA; gray circles, with BFA; open symbols, uptake; filled symbols, sulfation.
To simultaneously determine the kinetics for nanobody uptake and sulfation, cells expressing EGFP-CDMPR or -CIMPR were incubated with VHH-2xTS for various times up to 75 min while labeling with [35S]sulfate (Fig. 5 B–D). To generally disturb membrane traffic, the same experiment was also performed in the presence of brefeldin A (BFA), an inhibitor of guanine nucleotide exchange factors (GEFs; mainly GBF1 and BIG1/BIG2) of several Arf GTPases involved in vesicular transport and other functions (46). In the absence of BFA, sulfation started only after a lag time of ∼15 min and had not yet reached saturation after 75 min. Uptake and sulfation curves appeared shifted by almost 30 min, reflecting the transport time to the TGN. Both MPR fusion proteins behaved in a very similar manner. In the presence of BFA, sulfation was completely abolished, while uptake remained unaffected both in kinetics and in extent. This suggests a strong effect of BFA on transport of MPRs from endosomes to the compartment of sulfation, consistent also with previous observations for a CIMPR construct and cholera toxin (47, 48).
Uptake and sulfation of VHH-2xTS by EGFP-TGN46 were both slower, but similarly separated by ∼30 min (SI Appendix, Fig. S5). The intensity of sulfation, as apparent in Fig. 5A (lanes 9 and 12), is considerably higher in absolute terms than that with the EGFP-MPRs, most likely reflecting a prolonged residence time in the sulfation compartment.
Rapid Depletion of AP-1 Affects Retrograde Transport Kinetics of MPRs.
As an application of this tool of derivatized nanobodies, we analyzed the contribution of AP-1/clathrin coats to retrograde transport of the MPRs. Rather than measuring transport kinetics in normal cells vs. cells after knockdown or knockout of AP-1 subunits that might lead to compensatory effects of long-term protein silencing as documented before, we employed rapid AP-1 depletion using the knocksideways approach (20, 23). We recreated the AP-1 knocksideways system as described by Robinson et al. (20) in HeLa α cells by stably expressing γ-adaptin of AP-1 with FKBP inserted into the hinge region between the trunk and ear domains (γ-FKBP) and with a Mitotrap protein consisting of the signal and transmembrane domain of the mitochondrial outer membrane protein Tom70p, FRB, and a triple-FLAG epitope tag (SI Appendix, Fig. S6A).
Expression of recombinant γ-FKBP with or without siRNA-mediated silencing of endogenous γ-adaptin did not affect the protein levels of clathrin and AP-2 (SI Appendix, Fig. S6B). While Mitotrap always colocalized with mitochondria (SI Appendix, Fig. S6C), γ-FKBP (in the absence of endogenous γ) showed the expected distribution of AP-1 (perinuclear TGN region and peripheral endosomal structures) in the absence of rapamycin, but redistributed entirely to a mitochondrial pattern within 10 min of rapamycin addition (SI Appendix, Fig. S6D). Differential centrifugation and immunoblot analysis confirmed that most γ-FKBP/AP-1 was rerouted away from cytosol and light membranes to mitochondria after only 10 min of rapamycin treatment (SI Appendix, Fig. S6E), while AP-2 and clathrin were not affected, in agreement with Robinson et al. (20).
To assess the effect of rapamycin treatment on mTOR activity, we immunoblotted after different times of rapamycin addition for the mTOR kinase substrate S6 ribosomal protein, both total S6 and the phosphorylated form S6-P (phosphorylated at Ser235/236). While parental HeLa cells showed complete loss of S6 phosphorylation within 75 min, mTOR activity in HeLa-AP1ks cells was largely resistant to rapamycin (SI Appendix, Fig. S6F). The likely explanation is that endogenous FKBP12 was also rerouted to mitochondria by Mitotrap efficiently competing with mTOR for binding to the rapamycin–FKBP12 complex. In any case, effects of rapamycin via mTOR inhibition are not a concern within the time course of our transport experiments.
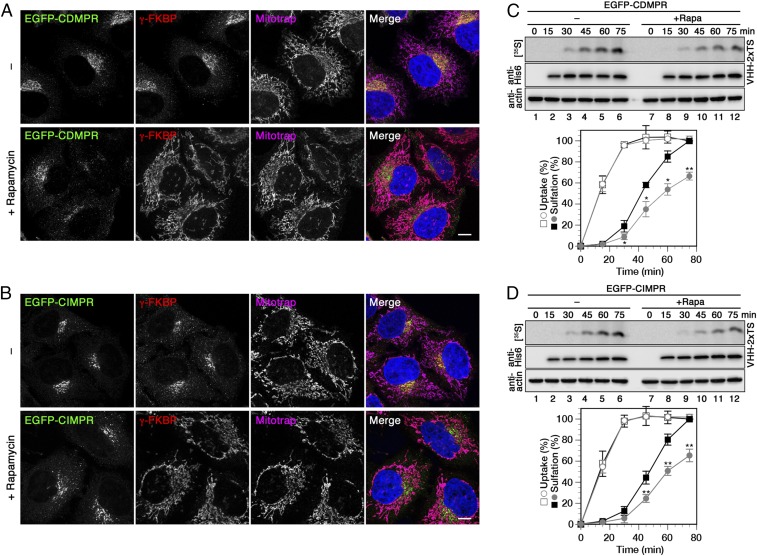
To study the functionality of the knocksideways system, HeLa-AP1ks cells stably expressing either EGFP-CDMPR or -CIMPR and silenced for endogenous γ-adaptin were incubated with or without rapamycin for 1 h (Fig. 6 A and B). In the absence of rapamycin, recombinant γ-FKBP showed the expected distribution of AP-1 on the TGN in the perinuclear region and on endosomes in the periphery, largely together with the MPRs. After rapamycin treatment, it was relocalized to mitochondria and the EGFP-MPRs showed an altered localization with an increased peripheral to juxtanuclear distribution. This redistribution of MPRs matches previous observations after AP-1 knockout (16, 49), knockdown, or knocksideways (20), thus confirming AP-1 inactivation.
Fig. 6.
Retrograde transport of MPRs is reduced upon rapid depletion of AP-1. (A and B) HeLa-AP1ks cells stably expressing EGFP-CDMPR or -CIMPR after silencing endogenous γ-adaptin were treated with or without 500 nM rapamycin for 1 h and processed for fluorescence microscopy to detect EGFP-MPR, recombinant γ-FKBP, and Mitotrap. (Scale bars, 10 µm.) (C and D) HeLa-AP1ks cells stably expressing EGFP-CDMPR or -CIMPR after silencing endogenous γ-adaptin were labeled with [35S]sulfate for up to 75 min in the presence of 2 μg/mL VHH-2xTS with or without 500 nM rapamycin. Cell-associated nanobodies were isolated and subjected to immunoblot analysis (anti-His6) and autoradiography ([35S]). Aliquots of cell lysates were immunoblotted for actin. Quantitation of VHH-2xTS uptake and sulfation is shown in percent of the value without rapamycin after 75 min (mean and SD of three independent experiments; two-sided Student’s t test: *P < 0.05; **P < 0.01). Black squares, without rapamycin; gray circles, with rapamycin; open symbols, uptake; filled symbols, sulfation.
For the analysis of retrograde transport with and without rapid AP-1 inactivation, HeLa-AP1ks cells stably expressing EGFP-CDMPR or -CIMPR were incubated with VHH-2xTS while simultaneously labeling them with [35S]sulfate in the presence or absence of rapamycin. Nanobody uptake and sulfation kinetics in knocksideways cells without rapamycin were virtually identical to those in normal HeLa cells (Fig. 6 C and D, square symbols, compared with Fig. 5D). In the presence of rapamycin, nanobody uptake was not affected at all, but the sulfation kinetics were significantly reduced to a similar extent for both MPRs (round symbols in Fig. 6 C and D). These results demonstrate a significant, but not exclusive, contribution of AP-1/clathrin in retrograde transport of MPRs.
Discussion
Because nanobodies are small, stable, monomeric, and thus noncross-linking, can be selected for high affinity, and—lacking disulfide bonds—can be expressed in the cytosol, they are beginning to be used in a growing range of applications in cell culture systems and organisms (26): for example as chromobodies to visualize intracellular target proteins in vivo (30), in deGradFP to inducibly and rapidly direct intracellular GFP fusion proteins to degradation [within 2–3 h of induction (50)], as Morphotrap on the cell surface to capture and thus block spreading of GFP-tagged morphogens (51, 52), or in the fluorescent three-hybrid system to assay in vivo interaction of fluorescently tagged proteins (53). Very recently, a comprehensive set of anti-mouse and anti-rabbit IgG nanobodies have been generated to be produced in bacteria as improved secondary antibodies (54).
In the present study, we have developed functionalized nanobodies produced by bacterial expression as tools to study retrograde traffic by different methods. We used an anti-GFP nanobody to make the tool versatile, able to target fusion proteins with extracellular GFP, YFP, or mCerulean that may already be available. Functionalization can of course be similarly applied to the rapidly growing number of nanobodies directed against untagged target proteins generated by immunization of camelids or by phage-display selection of synthetic VHH libraries (55).
We tested our tools first on cells expressing TfR-EGFP. The TfR is the best-characterized receptor cycling between plasma membrane and endosomes because of the unique properties of its ligand. It binds as ferro-Tf to the receptor and only releases the iron ion in acidic endosomes, but stays bound as apo-Tf, until it dissociates only after return to the neutral medium (36). The uptake kinetics we determined biochemically with VHH-std or by live-cell imaging with VHH-mCherry were in close agreement with the literature. Using VHH-tev containing a TEV protease cleavage site to cut off an epitope tag even allowed the reverse experiment of monitoring reappearance of intracellular receptor to the surface. The uptake kinetics determined with our tools for EGFP-MPRs and -TGN46 reflected their characteristics of intracellular distribution and for TGN46 its long residence time in the TGN. We used stable cell lines to avoid gross overexpression of the EGFP reporters as produced by transient transfection. In sensitive cases, however, genome editing will be an ideal way to produce GFP-tagged reporters.
By light microscopy, localization to the TGN is not easily distinguished from recycling and late endosomes that also accumulate in the same perinuclear region. Because our nanobodies are biotinylated, they can be easily modified by coupling of commercially available fluorescent streptavidin conjugates expanding the range of fluorescent colors for various costainings, but also for specific requirements, such as in superresolution microscopy. To make the step to the ultrastructural level, we generated a VHH-APEX2 fusion to use with DAB cytochemistry and EM. Even though HRP is more active and more stable to DAB cytochemistry (40), APEX2 nanobodies proved to be sufficient for localization in intracellular compartments by EM and for biochemical colocalization by ablation.
To go beyond localization to kinetic analysis of retrograde transport to the TGN, we produced nanobodies containing sulfation sites. Similarly, ricin and Shiga toxin subunits had previously been modified with sulfation sites to demonstrate a transport route through the TGN (e.g., refs. 56–58). TS peptides had also been chemically coupled to antibodies to assay retrograde transport of GFP-CIMPR and endogenous TGN46 to the TGN (19). The VHH-2xTS nanobodies have the advantage of easy and reproducible production and of a 1:1 stoichiometry with the target protein. Their high sensitivity upon longer labeling times is an excellent tool to test for retrograde transport of a protein of interest to the TGN. In addition, they allow us to determine kinetics of transport and therefore direct analysis of the effects of chemical inhibitors or genetic manipulations on transport.
As proof-of-principle, we applied VHH-2xTS nanobodies to study the role of AP-1 in retrograde transport of MPRs by direct kinetic analysis of nanobody sulfation in knocksideways cells in parallel with and without rapamycin-induced AP-1 depletion. Nanobody sulfation and thus retrograde transport of EGFP-MPRs was partially, but not completely blocked. The results support the notion that AP-1/clathrin contributes to MPR transport from early endosomes to the TGN. It thus also agrees with the conclusion derived from proteomic data of CCV contents upon AP-1 or GGA2 depletion by knocksideways (23). It is further consistent with other evidence of transport from late endosomes in a Rab9-dependent mechanism and from tubular early endosomes by SNX-BAR/retromer complexes. However, it is possible that the reduction of retrograde transport as observed by sulfation is underestimated, because nanobody–MPR complexes that entered the sulfation compartment in the presence of rapamycin by whichever mechanism likely stayed there longer before exiting again, and thus accumulated more radioactivity, because the exit route by AP-1/clathrin was inactive.
In the present study, we present a powerful and versatile nanobody-based tool to analyze GFP-reporter proteins from the cell surface. Nanobodies with other specificities may similarly be derivatized and alternate functionalization may allow the study of transport to other intracellular destinations beyond the TGN.
Materials and Methods
cDNAs and plasmids, bacterial expression and purification of functionalized nanobodies, cell culture and retroviral transduction, RNAi and DNA transfections, antibodies, uptake of functionalized nanobodies, gel electrophoresis and immunoblot analysis, fluorescence microscopy, live-cell imaging, sulfation analysis, compartment ablation, EM, differential centrifugation, and antibodies are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Paul Jenö, Nicole Beuret, the Biozentrum Imaging Core Facility, Janine Bögli from the FACS Core Facility, Adam Mazur from Research IT for support; the Affolter, Hall, and Scheiffele laboratories for reagents; and Scottie Robinson for materials and advice on the knocksideways system. This work was supported by Grant 31003A-162643 from the Swiss National Science Foundation. D.P.B. was supported by the Biozentrum Basel International PhD Program Fellowships for Excellence.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801865115/-/DCSupplemental.
References
- 1.Johannes L, Popoff V. Tracing the retrograde route in protein trafficking. Cell. 2008;135:1175–1187. doi: 10.1016/j.cell.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 3.Duncan JR, Kornfeld S. Intracellular movement of two mannose 6-phosphate receptors: Return to the Golgi apparatus. J Cell Biol. 1988;106:617–628. doi: 10.1083/jcb.106.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pallesen LT, Vaegter CB. Sortilin and SorLA regulate neuronal sorting of trophic and dementia-linked proteins. Mol Neurobiol. 2012;45:379–387. doi: 10.1007/s12035-012-8236-2. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, et al. WLS retrograde transport to the endoplasmic reticulum during Wnt secretion. Dev Cell. 2014;29:277–291. doi: 10.1016/j.devcel.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Sandvig K, Skotland T, van Deurs B, Klokk TI. Retrograde transport of protein toxins through the Golgi apparatus. Histochem Cell Biol. 2013;140:317–326. doi: 10.1007/s00418-013-1111-z. [DOI] [PubMed] [Google Scholar]
- 7.Gallon M, Cullen PJ. Retromer and sorting nexins in endosomal sorting. Biochem Soc Trans. 2015;43:33–47. doi: 10.1042/BST20140290. [DOI] [PubMed] [Google Scholar]
- 8.Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonetti B, Danson CM, Heesom KJ, Cullen PJ. Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR. J Cell Biol. 2017;216:3695–3712. doi: 10.1083/jcb.201703015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lombardi D, et al. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Díaz E, Pfeffer SR. TIP47: A cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- 13.Bulankina AV, et al. TIP47 functions in the biogenesis of lipid droplets. J Cell Biol. 2009;185:641–655. doi: 10.1083/jcb.200812042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: New twists in the tale. Nat Rev Mol Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 15.Puertollano R, et al. Morphology and dynamics of clathrin/GGA1-coated carriers budding from the trans-Golgi network. Mol Biol Cell. 2003;14:1545–1557. doi: 10.1091/mbc.02-07-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer C, et al. mu1A-adaptin-deficient mice: Lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsudaira T, Niki T, Taguchi T, Arai H. Transport of the cholera toxin B-subunit from recycling endosomes to the Golgi requires clathrin and AP-1. J Cell Sci. 2015;128:3131–3142. doi: 10.1242/jcs.172171. [DOI] [PubMed] [Google Scholar]
- 18.Hirst J, Miller SE, Taylor MJ, von Mollard GF, Robinson MS. EpsinR is an adaptor for the SNARE protein Vti1b. Mol Biol Cell. 2004;15:5593–5602. doi: 10.1091/mbc.E04-06-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saint-Pol A, et al. Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev Cell. 2004;6:525–538. doi: 10.1016/s1534-5807(04)00100-5. [DOI] [PubMed] [Google Scholar]
- 20.Robinson MS, Sahlender DA, Foster SD. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev Cell. 2010;18:324–331. doi: 10.1016/j.devcel.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson MS, Hirst J. Rapid inactivation of proteins by knocksideways. Curr Protoc Cell Biol. 2013;61:15.20.1–15.20.7. doi: 10.1002/0471143030.cb1520s61. [DOI] [PubMed] [Google Scholar]
- 22.Navarro Negredo P, et al. Contribution of the clathrin adaptor AP-1 subunit µ1 to acidic cluster protein sorting. J Cell Biol. 2017;216:2927–2943. doi: 10.1083/jcb.201602058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirst J, et al. Distinct and overlapping roles for AP-1 and GGAs revealed by the “knocksideways” system. Curr Biol. 2012;22:1711–1716. doi: 10.1016/j.cub.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Škrlec K, Štrukelj B, Berlec A. Non-immunoglobulin scaffolds: A focus on their targets. Trends Biotechnol. 2015;33:408–418. doi: 10.1016/j.tibtech.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Plückthun A. Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu Rev Pharmacol Toxicol. 2015;55:489–511. doi: 10.1146/annurev-pharmtox-010611-134654. [DOI] [PubMed] [Google Scholar]
- 26.Bieli D, et al. Development and application of functionalized protein binders in multicellular organisms. Int Rev Cell Mol Biol. 2016;325:181–213. doi: 10.1016/bs.ircmb.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 28.Greenberg AS, et al. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 29.Muyldermans S. Single domain camel antibodies: Current status. J Biotechnol. 2001;74:277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- 30.Rothbauer U, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 31.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Kubala MH, Kovtun O, Alexandrov K, Collins BM. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010;19:2389–2401. doi: 10.1002/pro.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waguri S, et al. Visualization of TGN to endosome trafficking through fluorescently labeled MPR and AP-1 in living cells. Mol Biol Cell. 2003;14:142–155. doi: 10.1091/mbc.E02-06-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anitei M, et al. A high-throughput siRNA screen identifies genes that regulate mannose 6-phosphate receptor trafficking. J Cell Sci. 2014;127:5079–5092. doi: 10.1242/jcs.159608. [DOI] [PubMed] [Google Scholar]
- 35.Hoflack B, Kornfeld S. Purification and characterization of a cation-dependent mannose 6-phosphate receptor from murine P388D1 macrophages and bovine liver. J Biol Chem. 1985;260:12008–12014. [PubMed] [Google Scholar]
- 36.Ciechanover A, Schwartz AL, Dautry-Varsat A, Lodish HF. Kinetics of internalization and recycling of transferrin and the transferrin receptor in a human hepatoma cell line. Effect of lysosomotropic agents. J Biol Chem. 1983;258:9681–9689. [PubMed] [Google Scholar]
- 37.Hirschmann DT, Kasper CA, Spiess M. Quantitative analysis of transferrin cycling by automated fluorescence microscopy. Methods Mol Biol. 2015;1270:365–378. doi: 10.1007/978-1-4939-2309-0_25. [DOI] [PubMed] [Google Scholar]
- 38.Beuret N, et al. Amyloid-like aggregation of provasopressin in diabetes insipidus and secretory granule sorting. BMC Biol. 2017;15:5. doi: 10.1186/s12915-017-0347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoorvogel W. Analysis of the endocytic system by using horseradish peroxidase. Trends Cell Biol. 1998;8:503–505. doi: 10.1016/s0962-8924(98)01380-4. [DOI] [PubMed] [Google Scholar]
- 40.Lam SS, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoorvogel W, Geuze HJ, Griffith JM, Strous GJ. The pathways of endocytosed transferrin and secretory protein are connected in the trans-Golgi reticulum. J Cell Biol. 1988;106:1821–1829. doi: 10.1083/jcb.106.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashiramoto M, James DE. Characterization of insulin-responsive GLUT4 storage vesicles isolated from 3T3-L1 adipocytes. Mol Cell Biol. 2000;20:416–427. doi: 10.1128/mcb.20.1.416-427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mihov D, Raja E, Spiess M. Chondroitin sulfate accelerates trans-Golgi-to-surface transport of proteoglycan amyloid precursor protein. Traffic. 2015;16:853–870. doi: 10.1111/tra.12294. [DOI] [PubMed] [Google Scholar]
- 44.Baeuerle PA, Huttner WB. Tyrosine sulfation is a trans-Golgi-specific protein modification. J Cell Biol. 1987;105:2655–2664. doi: 10.1083/jcb.105.6.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huttner WB. Tyrosine sulfation and the secretory pathway. Annu Rev Physiol. 1988;50:363–376. doi: 10.1146/annurev.ph.50.030188.002051. [DOI] [PubMed] [Google Scholar]
- 46.Casanova JE. Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 47.Mallard F, et al. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of Shiga toxin B-fragment transport. J Cell Biol. 1998;143:973–990. doi: 10.1083/jcb.143.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amessou M, et al. Syntaxin 16 and syntaxin 5 are required for efficient retrograde transport of several exogenous and endogenous cargo proteins. J Cell Sci. 2007;120:1457–1468. doi: 10.1242/jcs.03436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medigeshi GR, Schu P. Characterization of the in vitro retrograde transport of MPR46. Traffic. 2003;4:802–811. doi: 10.1034/j.1600-0854.2003.00136.x. [DOI] [PubMed] [Google Scholar]
- 50.Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol. 2011;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- 51.Harmansa S, Hamaratoglu F, Affolter M, Caussinus E. Dpp spreading is required for medial but not for lateral wing disc growth. Nature. 2015;527:317–322. doi: 10.1038/nature15712. [DOI] [PubMed] [Google Scholar]
- 52.Harmansa S, Alborelli I, Bieli D, Caussinus E, Affolter M. A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. eLife. 2017;6:e22549. doi: 10.7554/eLife.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herce HD, Deng W, Helma J, Leonhardt H, Cardoso MC. Visualization and targeted disruption of protein interactions in living cells. Nat Commun. 2013;4:2660. doi: 10.1038/ncomms3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pleiner T, Bates M, Gorlich D. A toolbox of anti-mouse and anti-rabbit IgG secondary nanobodies. J Cell Biol. 2017;217:1143–1154. doi: 10.1083/jcb.201709115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fridy PC, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johannes L, Tenza D, Antony C, Goud B. Retrograde transport of KDEL-bearing B-fragment of Shiga toxin. J Biol Chem. 1997;272:19554–19561. doi: 10.1074/jbc.272.31.19554. [DOI] [PubMed] [Google Scholar]
- 57.Mallard F, Johannes L. Shiga toxin B-subunit as a tool to study retrograde transport. Methods Mol Med. 2003;73:209–220. doi: 10.1385/1-59259-316-x:209. [DOI] [PubMed] [Google Scholar]
- 58.Utskarpen A, Slagsvold HH, Iversen TG, Wälchli S, Sandvig K. Transport of ricin from endosomes to the Golgi apparatus is regulated by Rab6A and Rab6A′. Traffic. 2006;7:663–672. doi: 10.1111/j.1600-0854.2006.00418.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.